Abstract
Microorganisms have a limited and highly adaptable repertoire of genes capable of encoding proteins containing single or variable multidomains. The phytopathogenic bacteria Xanthomonas citri subsp. citri (X. citri) (Xanthomonadaceae family), the etiological agent of Citrus Canker (CC), presents a collection of multidomain and multifunctional enzymes (MFEs) that remains to be explored. Recent studies have shown that multidomain enzymes that act on the metabolism of the peptidoglycan and bacterial cell wall, belonging to the Lytic Transglycosylases (LTs) superfamily, play an essential role in X. citri biology. One of these LTs, named XAC4296, apart from the Transglycosylase SLT_2 and Peptidoglycan binding-like domains, contains an unexpected aldose 1-epimerase domain linked to the central metabolism; therefore, resembling a canonical MFE. In this work, we experimentally characterized XAC4296 revealing its role as an MFE and demonstrating its probable gene fusion origin and evolutionary history. The XAC4296 is expressed during plant-pathogen interaction, and the Δ4296 mutant impacts CC progression. Moreover, Δ4296 exhibited chromosome segregation and cell division errors, and sensitivity to ampicillin, suggesting not only LT activity but also that the XAC4296 may also contribute to resistance to β-lactams. However, both Δ4296 phenotypes can be restored when the mutant is supplemented with sucrose or glutamic acid as a carbon and nitrogen source, respectively; therefore, supporting the epimerase domain’s functional relationship with the central carbon and cell wall metabolism. Taken together, these results elucidate the role of XAC4296 as an MFE in X. citri, also bringing new insights into the evolution of multidomain proteins and antimicrobial resistance in the Xanthomonadaceae family.
1. Introduction
Many Gram-negative phytopathogens relevant to agriculture belong to the Xanthomonadaceae family [1]. A critical species in this group is the Xanthomonas citri subsp. citri (X. citri), the causal agent of citrus canker (CC) [2]. CC is a severe disease that affects citrus crops and decreases fruit production, leading to economic losses [3]. Many efforts to understand CC mechanisms have been made since the disease was discovered in the early 1900s [4]. One of the hallmarks that led to several new insights into the plant-pathogen interactions was the analyses of the X. citri genome, revealing the genetic basis of bacterial pathogenicity [5]. Since then, most studies have been focused on Xanthomonas pathogenicity mechanisms, such as regulation and secretion of virulence factors, such as the Type 3 Secretion System [5,6]. Moreover, genetics studies were also conducted to understand chromosome segregation and cell division mechanism, aiming to better understand this phytopathogen’s cellular biology [7,8]. However, other possible genetic mechanisms related to X. citri pathogenicity remain unknown. For instance, multidomain and multifunctional enzymes (MFEs) are essential for bacterial cellular biology, virulence, and fitness.
The MFEs are ubiquitous in prokaryotes [9]. These proteins generally harbor more than one domain, each exhibiting distinct functions [10]. Therefore, the MFEs may simultaneously perform multiple physiologically biochemical or biophysical tasks in the cell [11,12]. These numerous functionalities might provide evolutionary advantages for the bacterium [13]. For instance, combining multiple functions enables the enzyme to catalyze different steps of a single metabolic pathway [14]. In addition, the MFEs can be considered a clever strategy for generating complexity from existing proteins without expanding the genome [13].
Besides MFEs, one interesting class of enzymes has gained attention for their relation to bacterial fitness and virulence. These are the Lytic Transglycosylases (LTs) related to peptidoglycan biosynthesis and recycling and cell-wall-antibiotic detection, also showing involvement with the bacterium septum division allowing cell separation and insertion of protein complexes like secretion systems, flagella, and pili [15,16,17,18,19,20,21]. Due to these features, LTs may also play a relevant role in the pathogenesis of many bacterial species, such as Neisseria gonorrhoeae [22] and Burkholderia pseudomallei [23].
Recently, we described the LT’s arsenal present in the X. citri genome (16 LTs from different families) [24]. Among those, we functionally revealed that two LTs from the 3B family: MltB2.1 and MltB2.2, are directly implicated in X. citri fitness [24]. We also identified another 3B-like LT named XAC4296 (NCBI locus_tag: XAC_RS21660). Notably, apart from the Transglycosylase SLT 2 (IPR031304) and Peptidoglycan binding-like (IPR002477) domains, XAC4296 contains an additional and unexpected aldose 1-epimerase domain (IPR015443) linked to carbohydrate metabolism, and potentially showing involvement with the bacterial cell wall metabolism and biosynthesis of a variety of cell surface polysaccharides [25]. Interestingly, the XAC4296 gene was previously identified exclusively in the Xanthomonas genus [24]. Moreover, in silico analyses revealed that XAC4296 appears to have been formed by a previous gene fusion event, which originated by two independent genes (a 3B family LT and D-hexose-6-phosphate mutarotase gene), commonly separated in distinct loci in other non-Xanthomonas species [24]. Therefore, XAC4296 resembles a canonical MFE, showing a multidomain architecture.
In this work, we performed in silico, fluorescence microscopy and pathogenicity assays to investigate the evolution and role of XAC4296 as a putative MFE. We also evaluated XAC4296 as a potential X. citri virulence and pathogenicity factor. Our results indicate that XAC4296 functions resemble a typical LT, mainly related to peptidoglycan biosynthesis. We also unveiled an additional role related to carbohydrate metabolism, compatible with the epimerase domain’s role and chromosome segregation during cell division. Taken together, these results demonstrate that XAC4296 behaves like a classic MFE, showing at least two unrelated and mechanistically different roles, both impacting X. citri fitness, a primary role related to enzymatic catalysis and a secondary role related to cell structural function.
2. Material and Methods
2.1. In Silico and Phylogenetic Analysis
Global comparisons were made based on sequenced Xanthomonadaceae genomes deposited in the National Center for Biotechnology Information (NCBI) repository. For XAC4296 homolog detection, we used as tblastn [26] parameters a query coverage and identity >90% and >60%, respectively, and including all three characteristics domains (Table S1 shows a complete list of genomes carrying XAC4296 homolog).
The XAC4296 three-dimensional structure was analyzed in two modules, the first considering the 420 amino acids located in the N-terminus of the protein, containing the SLT_2 (IPR031304) and PG_Binding 1 (IPR002477) domains, corresponding to the Lytic Transglycosylases from 3B family; the second module considering the last 309 amino acids located at the C-Terminus of the XAC4296 and corresponding to the D-hexose-6-phosphate mutarotase annotated gene, containing the aldose-1-epimerase domain (IPR015443). Molecular modeling was performed with the Robetta webserver [27]. The Chimera Tool [28] was used to generate the three-dimensional structures and interactive visualization of the entire XAC4296 protein. The three-dimensional structures of the PDB (Protein Data Bank) used as models for the LT and epimerase modules were, respectively, 5AO8 and 2HTA. These proteins were selected according to the ranking established by the alignment performed by software MAFFT 7309 [29]. The stereochemical quality of the generated models was evaluated by analyzing Ramachandran’s plot, carried out by Chimera Tool.
The XAC4296 homolog sequences were aligned with MAFFT 7.309 [29], and their best-fit evolutionary models were predicted with ProTest 3.2.4. A maximum-likelihood tree was reconstructed with RaxML 8.2.9 using a bootstrap value of 1000. The final tree was visualized in FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree (accessed on 20 November 2021)) and edited with Inkscape 0.92.4 (http://www.inkscape.org (accessed on 20 November 2021)). The Integrated Microbial Genomes & Microbiomes (IMG/M) system [30] was used for comparative analyses.
2.2. Strains and Growth Conditions
Bacterial strains and plasmid strains used in this study are shown in Table S2. The X. citri strains were grown in three different culture media: nutrient broth (NB: 0.5% peptone, 0.3% beef extract), nutrient agar (NA: 0.5% peptone, 0.3% beef extract, 0.15% agar) supplemented with L-arabinose (0.05% w/v) and sucrose (5% w/v) when required or, XVM2 (20 mM NaCl, 10 mM (NH4)2SO4, 5 mM MgSO4, 1 mM CaCl2, 0.16 mM KH2PO4, 0.32 mM K2HPO4, 0.01 mM FeSO4, 10 mM fructose, 10 mM sucrose, 0.03% casaminoacids, pH 6.7) at 29 °C. Escherichia coli strains were cultivated in Luria-Bertani medium (LB: 1% tryptone, 0.5% yeast extract, 0.10% NaCl, 0.15% agar; pH 7) and SOB media [31] at 37 °C. Antibiotics were used as needed at the following concentrations: kanamycin (Kn), 30 µg/mL; carbenicillin (Carb), 50 µg/mL; streptomycin (Str), 50 µg/mL; gentamycin (Gen), 10 µg/mL; ampicillin (Amp), 100 µg/mL.
2.3. RNA Extraction and cDNA Synthesis from XAC4296
X. citri total RNA was extracted using RNeasy protect bacteria Mini kit (Qiagen) according to the manufacturer. The first strand of complementary DNA was synthesized from 1 μg of total RNA using a qScript® cDNA SuperMix (Qiagen). Before cDNA synthesis, RNA samples were treated with DNaseI. The DNA and RNA quantification was performed using Qubit HS (High Sensitivity) (Thermo Fisher, Waltham, MA, USA). Primers F4296 (F) and pMAJIIc (R) were used for PCR reaction using cDNA as a template (Table S3). PCR products were checked by agarose gel electrophoresis.
2.4. Mutant Construction
Mutant of gene XAC4296 was generated using homologous suicide plasmid (pNPTS138) [32] integration through site-directed mutagenesis by PCR overlap extension approach [33]. To construct the deletion mutant of XAC4296 ORF, we used X. citri genomic DNA as a template and primers described in Table S3. The first PCR amplifications were made separately using pairs of primers A(F)–B(R) and C(F)–D(R) and Phusion high fidelity DNA polymerase (Thermo Fischer Scientific) to generate the products A–B and C–D, with self-complementary tails. The second PCR was performed using primers A(F)–D(R) and the products A–B and C–D as a template to obtain the A–D fragment, in which the XAC4296 sequence was deleted. The final PCR product A–D and pNPTS138 suicide vector were double digested with NheI/HindIII enzymes (New England BioLabs Inc.®, Ipswich, MA, USA). The ligation between vector and fragments was performed with T4 DNA Ligase (New England BioLabs Inc.®) according to the manufacturer’s instructions. The recombinant vector Δ4296-pNPTS138 was transformed into chemically competent E. coli DH10B [31], and transformant colonies were selected using antibiotics and Lac-Z promoter. The constructions were checked by agarose gel electrophoresis and sequencing on a 3730xI DNA analyzer (Thermo Fisher Scientific) using primers A–D. Finally, the Δ4296-pNPTS138 recombinant plasmid was used for X. citri electroporation [34], and colonies were selected by kanamycin resistance and sucrose susceptibility [35]. Mutant Δ4296 was confirmed by sequencing.
