Cyanobacterial Root Associations of Leafless Epiphytic Orchids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants
2.2. Sampling of Roots and Isolation of Cyanobacteria
2.3. Scanning Electron Microscopy
2.4. DNA Extraction, PCR Reactions, and DGGE Analysis
2.5. Phylogenetic Analyses
2.6. Data Analysis
3. Results
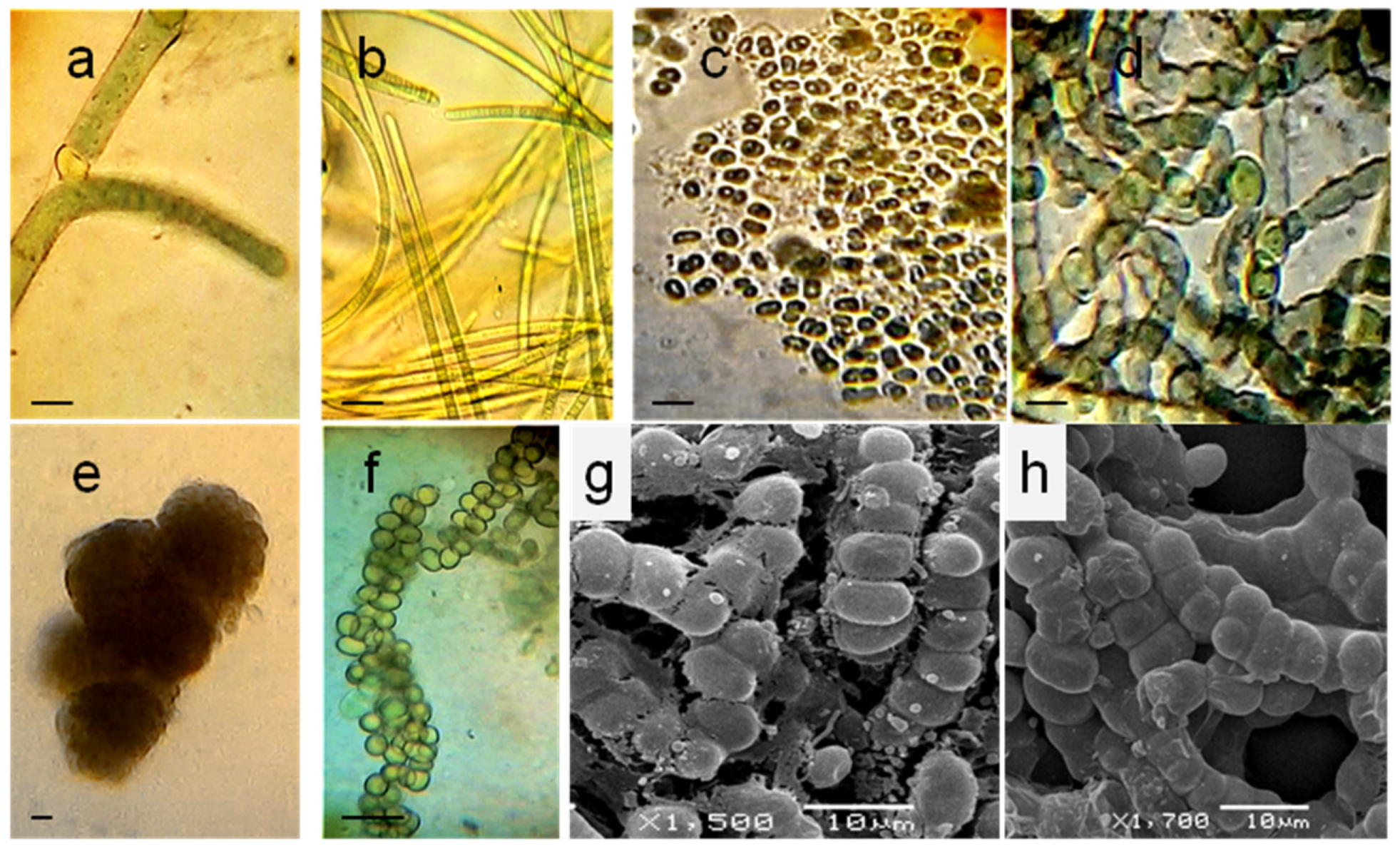
3.1. Localization and Diversity of Cyanobacterial Morphotypes on Roots
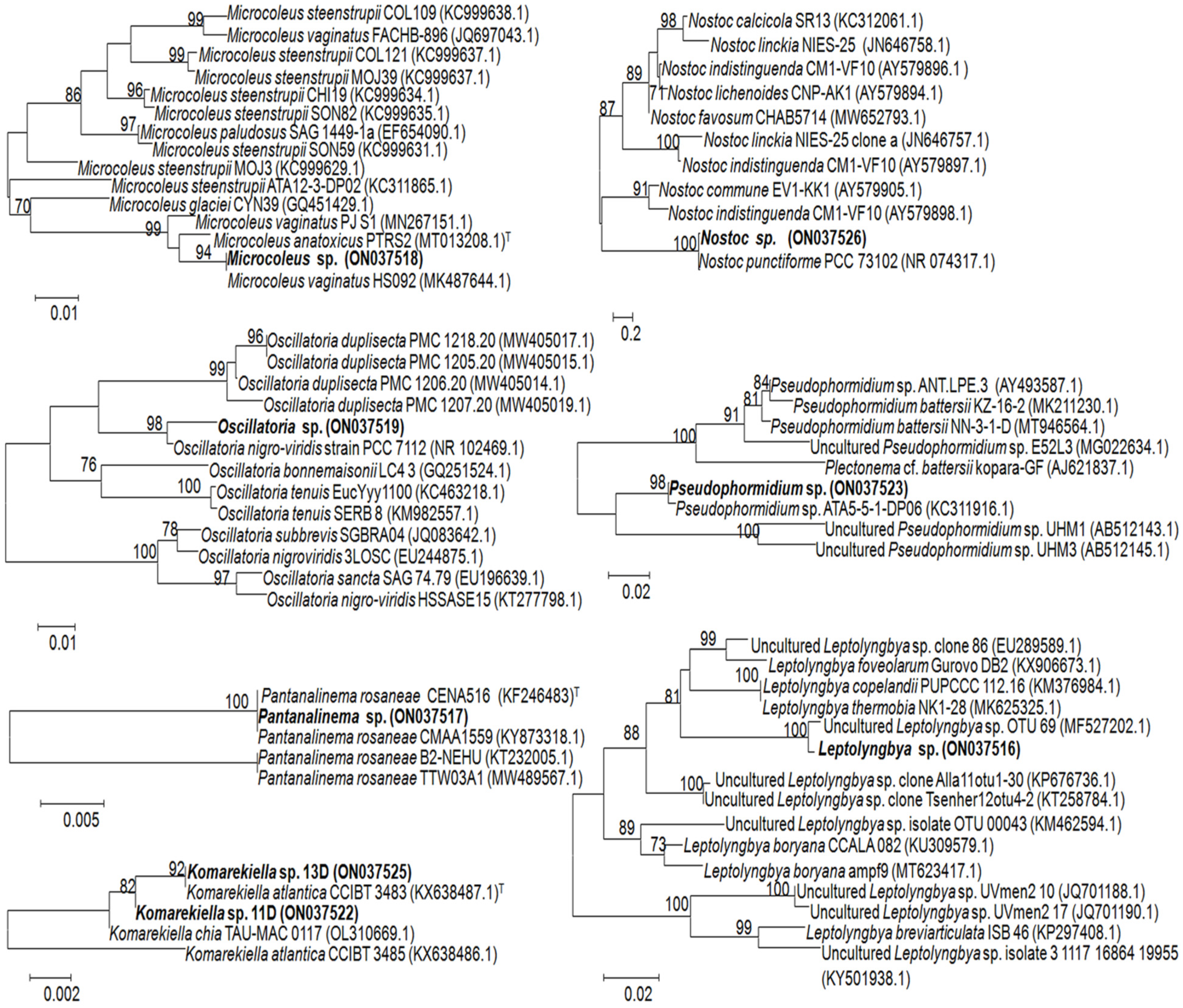
3.2. Identification of Associated Cyanobacteria
3.3. Cyanobacterial Isolates
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zotz, G. The systematic distribution of vascular epiphytes—A critical update. Bot. J. Linn. Soc. 2013, 171, 453–481. [Google Scholar] [CrossRef] [Green Version]
- Givinish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Doucette, A.; Caro, G.G.; McDaniel, J.; Clements, M.A.; et al. Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeogr. 2016, 43, 1905–1916. [Google Scholar] [CrossRef]
- Dycus, A.; Knudson, L. The role of the velamen of the aerial roots of orchids. Bot. Gaz. 1957, 119, 78–87. [Google Scholar] [CrossRef]
- Chomicki, G.; Bidel, L.P.R.; Ming, F.; Coiro, M.; Zhang, X.; Wang, Y.; Baissac, Y.; Jay-Allemand, C.; Renner, S.S. The velamen protects photosynthetic orchid roots against UV-B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytol. 2015, 205, 1330–1341. [Google Scholar] [CrossRef]
- Arditti, J. An history of orchid hybridization, seed germination and tissue culture. Bot. J. Linn. Soc. 1984, 89, 359–381. [Google Scholar] [CrossRef]
- Kwok-ki, H.; Hock-Hin, Y.; Choy-Sin, H. The presence of photosynthetic machinery in aerial roots of leafy orchids. Plant Cell Physiol. 1983, 24, 1317–1321. [Google Scholar]
- Silvera, K.; Santiago, L.S.; Cushman, J.C.; Winter, K. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 2009, 149, 1838–1847. [Google Scholar] [CrossRef] [Green Version]
- Benzing, D.H. Mycorrhizal infection of epiphytic orchids in South Florida. Am. Orch. Soc. Bull. 1982, 51, 618–622. [Google Scholar]
