A Porcine Model for the Development and Testing of Preoperative Skin Preparations
Abstract
:1. Introduction
2. Materials and Methods
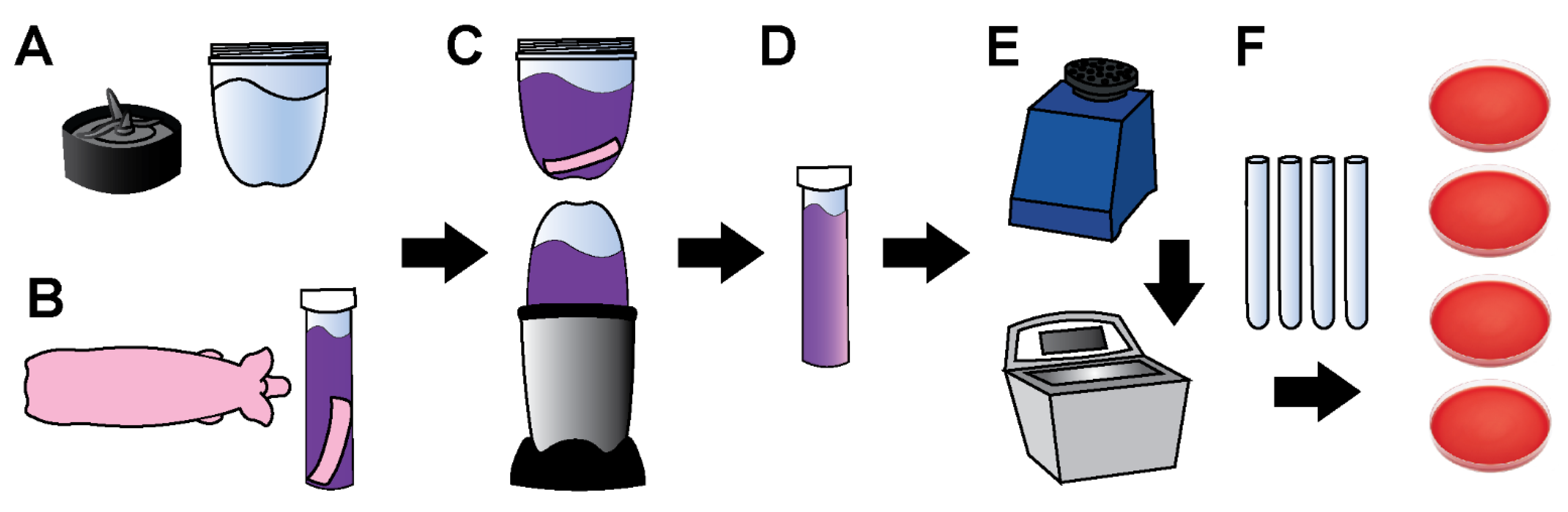
2.1. Supplies, Instruments, and Reagents
2.2. Tissue Sample Collection
2.3. Tissue Processing and Bioburden Quantification