The XAC4296 ORF was PCR amplified from X. citri genomic DNA using primers pMAJIIc (F)-pMAJIIc (R) (Table S3) and Phusion high fidelity DNA polymerase (Thermo Fisher Scientific). The PCR product and the integrative vector pMAJIIc [36] were double digested with NheI/XhoI enzymes (New England BioLabs Inc.®) and ligated with T4 DNA Ligase (New England BioLabs Inc.®), according to manufacturer’s instructions. The recombinant vector 4296-pMAJIIc was transformed into chemically competent E. coli DH10B [31]. Colonies were selected using kanamycin resistance. The recombinant plasmid DNA (pMAJIIc-XAC4296) was purified using Promega Wizard®Plus SV Minipreps DNA Purification System kit according to the manufacturer’s instructions and the inserted XAC4296 DNA sequence was confirmed by sequencing. The recombinant plasmid was used to transform both mutant strain 4296 and X. citri wild type strain by electroporation [34]. Colonies were selected by kanamycin resistance, and the integrative vector version was identified on NA plates supplemented with 0.2% soluble starch followed by iodine vapor crystals exposure [36]. The strain Δ4296-pMAJIIc-4296 (named Δ4296c) was used as complemented strain in the following assays, and the recombinant strain XccA-pMAJIIc-4296 was used for protein subcellular localization. The constructions were checked by agarose gel electrophoresis and DNA sequencing.
2.5. Pathogenicity Assay
We used two methods for pathogenicity assays. In the first method, bacterial strains were inoculated into the leaves’ surface only by spray [37]. X. citri and mutant Δ4296 were cultivated in NB medium for 16 h to O.D. 600-nm ~0.8 and diluted in fresh NB medium to O.D. 600 nm of 0.3. Cells were collected by centrifugation and resuspended in autoclaved tap water to an O.D. 600 nm of 0.3, equivalent to 108 CFU/mL. Three different “Pêra Rio” orange (Citrus sinensis L. Osbeck) plants were sprayed with each bacterial suspension until all leaves were thoroughly coated, then covered with a clear plastic bag for 24 h. After 25 days of inoculation (DAI), all leaves were quantified, those presenting citrus canker (CC) symptoms were photographed, the CC lesions were counted, and the results were analyzed and compared.
For pathogenicity assays by infiltration method, strains of X. citri and mutant Δ4296 were cultivated in NB medium for 16 h to O.D. 600 nm ~0.8 and diluted in fresh NB medium to O.D. 600 nm of 0.3. Cells were collected by centrifugation and resuspended in autoclaved tap water to an O.D. 600 nm of 0.3, equivalent to 108 CFU/mL. This inoculum was diluted 100-fold (106 CFU/mL) and infiltrated on the abaxial surface of three young leaves (technical replicates) in three different plants (biological replicates) of “Pera Rio” orange (C. sinensis L. Osbeck) using 1 mL needleless hypodermic syringes [38]. Symptoms were observed for 25 days, and photos were taken at 4, 8, 12, 15, and 21 days after inoculation (DAI) [38].
Inoculated plants were kept in a high-efficiency particulate air (HEPA) filtered plant laboratory with controlled environmental conditions (28–30 °C, 55% humidity, 12 h light cycle).
2.6. Ex Vivo Growth Curves
X. citri and Δ4296 mutant were cultivated in NB medium for 16 h and diluted in fresh NB medium to O.D. 600-nm of ~0.1. Cell cultures were distributed on 96 well plates and were incubated in a Synergy H1N1 microplate reader (BioTek®, Winooski, VT, USA) under constant agitation at 29 °C, and automated O.D. readings were taken every 30 min. Using GraphPad Prism 6 software, growth curves were generated based on three technical and three biological replicates [7].
2.7. In Planta Growth Curves
X. citri and Δ4296 mutant were cultivated in NB medium for 16 h until O.D. 600 nm got around 0.8 and diluted in fresh NB medium to O.D. 600 nm of 0.3. Cells were collected by centrifugation and resuspended in Falcon tubes containing 50 mL of autoclaved tap water to an O.D. 600 nm of 0.3, equivalent to 108 CFU/mL. This inoculum was diluted 100-fold (106 CFU/mL) and infiltrated on the abaxial surface of fifteen young leaves in four different plants (biological replicates) of “Pera Rio” orange (C. sinensis L. Osbeck) using 1 ML needleless hypodermic syringes. The strains were exuded from leaves at days 0, 1, 3, 6, and 10 DAI, and the number of cells per leave was achieved using the microculture strategy [38].
2.8. Microscopy
X. citri, Δ4296 mutant, and Δ4296c strains were cultivated in NB media until O.D. 600-nm reached around 0.3 ABS at 29 °C. We performed analysis in different conditions: with ampicillin (20 µg/mL), sucrose 2% (w/v) and glutamic acid 2% (w/v). For morphological analysis, strains were collected by centrifugation, and cells were resuspended in 0.85% NaCl. We used 4′,6-diamidino-2-phenylindole DAPI staining at a final concentration of 0.01% to visualize chromosome organization. Cultures were treated with propidium iodide (IP) for cell viability investigations at a 0.001 mg/mL final concentration. Cells were immobilized in agarose-covered slides for microscope observation [39]. We performed the assays three times and quantified cells individually (n = 800). Following treatments, cells were immediately visualized using an Olympus BX61 microscope equipped with a monochromatic camera OrcaFlash 2.8 (Hamamatsu, Japan). The software CellSens Version 11 (Olympus, Tokyo, Japan) was used for data collection and analysis.
2.9. Data Analysis
All results obtained were submitted to Welch’s ANOVA test-0.05 using GraphPad Prism 8.0.1. Graphics were generated using Microsoft Office Excel for Windows.
3. Results
3.1. XAC4296 Is Conserved in Xanthomonas, Pseudoxanthomonas, and Stenotrophomonas, Showing Two Distinct and Independent Domains Modules
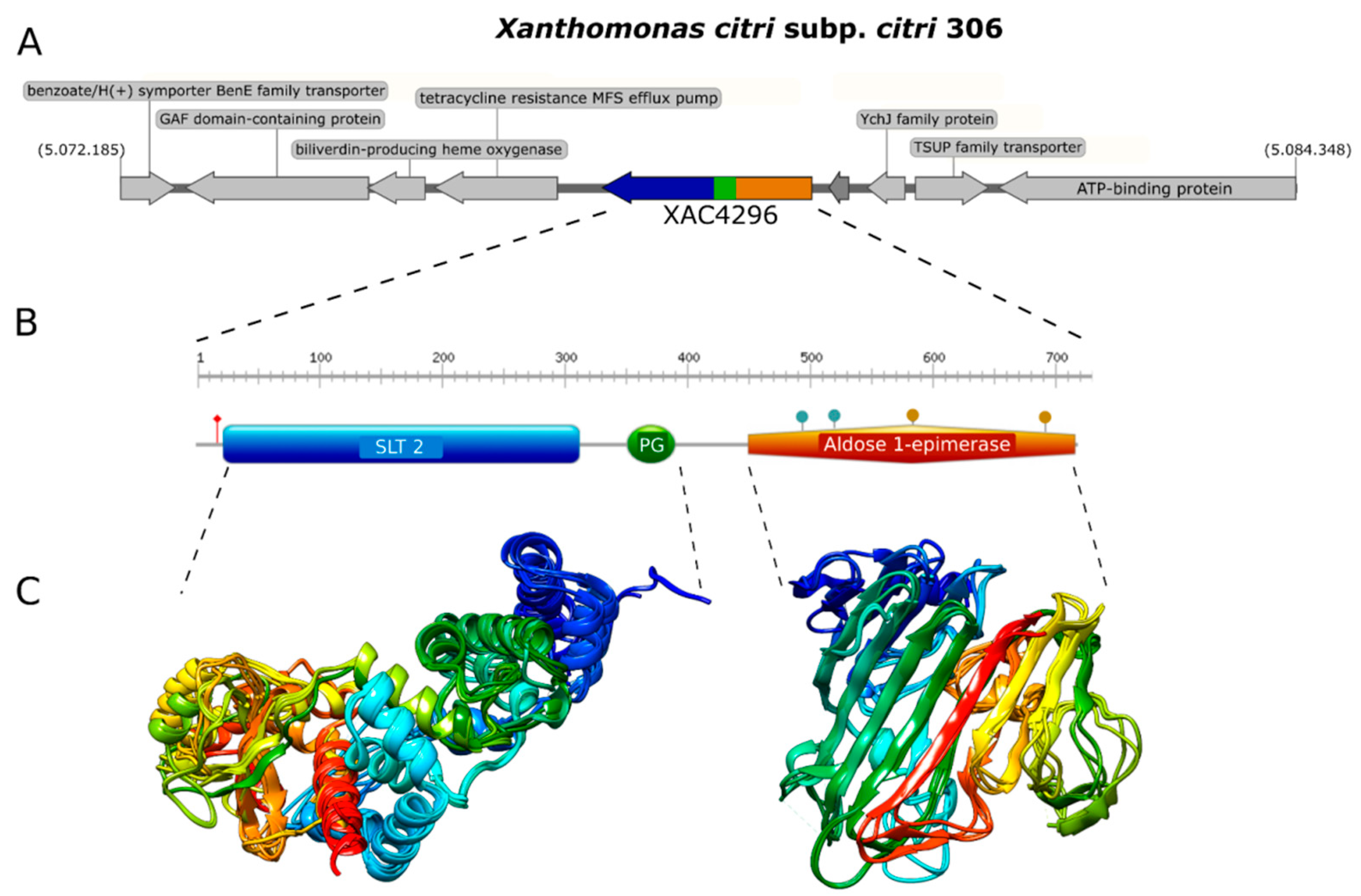
We first analyzed the XAC4296 gene (2163 bp) using molecular modeling approaches. The Robetta software failed to generate a unique 3D structure corresponding to the complete protein containing LT and epimerase domains. Therefore, both domains were modeled separately to unravel the XAC4296 (720 aa) structure. While the LT located on the N-terminus of XAC4296, containing the SLT_2 and PG_Binding 1 domains, was modeled based on a sequence of 408 amino acids, the XAC4296 C-terminus, having the epimerase bearing the aldose-1-epimerase domain, was modeled based on 312 amino acids sequence (Figure 1A,B).