- Goh, C.J.; Arditti, J.; Avadhani, P.N. Carbon fixation in orchid aerial roots. New Phytol. 1983, 95, 367–374. [Google Scholar] [CrossRef]
- Pridgeon, A.M. The velamen and exodermis of orchid roots. In Orchid Biology: Reviews and Persrectives; Arditti, J., Ed.; Cornell University Press: Ithaca, NY, USA, 1987; pp. 141–192. [Google Scholar]
- Zotz, G.; Winkler, U. Aerial roots of epiphytic orchids: The velamen radicum and its role in water and nutrient uptake. Oecologia 2013, 171, 733–741. [Google Scholar] [CrossRef]
- Chomicki, G.; Bidel, L.P.R.; Jay-Allemand, C. Exodermis structure controls fungal invasion in the leafless epiphytic orchid Dendrophylax lindenii (Lindl.) Benth. ex Rolfe. Flora 2014, 209, 88–94. [Google Scholar] [CrossRef]
- Carlsward, B.S.; Whitten, W.M.; Williams, N.H.; Bytebier, B. Molecular phylogenetics of Vandeae (Orchidaceae) and the evolution of leaflessness. Am. J. Bot. 2006, 93, 770–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Lobakova, E.S.; Kolomeitseva, G.L.; Netrusov, A.I. Microbiota of the orchid rhizoplane. Microbiology 2001, 70, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.A.; Lobakova, E.S.; Kolomeitseva, G.L.; Cherdyntseva, T.A.; Netrusov, A.I. Associative cyanobacteria isolated from the roots of epiphytic orchids. Microbiology 2003, 72, 92–97. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Lobakova, E.S.; Kolomeitseva, G.L.; Cherdyntseva, T.A.; Netrusov, A.I. Localization of associative cyanobacteria on the roots of epiphytic orchids. Microbiology 2003, 72, 99–104. [Google Scholar] [PubMed]
- Tsavkelova, E.A.; Egorova, M.A.; Leontieva, M.R.; Malakho, S.G.; Kolomeitseva, G.L.; Netrusov, A.I. Dendrobium nobile Lindl. seed germination in co-cultures with diverse associated bacteria. Plant Growth Regul. 2016, 80, 79–91. [Google Scholar] [CrossRef]
- Pavlova, A.S.; Leontieva, M.R.; Smirnova, T.A.; Kolomeitseva, G.L.; Netrusov, A.I.; Tsavkelova, E.A. Colonization strategy of the endophytic plant growth promoting strains of Pseudomonas fluorescens and Klebsiella oxytoca on the seeds, seedlings and roots of the epiphytic orchid, Dendrobium nobile Lindl. J. Appl. Microbiol. 2017, 123, 217–232. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Klimova, S.Y.; Shestakov, A.I.; Botina, S.G.; Netrusov, A.I. Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch. Microbiol. 2007, 188, 655–664. [Google Scholar] [CrossRef]
- Tsavkelova, E.; Oeser, B.; Oren-Young, L.; Israeli, M.; Sasson, Y.; Tudzynski, B.; Sharon, A. Identification and functional characterization of indole-3-acetamide-mediated IAA biosynthesis in plant-associated Fusarium species. Fungal Genet. Biol. 2012, 49, 48–57. [Google Scholar] [CrossRef]
- Averyanov, L.V.; Nguen, K.S.; Maisak, T.V. Chiloschista pulchella (Orchidaceae: Aeridinae) new orchid species from Lao PDR. Taiwania 2018, 63, 389–392. [Google Scholar]
- Mathew, J.M.; Jose, M.; Pichan, M.S.; Dariusz, L.S. Chiloschista confusa (Orchidaceae), a new species from the Southern Western Ghats, Kerala, India. Ann. Bot. Fennici 2021, 58, 347–353. [Google Scholar] [CrossRef]
- Deepthi, A.S.; Ray, J.G. Algal associates and the evidence of cyanobacterial nitrogen fixation in the velamen roots of epiphytic orchids. Global Ecol. Conserv. 2020, 22, e00946. [Google Scholar] [CrossRef]
- Ram, A.T.; Shamina, M. Identification of cyanobacterial association from the roots of the terrestrial orchid, Spathoglottis plicata Blume. Int. J. Adv. Res. 2015, 3, 1477–1479. [Google Scholar]
- Pankratova, E.M.; Trefilova, L.V.; Zyablykh, R.Y.; Ustyuzhanin, I.A. Cyanobacterium Nostoc paludosum Kütz as a basis for creation of agriculturally useful microbial associations by the example of bacteria of the genus Rhizobium. Microbiology 2008, 77, 228–234. [Google Scholar] [CrossRef]
- Pereira, I.; Ortega, R.; Barrientos, L.; Moya, M.; Reyes, G.; Kramm, V. Development of a biofertilizer based on filamentous nitrogen-fixing cyanobacteria for rice crops in Chile. J. Appl. Phycol. 2009, 21, 135–144. [Google Scholar] [CrossRef]