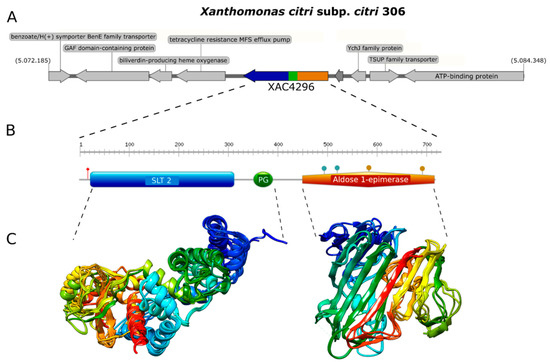
Figure 1.
Genome context, protein domain, and structure of XAC4296. (A). Genome context of XAC4296 from X. citri genome. (B). Protein domain and structure of XAC4296. XAC4296 has 720 aa with the LT domain associated with 3B family: Transglycosylase SLT domain (SLT_2) (IPR031304.) and Peptidoglycan binding (PG_binding_1) (IPR002477) domains, and the Aldose 1-epimerase (IPR015443) domain. (C). Molecular modeling cartoon representation of the LT and epimerase XAC4296 domains.
The first module resembles a classic 3B LT showing the 3D structure composed mainly of alpha-helices (Figure 1C). In contrast, the second module shows mainly beta-sheets, exhibiting high identity to other well-characterized D-hexose-6-phosphate mutarotase (Figure 1C). Therefore, it is presumed that the aldose 1-epimerase domain might perform the epimerization by ring-opening or mutarotation [40]. The confidence value for both models was 0.86 and 0.81, respectively, supporting a reasonable quality of both 3D structure predictions. These findings suggest that XAC4296 may have two completely independent domains that might preserve the transglycosylase and epimerase activities separately.
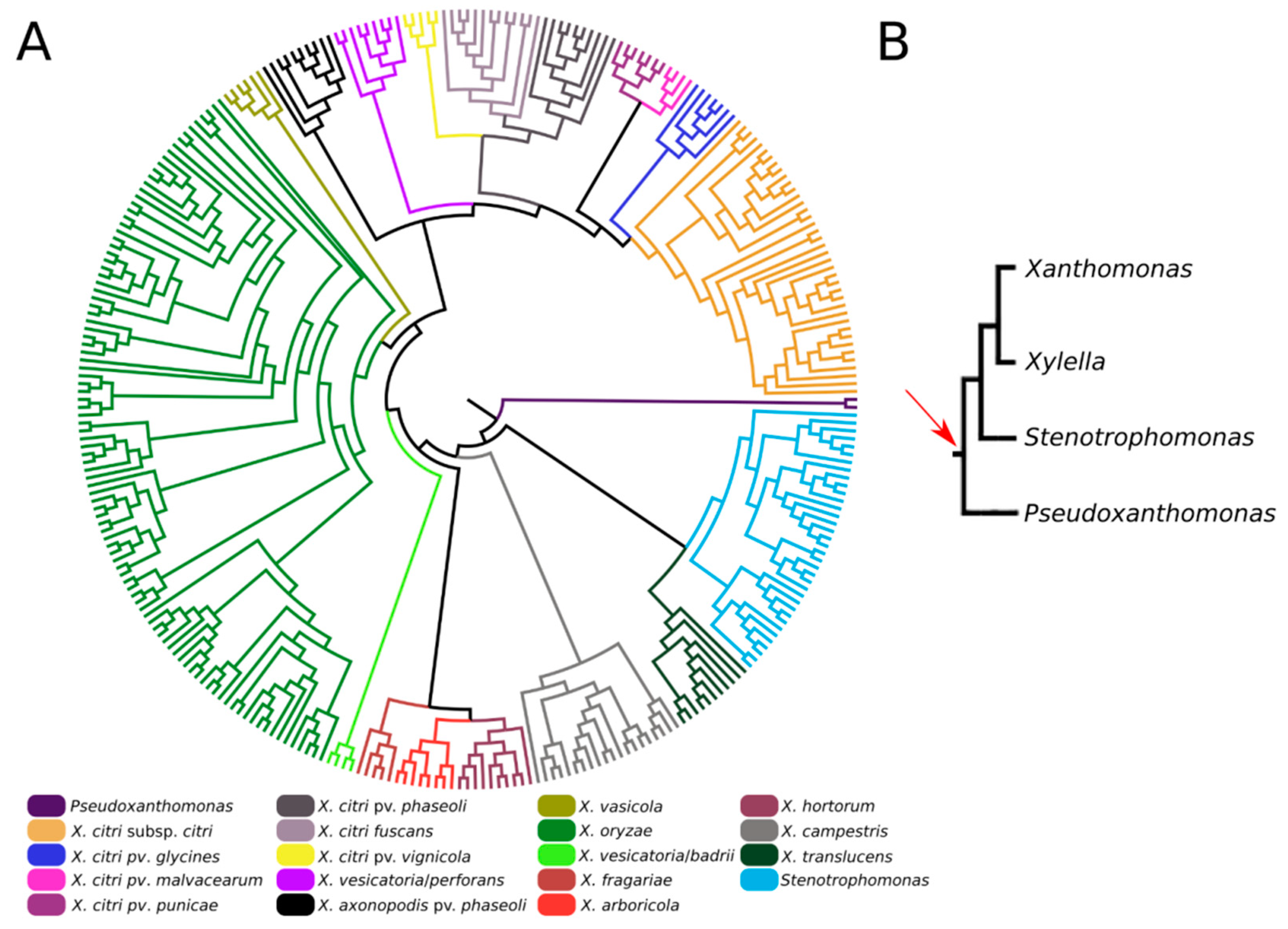
We also investigated the origin and evolution of XAC4296, revealing that apart from different species of Xanthomonas, this gene can also be found in different members of the Xanthomonadaceae family, such as Pseudoxanthomonas and Stenotrophomonas, accounting for at least 308 complete sequenced genomes available in the GenBank database (Figure 2A and Table S1). Moreover, considering these three genera, the XAC4296 homologs are generally located in a conserved genomic context (Figure S1). The XAC4296 and their homologs are not associated with common mobile genetic elements, such as prophages, Insertion Sequences, Transposons, Integrons, and Genomic Islands. Therefore, strongly supports that the XAC4296 origin is not associated with common lateral gene transfer mechanisms. It is noteworthy to mention that the XAC4296 homolog is not present in the Xanthomonas albilineans and the phylogenetically closely-related Xylella genus (Figure 2A). Xylella carries an independent epimerase gene (i.e., WP_010894718.1) and a least 4 LTs genes, all located at distant genomic loci. Conversely, in addition to an aldose 1-epimerase domain-containing gene (i.e., XALC_0947 and XaFJ1_GM000925), the X. albilineans species (e.g., GPE PC73 and Xa-FJ1 strains), also carries the two modules (LT and Epimerase) as separated and overlapping genes (21 nucleotides of overlapping), each gene showing the domains modules in different frames, and resembling a degenerated XAC4296 homolog (Figure S2). Moreover, the other Xanthomonas species, Pseudoxanthomonas and Stenotrophomonas carry their own set of LTs (varying in number and diversity of families). However, the aldose-1-epimerase domain is exclusive for each XAC4296 homolog and thus does not exist as an alone or duplicated feature such as the D-hexose-6-phosphate mutarotase gene in these genomes, as observed in Xylella and X. albilineans.
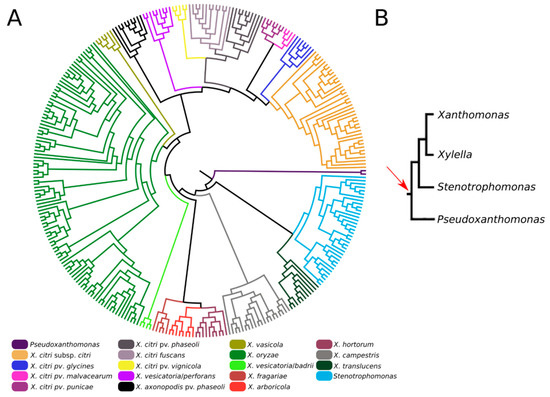
Figure 2.
(A). Maximum-likelihood phylogenetic tree of XAC4296 homologs across the Xanthomonadaceae family supports the XAC4296 potential origin before Xanthomonas, Xylella, Pseudoxanthomonas, and Stenotrophomonas differentiation. The closely-related Xylella genus lost the XAC4296 homolog. (B). Phylogenetic construction of the Xanthomonadaceae family phylogroup formed by Xanthomonas, Xylella, Pseudoxanthomonas, and Stenotrophomonas (based on [41]). The red arrow indicates the potential gene fusion event that originated the XAC4296 ancestor.
3.2. X. citri Expresses XAC4296 in Citrus sinensis L. Osbeck, and Δ4296 Mutant Does Not Affect Bacterial Growth but Impacts the CC Progression
We further investigated the role of XAC4296 on Xanthomonas citri virulence and pathogenicity. To confirm the XAC4296 expression during plant-pathogen interaction, Citrus sinensis L. Osbeck (considered a moderately resistant host) was inoculated with X. citri. The pathogen was exuded five days after inoculation. The synthesis of double-stranded cDNA from total RNA was performed and used as a template for PCR reaction. The fragment with approximately 2200 pb was obtained, indicating that XAC4296 is entirely expressed during plant-pathogen interaction (Figure S3).
Furthermore, the XAC4296 gene was deleted by site-directed mutagenesis to generate mutant X. citri Δ4296, and the integrative plasmid pMAJIIc [36] was employed to construct the complemented strain Δ4296c. Different bacterial inoculation methods (spray and infiltration) (Table 1 and Figure S4) and growth curve analyses (Figure S5) indicated that the wild-type and mutant exhibited similar growth patterns; pointing to the fact, however, that XAC4296 can be related to CC symptoms enhancement in different hosts (e.g., Citrus latifolia Tan, Citrus sinensis, and Citrus reticulata) regarding only the spray method.

Table 1.
Citrus canker quantification of “Pera Rio” orange leaves (Citrus sinensis L. Osbeck) by spray method. Strains were inoculated in a total of 65 leaves. The number of lesions was quantified on the abaxial surface of the leaves.