- Burgeff, H. Mycorrhiza of orchids. In The Orchids: A Scientific Survey; Wither, K., Ed.; The Ronald Press Company: New York, NY, USA, 1959; pp. 361–395. [Google Scholar]
- Beyrle, H.F.; Smith, S.E.; Peterson, R.L.; Franco, C.M.M. Colonization of Orchis morio protocorms by mycorrhizal fungus: Effects of nitrogen nutrition and glyphosate in modifying the responses. Can. J. Bot. 1995, 73, 1128–1140. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.J. Genetic assignements, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar]
- Komárek, J.; Johansen, J. Filamentous Cyanobacteria. In Freshwater Algae of North America, 2nd ed.; Wehr, J.D., Sheath, R.G., Kociolek, P., Eds.; Academic Press: Amsterdam, The Netherlands, 2015; pp. 75–235. [Google Scholar]
- Hauer, T.; Bohunická, M.; Johansen, J.; Mareš, J.; Berrendero-Gomez, E. Reassessment of the cyanobacterial family Microchaetaceae and establishment of new families Tolypothrichaceae and Godleyaceae. J. Phycol. 2014, 50, 1089–1100. [Google Scholar] [CrossRef]
- Gama, W.A., Jr.; Laughinghouse, H.D., IV; Sant’Anna, C.L. How diverse are coccoid cyanobacteria? A case study of terrestrial habitats from the Atlantic Rainforest (São Paulo, Brazil). Phytotaxa 2014, 178, 61–97. [Google Scholar] [CrossRef]
- Casamatta, D.; Hašler, P. Blue-green algae (Cyanobacteria) in rivers. In River Algae; Necchi, O., Jr., Ed.; Springer: Cham, Switzerland, 2016; pp. 5–34. [Google Scholar]
- Hentschke, G.; Johansen, J.; Pietrasiak, N.; Rigonato, J.; Fiore, M.; Sant’Anna, C. Komarekiella atlantica gen. et sp. nov. (Nostocaceae, Cyanobacteria): A new subaerial taxon from the Atlantic rainforest and Kauai, Hawaii. J. Czech Phycol. Soc. 2017, 17, 178–190. [Google Scholar] [CrossRef] [Green Version]
- Hanshew, A.S.; Mason, C.J.; Raffa, K.F.; Currie, C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 2013, 95, 149–155. [Google Scholar] [CrossRef] [Green Version]