3.3. Δ4296 Cells Displayed Abnormal Nucleoid Distribution, Chains, and Short Filaments
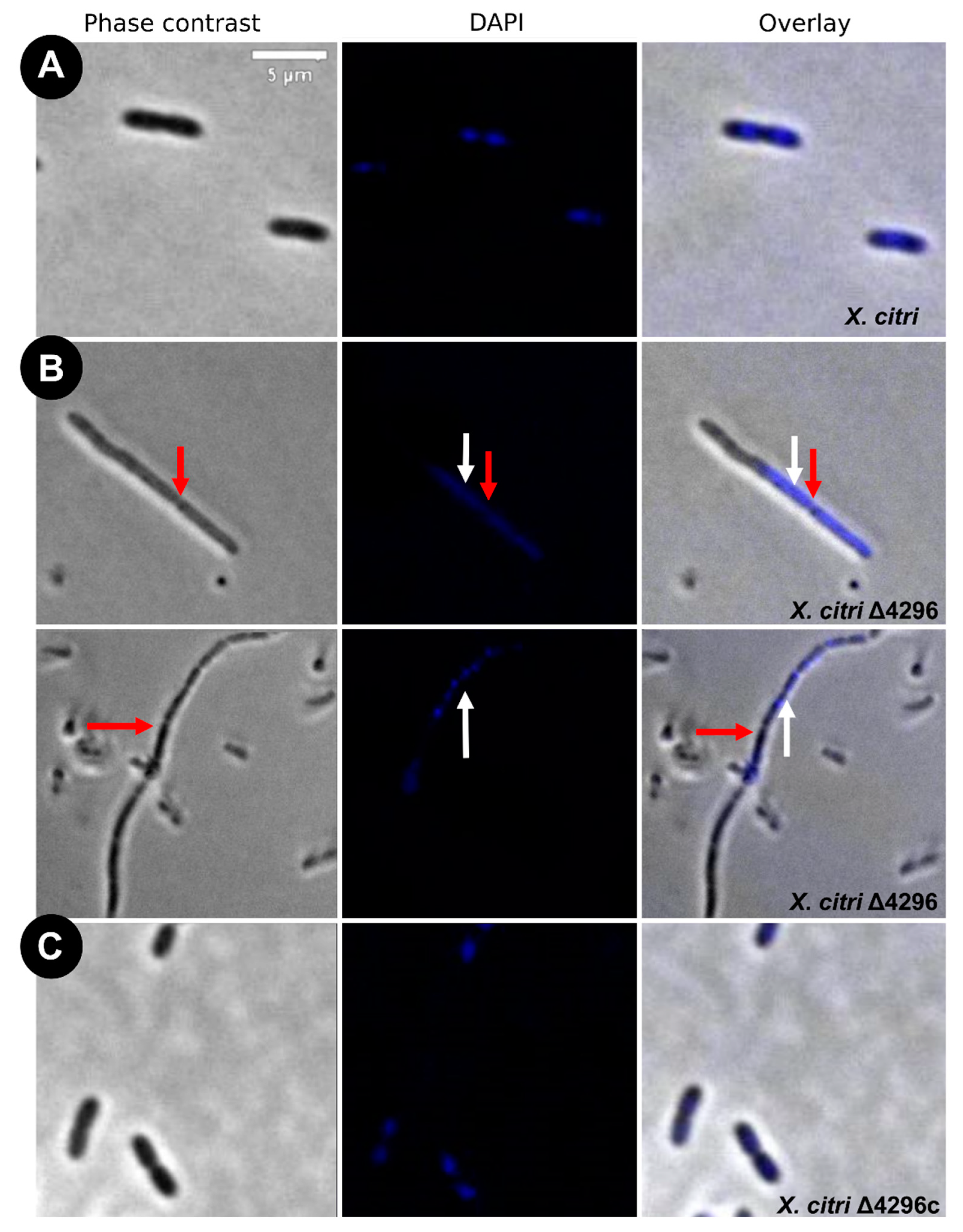
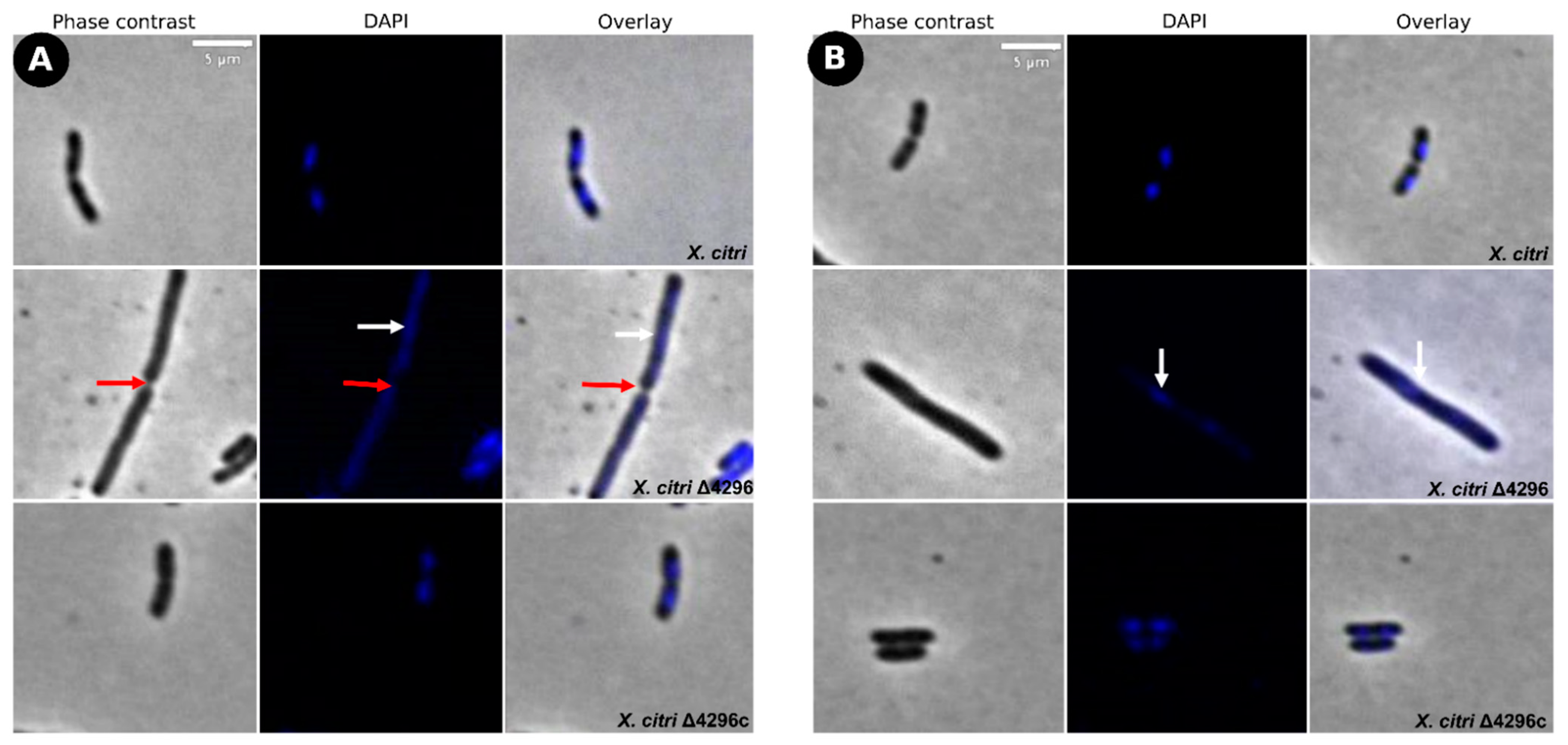
To evaluate the potential role of the XAC4296 LT domain with bacterial Peptidoglycan metabolism, the cell morphology and chromosome organization of wild-type X. citri and the Δ4296 mutant were investigated using DAPI-staining (Figure 3 and Table 2). The wild-type X. citri displays an average cell length of approximately 1.44 ± 0.31 µm [8]. Moreover, in standard growth conditions, X. citri exhibits a bilobed nucleoid centrally located in a single compartment (newborn or not-dividing rods) (Figure 3A) or evenly distributed, one per cell half, when division constriction is present.
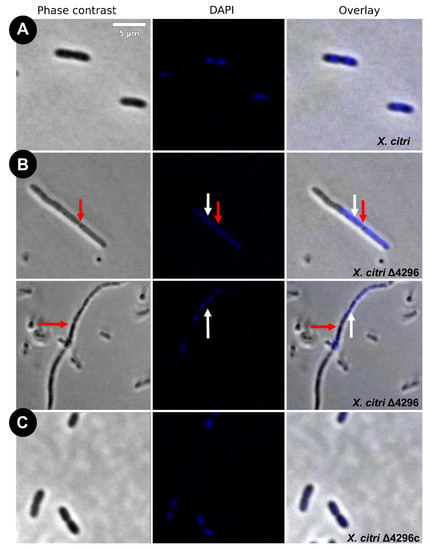
Figure 3.
Morphological analysis and errors/aberrant nucleoid distribution of X. citri, Δ4296, and Δ4296c strains. Short filaments, chain phenotype, and errors/unusual nucleoid distribution were intensified without XAC4296. The figure shows microscopy phase contrast, DAPI, and overlay of the two filters for (A) X. citri WT; (B) Δ4296, and (C) Δ4296c (pMAJIIc-XAC4296) (magnification of 100×). White arrows indicate nucleoid distribution; red arrows indicate septum constriction (magnification of 100×)—scale = 5 µm.

Table 2.
Statistics of morphotypes and nucleoid distribution of cells cultivated in rich medium (NB).
Different from the wild type, the Δ4296 mutant formed chains and sometimes a phenotype resembling short filaments, accompanied by irregular distribution of the chromosomal mass (Figure 3B, white arrows). The proportions of short filaments and chains measured in a given mutant culture were 30.6% and 15%, respectively (Table 3). Close inspection of the chains with clear septal constrictions (red arrows in Figure 3) showed no apparent nucleoid bisection. However, the Δ4296 mutant seems perfectly competent in chromosome segregation because many cells in a culture display normal nucleoid distribution (Figure 3B). However, in successive cellular cycles, errors in late cell division lead to the accumulation of chromosomal mass generated by subsequent events of replication. The short filaments morphotype that happened nearly twice as much as the chains could be caused by triggering the division error in an early stage of the cell cycle or converting chains into short filaments. At the moment, we cannot pinpoint which of these options is taking place. Finally, complementation of X. citri Δ4296 with pMAJIIc (Δ4296c) expressed from an ectopic site completely restored the wild-type phenotype (Figure 3C).

Table 3.
Statistics of morphotypes and nucleoid distribution of cells cultivated in rich medium (NB) and ampicillin.
These findings suggest that the first cell error probably occurs mainly during late cell division. The filamentation seems to happen due to the cell losing the ability to form septa in successive cellular cycles.
3.4. XAC4296 Is Required for Proper β-Lactam Resistance and May Affect Cell Wall Synthesis
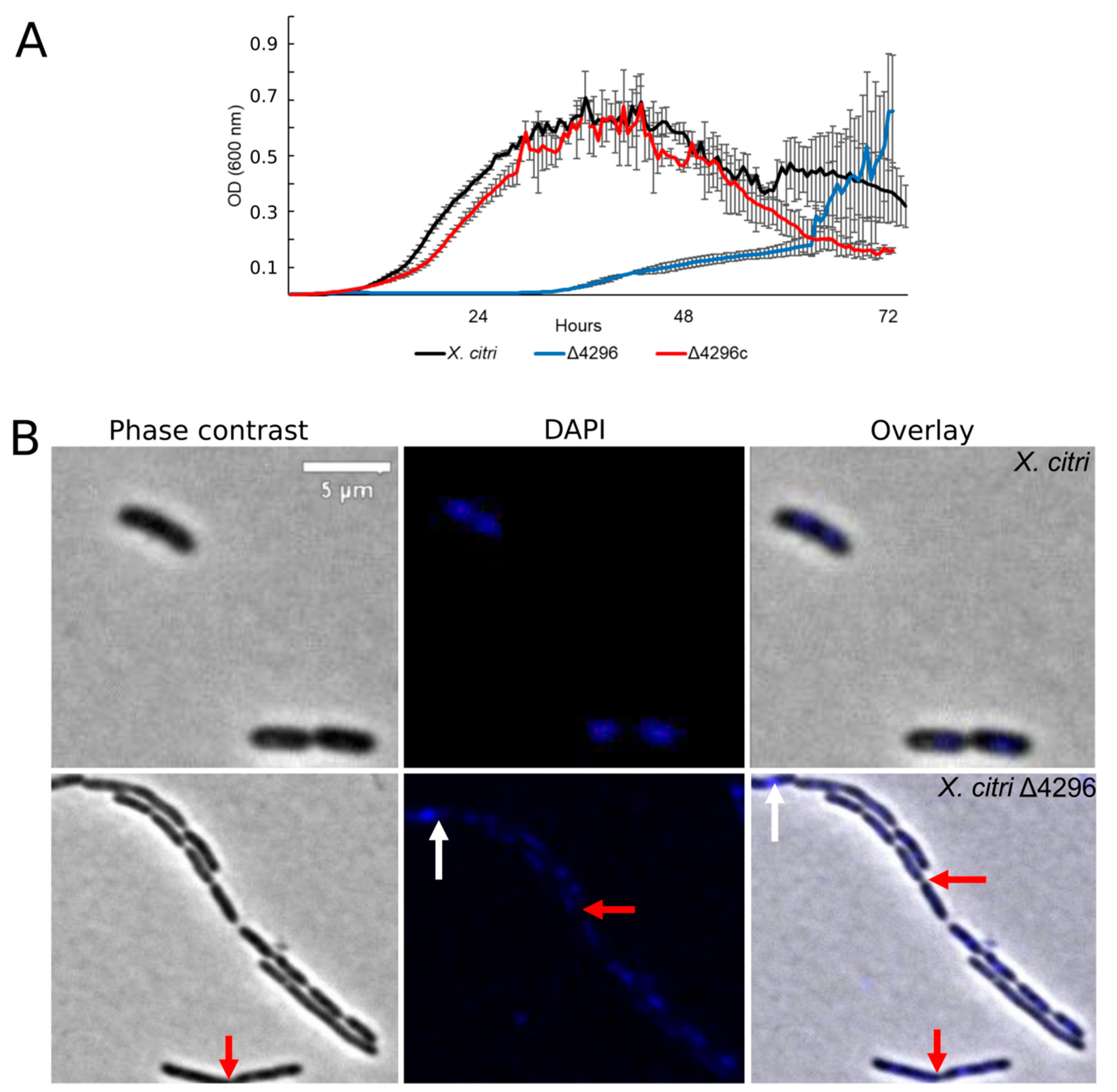
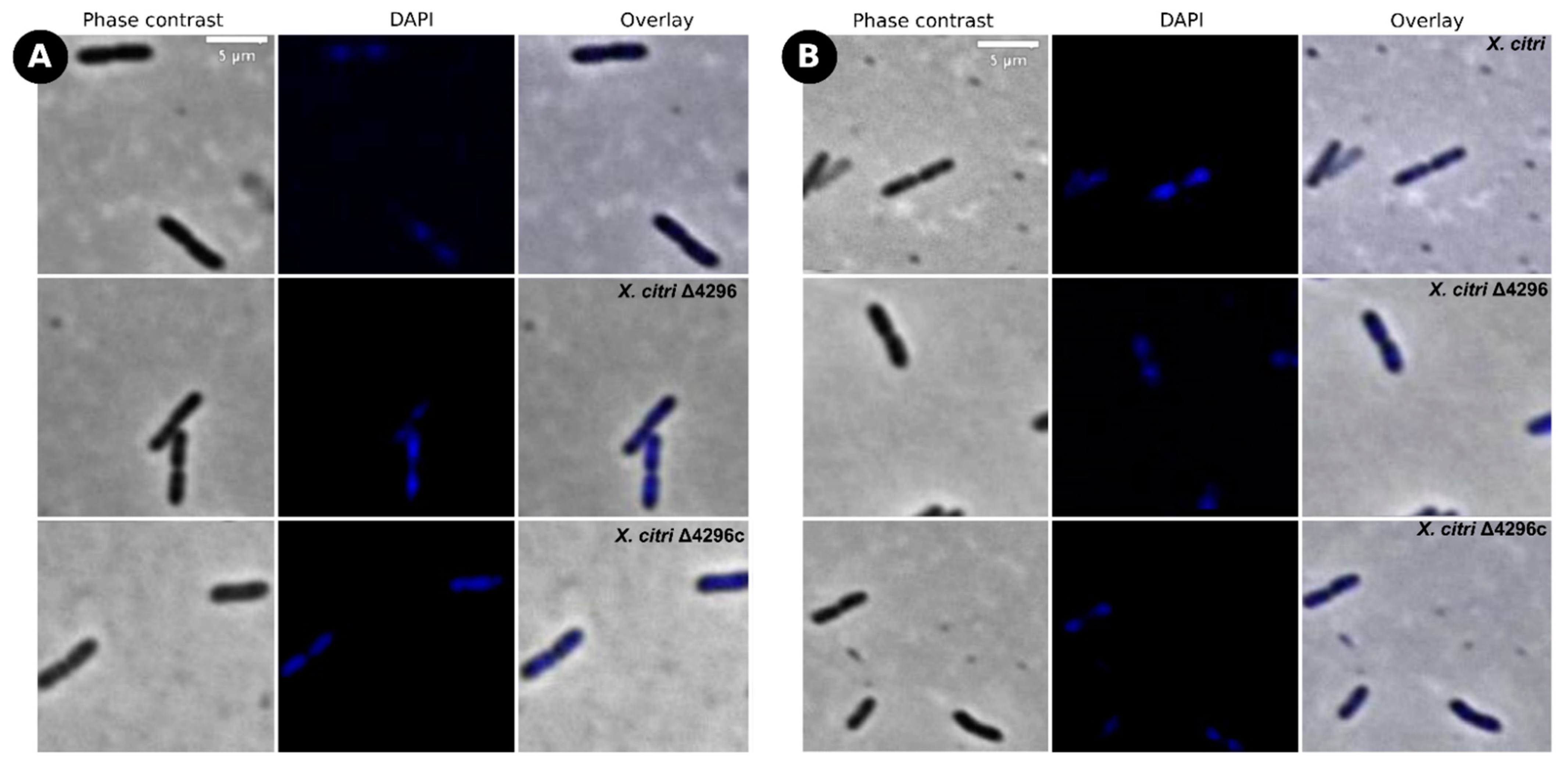
Considering that other members from the Xanthomonas genus are naturally resistant to ampicillin [42] (in X. citri the XAC_RS19350/XAC3833 gene provides the resistance), and the cell wall synthesis is closely associated with cell morphology, we further evaluated whether the XAC4296 deletion would affect the sensitivity to ampicillin, the cell morphology and the chromosome organization of wild type X. citri and the Δ4296 mutant in the presence of ampicillin antibiotic using DAPI-staining (Figure 4 and Table 3). The Δ4296 mutant showed a significant growth delay in the presence of ampicillin, suggesting that antibiotic sensitivity is pronounced (Figure 4A). All cells of the Δ4296 cultures displayed short filaments and chains with the abnormal distribution of the nucleoids (Figure 4B and Table 3), indicating an altered or disturbed nucleoid segregation. Permeability analysis showed that membrane disruption was not detected (Figure S6 and Table S4).
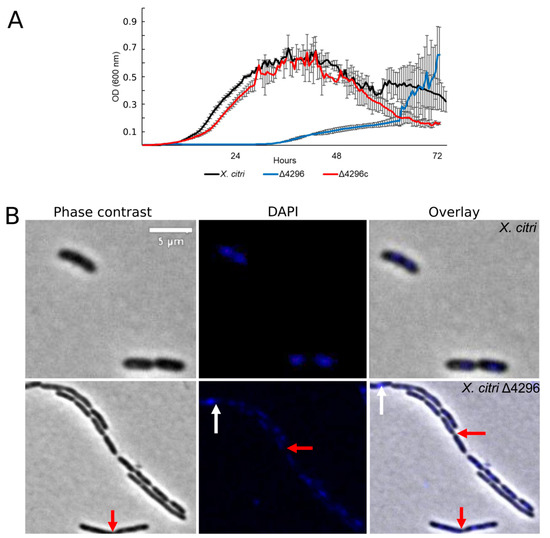
Figure 4.
XAC4296 may be related to β-lactam antibiotic resistance in X. citri (A) Ex planta bacterial growth curves performed on rich medium and ampicillin 20 µg/mL for X. citri, Δ4296, and Δ4296c. Δ4296 growth is affected in the presence of ampicillin. Error bars indicate the standard error of three independent biological and technical replicates. (B) Morphological analysis and errors/aberrant nucleoid distribution of X. citri, Δ4296, and Δ4296c strains of X. citri and Δ4296 strains on NB supplemented with ampicillin 20 µg/mL. All cultures of mutant Δ4296 exhibit short filaments and chain phenotype. The figure shows the phase contrast, DAPI, and overlay of the two images for X. citri and Δ4296 (magnification of 100×). White arrows indicate the chromosome position, and red arrows indicate septum constriction (magnification of 100×)—scale = 5 µm.
3.5. XAC4296 Is Related to Bacterial Central Carbon Metabolism
To evaluate the potential role of the XAC4296 epimerase domain, two different carbon and nitrogen sources were used to supplement the NB medium with ampicillin. In the presence of sucrose, the Δ4296 mutant cell shape and chromosome organization were partially restored (Figure 5 and Table 4), but cells still showed short filaments, chains, and unusual chromosome organization/segregation (Figure 5A,B and Figure S7, Table 4). In contrast, the addition of glutamic acid-enhanced Δ4296 growth to the point where it matches that of the wild type X. citri and fully reversing back to normal chromosome organization/segregation and cell division (Figure 6A,B and Figure S7, Table 4). Considering that: (a) D-hexose-6-phosphate mutarotase gene containing the aldose 1-epimerase domain can be related to epimerization by ring-opening or mutarotation acting in the central carbon metabolism and impacting the cell-wall metabolism, and (b) the supplementation with different carbon sources restores the Δ4296 mutant to the wild-type X. citri phenotype; these results support that the XAC4296 epimerase domain might have an essential relation with the central carbon metabolism.