- Muyzer, G.; Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 1998, 73, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutte, C.; Grubisic, S.; Balthasart, P. Testing of primers for the study of cyanobacterial Wilmotte molecular diversity by DGGE. J. Microbiol. Methods 2006, 65, 542–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, A.N.; Soderback, E.; Bergman, B. Cyanobacterium–plant symbioses. New Phytol. 2000, 147, 449–481. [Google Scholar] [CrossRef]
- Nilsson, M.; Bhattacharya, J.; Rai, A.N.; Bergman, B. Colonization of roots of rice (Oryza sativa) by symbiotic Nostoc strains. New Phytol. 2002, 156, 517–525. [Google Scholar] [CrossRef] [Green Version]
- Rana, A.; Joshi, M.; Prasanna, R.; Shivay, Y.S.; Nain, L. Biofortification of wheat through inoculation of plant growth promoting rhizobacteria and cyanobacteria. Eur. J. Soil. Biol. 2012, 50, 118–126. [Google Scholar] [CrossRef]
- Neustupa, J.; Škaloud, P. Diversity of subaerial algae and cyanobacteria on tree bark in tropical mountain habitats. Biologia 2008, 63, 806–812. [Google Scholar] [CrossRef]
- Toledo, G.; Bashan, Y.; Soeldner, A. Cyanobacteria and black mangroves in Northwestern Mexico: Colonization, and diurnal and seasonal nitrogen fixation on aerial roots. Can. J. Microbiol. 1995, 41, 999–1011. [Google Scholar] [CrossRef]
- Ram, A.T.; Shamina, M. New reports of the cyanobacterial association on the roots of epiphytic orchid, Dendrobium crumenatum Sw. Int. J. Chem. Biol. Sci. 2015, 1, 19–21. [Google Scholar]


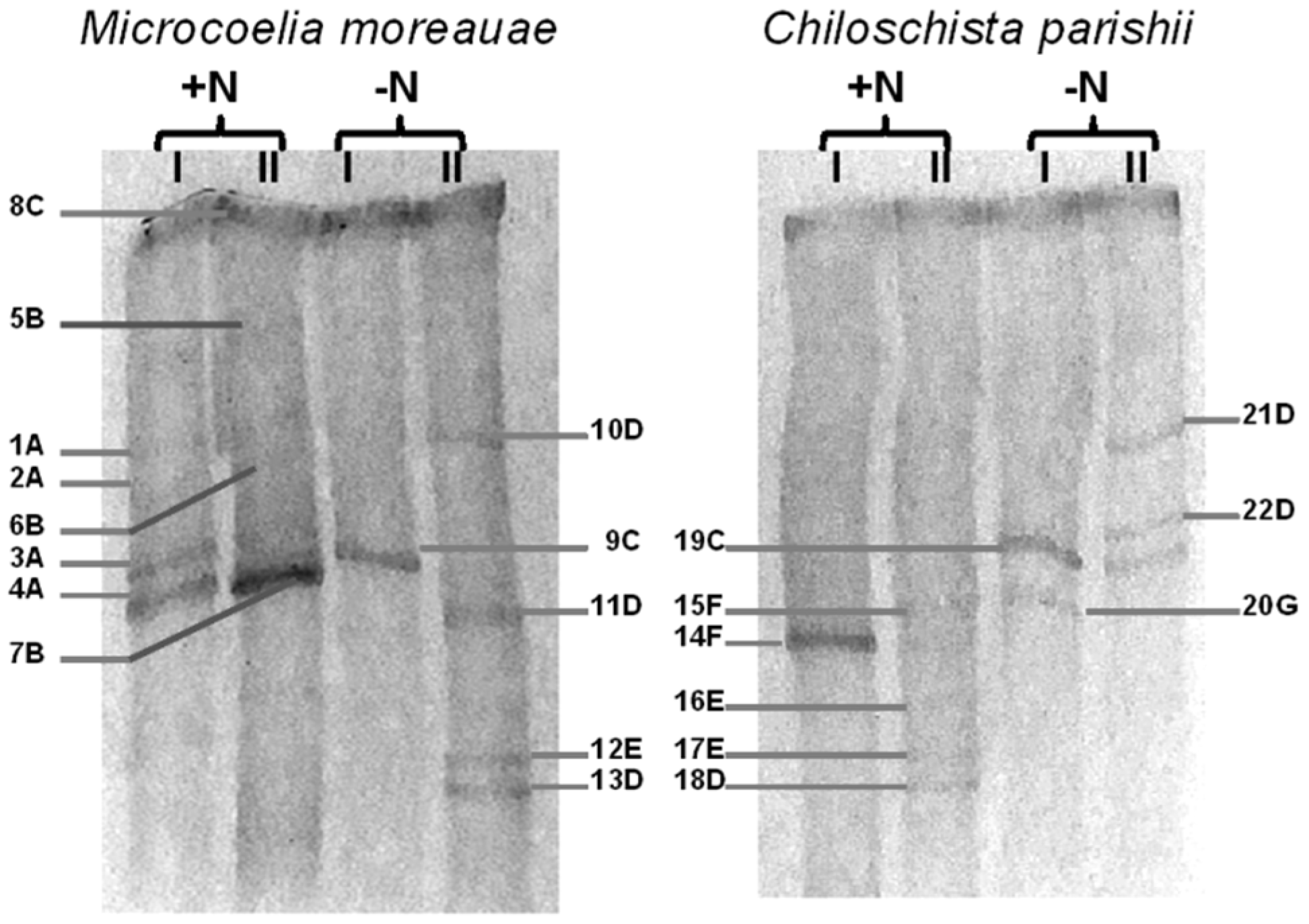
| Band’s Number | Closest Match in GenBank, NCBI | Affiliation | |
|---|---|---|---|
| Affiliation and Identity (%), BLASTn ID Accession Number | Genus (ID Accession Number, in GenBank, NCBI) | Order | |
| 1A–4A | Microcoleus vaginatus HS092 16S rRNA gene, partial sequence (100%); MK487644.1 | Microcoleus sp. (ON037518) | Oscillatoriales |
| 5B–7B | Oscillatoria prolifera 16S rRNA gene, partial sequence (100%); MK771147 | Oscillatoria sp. (ON037519) | Oscillatoriales |