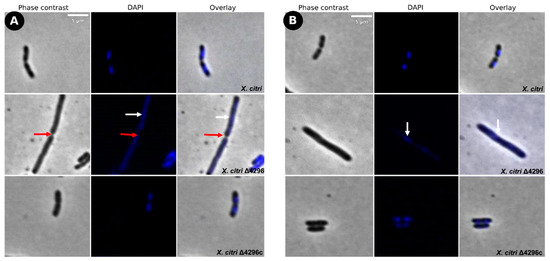
Figure 5.
Short filaments and chain phenotype show partial reversion in sucrose’s presence as carbon source (A) Morphological analysis of X. citri, Δ4296, and Δ4296c strains on NB supplemented with sucrose 0.1% (w/v). (B) NB supplemented with sucrose 0.1% (w/v) and ampicillin 20 µg/mL. The figure shows the phase contrast, DAPI, and overlay of the two filters for X. citri; Δ4296 and Δ4296c. White arrows indicate the chromosome distribution, and red arrows indicate septum constriction (magnification of 100×)—scale = 5 µm.

Table 4.
Short filaments, chain, and aberrant nucleoid organization phenotype were restored to normal on NB medium with sucrose or ampicillin and NB medium with glutamic acid or ampicillin.
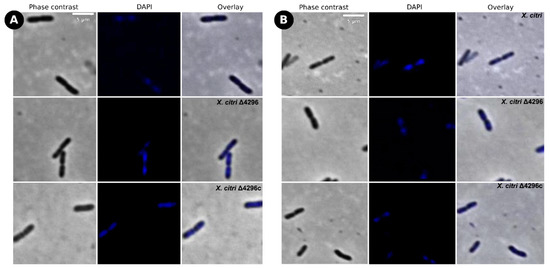
Figure 6.
Short filaments, chain phenotype, and nucleoid organization show full reversion in glutamate’s presence as carbon source (A) Morphological analysis of X. citri, Δ4296, and Δ4296c strains on NB supplemented with glutamic acid 0.1% (w/v). (B) NB supplemented with glutamic acid 0.1% (w/v) and ampicillin 20 µg/mL. The figure shows the phase contrast, DAPI, and overlay of the two filters for X. citri, Δ4296, and Δ4296c (magnification of 100×)—Scale = 5 µm.
4. Discussion
In this work, we investigate the role of the XAC4296 in Xanthomonas citri metabolism and virulence using in silico and molecular approaches. The XAC4296 protein contains two distinct modules: the first containing the SLT_2 (IPR031304) and PG_Binding1 (IPR002477) domains, homologs to LTs 3B Family. The second module contains the aldose-1-epimerase domain (IPR015443), classified as an epimerase superfamily. Proteins belonging to the LTs (Lytic Murein Transglycosylases) family cleave the polysaccharide of the peptidoglycan at the NAM-NAG glycosidic bond by intramolecular cyclization of the n-acetylmuramyl moiety to yield a 1,6-anhydro-n-acetyl-β-d-muramyl (1,6-anhydroMurNAc) product during the peptidoglycan biosynthesis [16]. On the other hand, epimerases are usually involved in metabolic pathways such as inversion of D-alanine and D-glutamate for bacterial cell wall metabolism [25]; biosynthesis of a variety of cell surface polysaccharides; biosynthesis of LPS and capsular sugar precursors [43]; and complex biosynthetic pathways, such as Glycolysis, Entner-Doudoroff, Leloir and others that present several chemical steps [44,45]. Epimerases are also involved in oxidation, acetylation, dehydration, and carbohydrate reduction (reviewed by [40]). Interestingly, we did not find any difference in plate colony appearance or biofilm production of the Δ4296 mutant compared to the X. citri wild type strain, indicating that, in principle, the XAC4296 epimerase domain is not related to cell surface polysaccharides production (data not shown). Indeed, the presented results indicated that the XAC4296 epimerase domain might be associated with other metabolic pathways, such as those related to bacterial cell wall metabolism. Our molecular modeling results also support that the LT and epimerase modules can act independently and may synergistically function as a canonical MFE, thus, performing multiple physiologically biochemical or biophysical functions simultaneously in the cell.
Previous studies revealed that the XAC4296 modules exist as separate and independent genes in other γ-proteobacteria [24]. Our results indicate that XAC4296 homolog is widespread in different Xanthomonas species and exists in other members of the Xanthomonadaceae family, such as Stenotrophomonas and Pseudoxanthomonas. It is worth mentioning that the Xanthomonas, Stenotrophomonas, Xylella, and Pseudoxanthomonas genera from the Xanthomonadaceae family are closely related and form a phylogroup [41]. Therefore, our results support that the XAC4296 origin may be related to a previous gene fusion origin before or during this phylogroup differentiation. However, Xylella and Xanthomonas albilineans do not have the XAC4296 homolog. For instance, Xylella carries an independent D-hexose-6-phosphate mutarotase gene containing the aldose-1-epimerase domain, and their LT gene repertoire is in a distinct genomic locus. In contrast, the X. albilineans show both protein modules, the LT and epimerase, homologs to XAC4296, as separate but overlapping genes and an additional and independent D-hexose-6-phosphate mutarotase gene. It is known that the Xylella genus has undergone drastic genome reduction since diverging from the Xanthomonas genus [46,47] and that X. albilineans has experienced significant genomic erosion, having unique genomic features in comparison to other Xanthomonas species [48]. Therefore, the most parsimonious hypothesis to explain the absences of XAC4296 homologs in Xylella and X. albilineans is that while Xylella lost their XAC4296 homolog during the genome reduction process, but maintaining their epimerase gene alone, and their own LTs repertoire, the X. albilineans genome is accumulating mutations and nucleotide deletions that led to the formation of two separated genes, and thus, suggesting a current process of gene decay. This hypothesis is based on the current knowledge that the genome reduction or erosion process is currently shaping Xyella and X. albilineans and adapting both pathogens to a restricted host range [46,47,48].
This work further evaluated XAC4296 role in X. citri fitness, virulence, and pathogenicity. Our results indicate that the XAC4296 gene is not essential for X. citri survival or in planta CC development but may play a role in bacterial fitness. Our findings also support XAC4296 direct relationship with X. citri pathogenicity, particularly CC progression. However, this result is not surprising since previous studies revealed a role of the other LT from the 3B family with X. citri pathogenicity and fitness [24].
To explore the involvement of the XAC4296 LT domain with the peptidoglycan metabolism, the cell morphology was examined under the microscope using DAPI-staining. X. citri lacking XAC4296 formed chains related to late cell division errors. In these cells, the early division is normal, forming constriction but does not progress until complete closure of the septum and cell separation. This is not an unexpected result since many studies based on LTs mutants (from different families) reported similar phenotypes [22,49,50,51]. Indeed, cell-wall biosynthesis stays in a homeostatic balance between construction and demolition [52]. In the absence of the activity of the LT domain provided by XAC4296, the peptidoglycan maintenance is perturbed, leading to these morphological defects observed in X. citri. This finding supports the hypothesis that the XAC4296 function is related to the 1,6-anhydroMurNAc-containing muropeptides production, the hallmark of LT catalysis [16]. These muropeptides may be transported from the periplasm to the cytoplasm through the transmembrane protein AmpG [53]. Next, these muropeptides are degraded in the cytoplasm, and their components are used for Lipid II biosynthesis that is assembled in the cytoplasm and, again, delivered to the periplasm for de novo synthesis of the peptidoglycan [54,55,56,57,58,59,60,61,62,63,64]. Therefore, our results support the XAC4296 LT role, acting in the peptidoglycan synthesis and dynamics, consequently influencing cell shape as previously described for this class of enzymes [16].
In this work, we observed that the short filaments phenotype intensified when ampicillin was added to the Δ4296 mutant culture, impacting cell growth. Ampicillin is a β-lactam antibiotic that blocks the activity of penicillin-binding proteins [65]. Although other Xanthomonas members (e.g., Xanthomonas campestris) appears to express β-lactamase constitutively [42], in the Δ4296 strain, the absence of XAC4296 protein seems to hinder their ability to reconstruct cell wall making the bacteria susceptible to this antibiotic. Indeed, the bacterial exposure to β-lactams leads to disturbs in peptidoglycan recycling and accumulation of MurNAc-peptides in the cytoplasm [66]. In addition, during LT catalysis, the product 1,6-anhydroMurNAc-containing muropeptides are transported from the periplasm to the cytoplasm [54]. These muropeptides are transported through the transmembrane protein AmpG [54]. The muropeptides may be metabolized further through multiple routes to yield UDP-MurNAc-pentapeptide, a precursor of peptidoglycan biosynthesis [21,67], or may bind to AmpR and convert it into an activator of ampC transcription [66,68]. Here, we hypothesize that the observed behavior of Δ4296 may be explained due to the bacterial decreasing of the pool of muropeptides in the periplasm, consequently in the cytoplasm, leading to peptidoglycan synthesis imbalance and interrupting ampC transcription, and consequently, increasing β-lactams susceptibility. Therefore, our results strongly support that XAC4296 protein may also contribute to β-lactam antibiotic resistance in X. citri.
However, the morphological defects observed in the XAC4296 mutant (with or without the addition of ampicillin) can be directly related to the epimerase domain since our results also indicate that the late cell division errors observed in the chain and short filaments phenotypes can be wholly restored with sucrose and glutamate supplementation. One possible way to interpret these results is that the XAC4296 epimerase domain may contribute to many reactions in carbohydrate metabolism, such as the D-hexose-6-phosphate mutarotase. Its absence leads to an imbalance of metabolic precursors related to anabolism pathways, indirectly affecting bacterial cell division and chromosome segregation in X. citri. In the absence of XAC4296, glutamate and sucrose supplement may probably provide substrate for alpha-glucose-6-phosphate production for the central carbon source metabolism, and together with the activity of the other LTs from the 3B family present in the X. citri genome and their known functional redundancy mechanism [16,24] restored the bacterial metabolism and, consequently, the cell cycle. In addition, the glucose-6 phosphate is the primary metabolic substrate present for the glycolysis, phosphogluconate, and Entner–Doudoroff pathways. Nonetheless, the glutamate and sucrose supplement may provide a comprehensive carbon source to the bacteria, facilitating energy metabolism and bacterial adaptation and survival to stressful conditions, free-living, and wide host ranges. However, further studies such as protein purification and in vitro tests to check XAC4296 enzymatic activity are still needed to determine its role thoroughly.
Finally, previous studies raised the possibility of combining an LT inhibitor with β-lactam antibiotics as an alternative for future antibiotic development [69]. For example, the possible interaction between the LT Slt35 (3B family) and Bulgecin A [70], a potent LTs inhibitor that restores the efficiency of β-lactam antibiotics against resistant bacteria [69]. Taken together, our results strongly support these previous studies, suggesting the future development of 3B LT and epimerase inhibitors as potential new tools, not only for the control of the disease caused by Xanthomonas and other phytopathogens but also for antimicrobial resistance in general.
5. Conclusions
The XAC4296 protein seems to be related to late cell division leading to chromosome segregation errors and ampicillin susceptibility. Moreover, we have shown that XAC4296 mutants display a metabolism-dependent phenotype, resulting from the imbalance of metabolic precursors related to anabolism pathways, suggesting that XAC4296 also acts in central carbon metabolism. In conclusion, our results strongly indicate that XAC4296 encodes a multifunctional protein, playing a role as a transglycosylase as much as an epimerase, impacting bacterial fitness and bringing new insights into X. citri and other Xanthomonadaceae metabolism, evolution, and antimicrobial resistance emergence.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10051008/s1, Figure S1: Genome context analysis of XAC4296 homologs across Xanthomonas, Pseudoxanthomonas, and Stenotrophomonas spp. The central gray rectangle represents the XAC4296 position for each Xanthomonadaceae genome; Figure S2: Comparative analyses of XAC4296 gene nucleotide structure and domains localization against the Xanthomonas albilineans strain Xa-FJ1 and GPE PC73 respective locus, showing the presence of the Epimerase and LT 3B as separated but overlapping genes; Figure S3: XAC4296 expression. 1% agarose gels showing expression of the XAC4296 in X. citri (M) 1 Kb Fermentas DNA Ladder marker. (1) PCR product from genomic DNA from X. citri (~2400 bp) used as a positive control (non-infecting condition). (2, 3, 4) cDNA from X. citri representing the in vitro expression of XAC4296; Figure S4: Pathogenicity tests. (A) Spray method. Citrus canker disease progression among X. citri and the mutant Δ4296 after 25 days of inoculation. Each bacterial suspension was sprayed at 108 CFU/mL into a whole moderately susceptible cultivar, “Pera Rio” orange (Citrus sinensis L. Osbeck) tree. Leaves of each treatment with the highest disease severity were photographed, and symptoms were quantified. (B) Infiltration method. Citrus canker disease progression among X. citri, Δ4296, Δ4296c, and autoclaved tap water as negative control after 25 days of inoculation. The inoculum was infiltrated at 106 CFU/mL at two points on the abaxial surface of three young leaves (technical replicates) in three different plants (Biological replicates) of the most susceptible cultivar, “Galego” acid lime (Citrus latifolia Tan), moderately susceptible cultivar “Pêra Rio” orange (Citrus sinensis L. Osbeck) and least susceptible “Ponkan” orange variety (Citrus reticulata) using 1 mL needleless hypodermic syringes. On each leaf, X. citri and Δ4296c were infiltrated on the central vein’s left-hand side, while mutant and negative control were infiltrated on the right-hand side; Figure S5: Comparison of the growth curves of ex planta bacterial growth and in planta bacterial growth. (A) Ex planta bacterial growth curve performed on rich medium NB for 72 h. (B) Ex planta bacterial growth was achieved on XVM2 defined medium for 72 h. (C) In planta bacterial growth curve. Error bars indicate the standard error of three independent biological and technical replicates; Figure S6: Live-dead assay for X. citri and mutant strain Δ4296. Quantitative analysis of live-dead cells showed 10% mortality for X. citri strain and 12% for Δ4296 mutant. The figure shows the phase contrast, DAPI, Propidium iodide (IP), and an overlay of the two filters, respectively (magnification of 100×). Scale = 5 μm; Figure S7: Ex planta growth curve for X. citri, Δ4296 mutant, and Δ4296c in different conditions. A: NB medium supplemented with sucrose 0.1%; B: NB medium supplemented with sucrose 0.1% (w/v) and ampicillin 20 μg/mL; C: NB medium supplemented with glutamic acid 2% (w/v); D: NB medium supplemented with glutamic acid 0.1% (w/v) and ampicillin 20 μg/mL. Error bars indicate the standard error of three independent biological and technical replicates. Table S1: Xanthomonadaceae family genomes exhibiting XAC4296 homologs and used for comparative genomics. In this table, there are 2 species for the Pseudoxanthomonas genus, 36 species for the Stenotrophomonas genus, and 270 species for the Xanthomonas genus; Table S2: List of strains and plasmids used in this study; Table S3: Oligonucleotides utilized in this study; Table S4: Live-dead quantification for X. citri constructions on rich medium with ampicillin 20 μg/mL.
Author Contributions
A.C.P.d.O. conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft. R.M.F. conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft. M.I.T.F. contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, and approved the final draft. J.A.F. conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, and approved the final draft. C.Z. performed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft. H.F. conceived and designed the ex planta growth curves and microscopy assays experiments and approved the final draft. A.M.V. conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES)-Finance Code 001. A.M.V., H.F., J.A.F. and M.I.T.F. received a productivity fellowship from the Brazilian National Research Council (CNPq), and RMF is a recipient of a post-doctoral PNPD fellowship from CAPES and provided by the Agronomy (Genetics and Plant Breeding) Graduate Program of the Faculty of Agricultural and Veterinary Sciences (FCAV-UNESP-Jaboticabal).
Acknowledgments
We thank the Center for Biological Resources and Genomic Biology (CREBIO) from São Paulo State University (UNESP) at Jaboticabal Campus, Brazil, for the molecular biology laboratory support. We also thank the lab team at the Laboratory of Bacterial Genetics of the Department of General and Applied Biology, Biosciences Institute of São Paulo State University (UNESP) at Rio Claro Campus, Brazil, and all the members of the Laboratory of Molecular Biology of the Department of Agricultural and Environmental Sciences of São Paulo State University (UNESP) at Jaboticabal Campus, Brazil for all the laboratory support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- An, S.Q.; Potnis, N.; Dow, M.; Vorhölter, F.J.; He, Y.Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2019, 44, 1–32. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Grajales, A.; Arrieta-Ortiz, M.L.; Salazar, C.; Restrepo, S.; Bernal, A. Genomes-based phylogeny of the genus Xanthomonas. BMC Microbiol. 2012, 12, 43. [Google Scholar] [CrossRef]
- Gottwald, T.R.; Graham, S.J.; Schubert, T.S. Citrus canker: The Pathogen and Its Impact. APS Publ. 2002, 3, 36. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef]
- Ryan, R.P.; Vorhölter, F.J.; Potnis, N.; Jones, J.B.; Van Sluys, M.A.; Bogdanove, A.J.; Dow, J.M. Pathogenomics of Xanthomonas: Understanding bacterium-plant interactions. Nat. Rev. Microbiol. 2011, 9, 344–355. [Google Scholar] [CrossRef]
- Büttner, D.; Bonas, U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 2010, 34, 107–133. [Google Scholar] [CrossRef]
- Lacerda, L.A.; Cavalca, L.B.; Martins, P.M.M.; Govone, J.S.; Bacci, M.; Ferreira, H. Protein depletion using the arabinose promoter in Xanthomonas citri subsp. citri. Plasmid 2017, 90, 44–52. [Google Scholar] [CrossRef]
- Ucci, A.P.; Martins, P.M.M.; Lau, I.F.; Bacci, M.; Belasque, J.; Ferreira, H. Asymmetric chromosome segregation in Xanthomonas citri ssp. citri. Microbiologyopen 2014, 3, 29–41. [Google Scholar] [CrossRef]
- Sriram, G.; Martinez, J.A.; McCabe, E.R.B.; Liao, J.C.; Dipple, K.M. Single-gene disorders: What role could moonlighting enzymes play? Am. J. Hum. Genet. 2005, 76, 911–924. [Google Scholar] [CrossRef]
- Hult, K.; Berglund, P. Enzyme promiscuity: Mechanism and applications. Trends Biotechnol. 2007, 25, 231–238. [Google Scholar] [CrossRef]
- Moore, B.D. Bifunctional and moonlighting enzymes: Lighting the way to regulatory control. Trends Plant Sci. 2004, 9, 221–228. [Google Scholar] [CrossRef]
- Vogel, C.; Bashton, M.; Kerrison, N.D.; Chothia, C.; Teichmann, S.A. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 2004, 14, 208–216. [Google Scholar] [CrossRef]
- Aharoni, A.; Gaidukov, L.; Khersonsky, O.; Gould, S.M.Q.; Roodveldt, C.; Tawfik, D.S. The “evolvability” of promiscuous protein functions. Nat. Genet. 2005, 37, 73–76. [Google Scholar] [CrossRef]
- Jeffery, C.J. Multifunctional proteins: Examples of gene sharing. Ann. Med. 2003, 35, 28–35. [Google Scholar] [CrossRef]
- Alcorlo, M.; Martínez-Caballero, S.; Molina, R.; Hermoso, J.A. Carbohydrate recognition and lysis by bacterial peptidoglycan hydrolases. Curr. Opin. Struct. Biol. 2017, 44, 87–100. [Google Scholar] [CrossRef]
- Dik, D.A.; Marous, D.R.; Fisher, J.F.; Mobashery, S. Lytic transglycosylases: Concinnity in concision of the bacterial cell wall. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 503–542. [Google Scholar] [CrossRef]
- Höltje, J.V. From growth to autolysis: The murein hydrolases in Escherichia coli. Arch. Microbiol. 1995, 164, 243–254. [Google Scholar] [CrossRef]
- Koraimann, G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 2003, 60, 2371–2388. [Google Scholar] [CrossRef]
- Scheurwater, E.; Reid, C.W.; Clarke, A.J. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 2008, 40, 586–591. [Google Scholar] [CrossRef]