| 8C, 9C, 19C | Uncultured Leptolyngbya sp. clone OTU_69 16S rRNA gene, partial sequence (99.8%); MF527202.1 | Leptolyngbya sp. (ON037516) | Synechococcales |
| 10D, 11D, 13D, 18D, 21D, 22D | Komarekiella atlantica HA4396-MV6 partial sequence; 16S-23S rRNA intergenic spacer, and 23S rRNA gene, partial sequence (100%); KX646832.1 | Komarekiella sp. (ON037522) | Nostocales |
| 12E, 16E, 17E | Nostoc sp. 9E-03 partial 16S rRNA gene, strain 9E-03 (99.7%); FR798938.1 | Nostoc sp. (ON037526) | Nostocales |
| 14F, 15F | Pantanalinema rosaneae CMAA1559 16S rRNA gene, partial sequence (100%); KY873318.1 | Pantanalinema sp. (ON037517) | Synechococcales |
| 20G | Pseudophormidium sp. ATA5-5-1-DP06 16S rRNA gene, complete sequence (99.7%); KC311916.1 | Pseudophormidium sp. (ON037523) | Oscillatoriales |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsavkelova, E.A.; Glukhareva, I.D.; Volynchikova, E.A.; Egorova, M.A.; Leontieva, M.R.; Malakhova, D.V.; Kolomeitseva, G.L.; Netrusov, A.I. Cyanobacterial Root Associations of Leafless Epiphytic Orchids. Microorganisms 2022, 10, 1006. https://doi.org/10.3390/microorganisms10051006
Tsavkelova EA, Glukhareva ID, Volynchikova EA, Egorova MA, Leontieva MR, Malakhova DV, Kolomeitseva GL, Netrusov AI. Cyanobacterial Root Associations of Leafless Epiphytic Orchids. Microorganisms. 2022; 10(5):1006. https://doi.org/10.3390/microorganisms10051006
Chicago/Turabian StyleTsavkelova, Elena A., Irina D. Glukhareva, Elena A. Volynchikova, Maria A. Egorova, Maria R. Leontieva, Dina V. Malakhova, Galina L. Kolomeitseva, and Alexander I. Netrusov. 2022. "Cyanobacterial Root Associations of Leafless Epiphytic Orchids" Microorganisms 10, no. 5: 1006. https://doi.org/10.3390/microorganisms10051006
APA StyleTsavkelova, E. A., Glukhareva, I. D., Volynchikova, E. A., Egorova, M. A., Leontieva, M. R., Malakhova, D. V., Kolomeitseva, G. L., & Netrusov, A. I. (2022). Cyanobacterial Root Associations of Leafless Epiphytic Orchids. Microorganisms, 10(5), 1006. https://doi.org/10.3390/microorganisms10051006