- Scheurwater, E.M.; Burrows, L.L. Maintaining network security: How macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 2011, 318, 1–9. [Google Scholar] [CrossRef]
- Uehara, T.; Park, J.T. Growth of Escherichia coli: Significance of peptidoglycan degradation during elongation and septation. J. Bacteriol. 2008, 190, 3914–3922. [Google Scholar] [CrossRef]
- Cloud, K.A.; Dillard, J.P. Mutation of a Single Lytic Transglycosylase Causes Aberrant Septation and Inhibits Cell Separation of Neisseria gonorrhoeae. J. Bacteriol. 2004, 186, 7811–7814. [Google Scholar] [CrossRef]
- Jenkins, C.H.; Wallis, R.; Allcock, N.; Barnes, K.B.; Richards, M.I.; Auty, J.M.; Galyov, E.E.; Harding, S.V.; Mukamolova, G.V. The lytic transglycosylase, LtgG, controls cell morphology and virulence in Burkholderia pseudomallei. Sci. Rep. 2019, 9, 11060. [Google Scholar] [CrossRef]
- Oliveira, A.C.P.; Ferreira, R.M.; Ferro, M.I.T.; Ferro, J.A.; Chandler, M.; Varani, A.M. Transposons and pathogenicity in Xanthomonas: Acquisition of murein lytic transglycosylases by TnXax1 enhances Xanthomonas citri subsp. citri 306 virulence and fitness. PeerJ 2018, 6, e6111. [Google Scholar] [CrossRef]
- Sala, R.F.; Morgan, P.M.; Tanner, M.E. Enzymatic formation and release of a stable glycal intermediate: The mechanism of the reaction catalyzed by UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 1996, 118, 3033–3034. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, 526–531. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Chen, I.M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, 115–122. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Bueno, D.; Pedrolli, D.B.; Martins, P.M.M.; Bocchini, D.A.; Moraes, K.C.M.; Facincani, A.P.; Ferro, J.A.; Varani, A.M.; Pena, M.M.; Ferreira, H. Riboswitch theo/mete as a transcription regulation tool for Xanthomonas citri subsp. citri. Microorganisms 2021, 9, 329. [Google Scholar] [CrossRef]
- Lee, J.; Lee, H.J.; Shin, M.K.; Ryu, W.S. Versatile PCR-mediated insertion or deletion mutagenesis. Biotechniques 2004, 36, 398–400. [Google Scholar] [CrossRef]
- Amaral, A.M.D.; Toledo, C.P.; Baptista, J.C.; Machado, M.A. Transformation of Xanthomonas axonopodis pv. citri by electroporation. Fitopatol. Bras. 2005, 30, 292–294. [Google Scholar] [CrossRef]
- Kaniga, K.; Delor, I.; Cornelis, G.R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: Inactivation of the blaA gene of Yersinia enterocolitica. Gene 1991, 109, 137–141. [Google Scholar] [CrossRef]
- Pena, M.M.; Teper, D.; Ferreira, H.; Wang, N.; Sato, K.U.; Ferro, M.I.T.; Ferro, J.A. mCherry fusions enable the subcellular localization of periplasmic and cytoplasmic proteins in Xanthomonas sp. PLoS ONE 2020, 15, e0236185. [Google Scholar] [CrossRef]
- Li, J.; Wang, N. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol. Plant Pathol. 2011, 12, 381–396. [Google Scholar] [CrossRef]
- Laia, M.L.; Moreira, L.M.; Dezajacomo, J.; Brigati, J.B.; Ferreira, C.B.; Ferro, M.I.T.; Silva, A.C.R.; Ferro, J.A.; Oliveira, J.C.F. New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC Microbiol. 2009, 9, 12. [Google Scholar] [CrossRef]
- Martins, P.M.M.; Lau, I.F.; Bacci, M.; Belasque, J.; Do Amaral, A.M.; Taboga, S.R.; Ferreira, H. Subcellular localization of proteins labeled with GFP in Xanthomonas citri ssp. citri: Targeting the division septum. FEMS Microbiol. Lett. 2010, 310, 76–83. [Google Scholar] [CrossRef]
- Allard, S.T.M.; Giraud, M.F.; Naismith, J.H. Epimerases: Structure, function and mechanism. Cell. Mol. Life Sci. 2001, 58, 1650–1665. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S.; Kaur, A.; Singh, A.; Patil, P.B. Deep phylo-taxono genomics reveals Xylella as a variant lineage of plant associated Xanthomonas and supports their taxonomic reunification along with Stenotrophomonas and Pseudoxanthomonas. Genomics 2021, 113, 3989–4003. [Google Scholar] [CrossRef]
- Weng, S.F.; Lin, J.W.; Chen, C.H.; Chen, Y.Y.; Tseng, Y.H.; Tseng, Y.H. Constitutive Expression of a Chromosomal Class A (BJM Group 2) β-Lactamase in Xanthomonas campestris. Antimicrob. Agents Chemother. 2004, 48, 209–215. [Google Scholar] [CrossRef]
- McNeil, M.; Daffe, M.; Brennan, P.J. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 1990, 265, 18200–18206. [Google Scholar] [CrossRef]
- Nowitzki, U.; Wyrich, R.; Westhoff, P.; Henze, K.; Schnarrenberger, C.; Martin, W. Cloning of the amphibolic Calvin cycle/OPPP enzyme d-ribulose-5-phosphate 3-epimerase (EC 5.1.3.1) from spinach chloroplasts: Functional and evolutionary aspects. Plant Mol. Biol. 1995, 29, 1279–1291. [Google Scholar] [CrossRef]
- Teige, M.; Kopriva, S.; Bauwe, H.; Süss, K.H. Chloroplast pentose-5-phosphate 3-epimerase from potato: Cloning, cDNA sequence, and tissue-specific enzyme accumulation. FEBS Lett. 1995, 377, 349–352. [Google Scholar] [CrossRef]
- Firrao, G.; Scortichini, M.; Pagliari, L. Orthology-based estimate of the contribution of horizontal gene transfer from distantly related bacteria to the intraspecific diversity and differentiation of Xylella fastidiosa. Pathogens 2021, 10, 46. [Google Scholar] [CrossRef]
- Pieretti, I.; Royer, M.; Barbe, V.; Carrere, S.; Koebnik, R.; Cociancich, S.; Couloux, A.; Darrasse, A.; Gouzy, J.; Jacques, M.A.; et al. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genom. 2009, 10, 616. [Google Scholar] [CrossRef]
- Pieretti, I.; Pesic, A.; Petras, D.; Royer, M.; Süssmuth, R.D.; Cociancich, S. What makes Xanthomonas albilineans unique amongst xanthomonads? Front. Plant Sci. 2015, 6, 289. [Google Scholar] [CrossRef]
- Heidrich, C.; Ursinus, A.; Berger, J.; Schwarz, H.; Höltje, J.V. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 2002, 184, 6093–6099. [Google Scholar] [CrossRef]
- Jorgenson, M.A.; Chen, Y.; Yahashiri, A.; Popham, D.L.; Weiss, D.S. The Bacterial Septal Ring Protein RlpA is a Lytic Transglycosylase that Contributes to Rod Shape and Daughter Cell Separation in Pseudomonas aeruginosa. Mol. Microbiol. 2015, 93, 319–335. [Google Scholar] [CrossRef]
- Lommatzsch, R.; Templin, M.F.; Kraft, A.R.; Vollmer, W.; Biochemie, A. Outer Membrane Localization of Murein Hydrolases: MltA, a Third Lipoprotein Lytic Transglycosylase in Escherichia coli. J. Bacteriol. 1997, 179, 5465–5470. [Google Scholar] [CrossRef]
- Johnson, J.W.; Fisher, J.F.; Mobashery, S. Bacterial cell-wall recycling. Ann. N. Y. Acad. Sci. 2014, 1277, 54–75. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.; Huang, L.J.; Bartowsky, E.; Normark, S.; Park, J.T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994, 13, 4684–4694. [Google Scholar] [CrossRef] [PubMed]
- Barreteau, H.; Kovač, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef] [PubMed]
- Bouhss, A.; Trunkfield, A.E.; Bugg, T.D.H.; Mengin-Lecreulx, D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 2008, 32, 208–233. [Google Scholar] [CrossRef]
- Butler, E.K.; Davis, R.M.; Bari, V.; Nicholson, P.A.; Ruiz, N. Structure-Function Analysis of MurJ Reveals a Solvent-Exposed Cavity Containing Residues Essential for Peptidoglycan Biogenesis in Escherichia coli. J. Bacteriol. 2013, 195, 4639–4649. [Google Scholar] [CrossRef]
- Kuk, A.C.Y.; Mashalidis, E.H.; Lee, S.; Carolina, N. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat. Struct. Mol. Biol. 2017, 24, 171–176. [Google Scholar] [CrossRef]
- Leclercq, S.; Derouaux, A.; Olatunji, S.; Fraipont, C.; Egan, A.J.F.; Vollmer, W.; Breukink, E.; Terrak, M. Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci. Rep. 2017, 7, 43306. [Google Scholar] [CrossRef]
- Meeske, A.J.; Sham, L.; Kimsey, H.; Koo, B.; Gross, C.A.; Bernhardt, T.G. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2015, 112, 6437–6442. [Google Scholar] [CrossRef]
- Mohammadi, T.; Sijbrandi, R.; Lutters, M.; Verheul, J.; Martin, N.I.; Den Blaauwen, T.; De Kruijff, B.; Breukink, E. Specificity of the Transport of Lipid II by FtsW in Escherichia coli. J. Biol. Chem. 2014, 289, 14707–14718. [Google Scholar] [CrossRef]
- Scheffers, D.J.; Tol, M.B. LipidII: Just Another Brick in the Wall? PLoS Pathog. 2015, 11, e1005213. [Google Scholar] [CrossRef]
- Sham, L.-T.; Butler, E.K.; Lebar, M.D.; Kahne, D.; Bernhardt, T.G.; Ruiz, N. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 2014, 345, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Sieger, B.; Schubert, K.; Donovan, C.; Bramkamp, M. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol. Microbiol. 2013, 90, 966–982. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Bertsche, U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 1714–1734. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta-Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.; Fre, J. Cytosolic Intermediates for Cell Wall Biosynthesis and Degradation Control Inducible β-Lactam Resistance in Gram-Negative Bacteria. Cell 1997, 88, 823–832. [Google Scholar] [CrossRef]
- Gisin, J.; Schneider, A.; Nägele, B.; Borisova, M.; Mayer, C. A cell wall recycling shortcut that bypasses peptidoglycan de novo biosynthesis. Nat. Chem. Biol. 2013, 9, 491–493. [Google Scholar] [CrossRef]
- Dietz, H.; Pfeifle, D.; Wiedemann, B. The Signal Molecule for β-Lactamase Induction in Enterobacter cloacae Is the Anhydromuramyl-Pentapeptide. Antimicrob. Agents Chemother. 1997, 41, 2113–2120. [Google Scholar] [CrossRef]
- Williams, A.H.; Wheeler, R.; Thiriau, C.; Haouz, A.; Taha, M.K.; Boneca, I.G. Bulgecin a: The key to a broad-spectrum inhibitor that targets lytic transglycosylases. Antibiotics 2017, 6, 8. [Google Scholar] [CrossRef]
- Van Asselt, E.J.; Dijkstra, A.J.; Kalk, K.H.; Takacs, B.; Keck, W.; Dijkstra, B.W. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure 1999, 7, 1167–1180. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).