2.4. Processing Controls
2.5. Bacterial Characterization
2.6. Histology
2.7. Statistics
2.8. Calculating Approximate Penetration Depth
3. Results
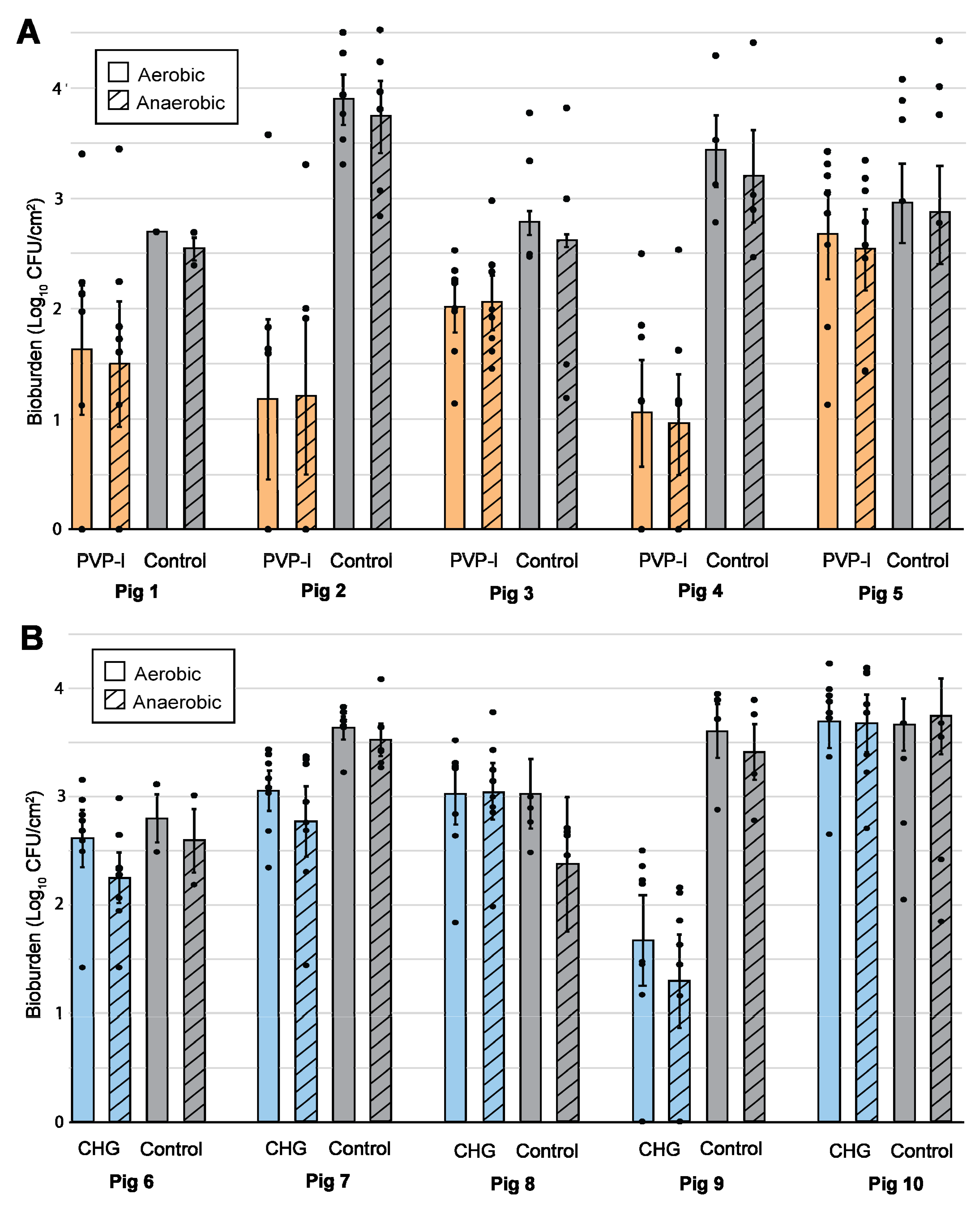
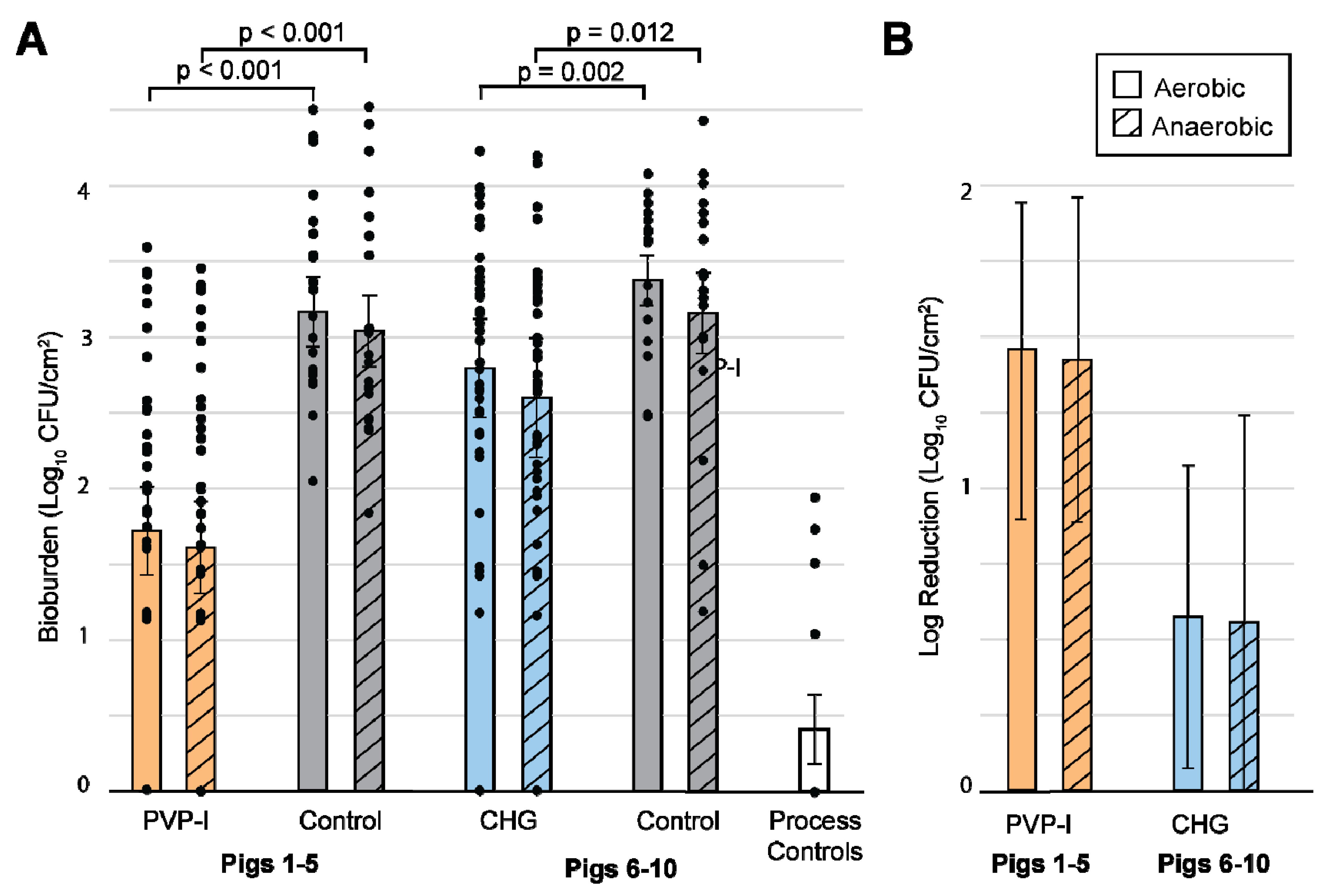
3.1. Bioburden Quantification
3.2. Statistics
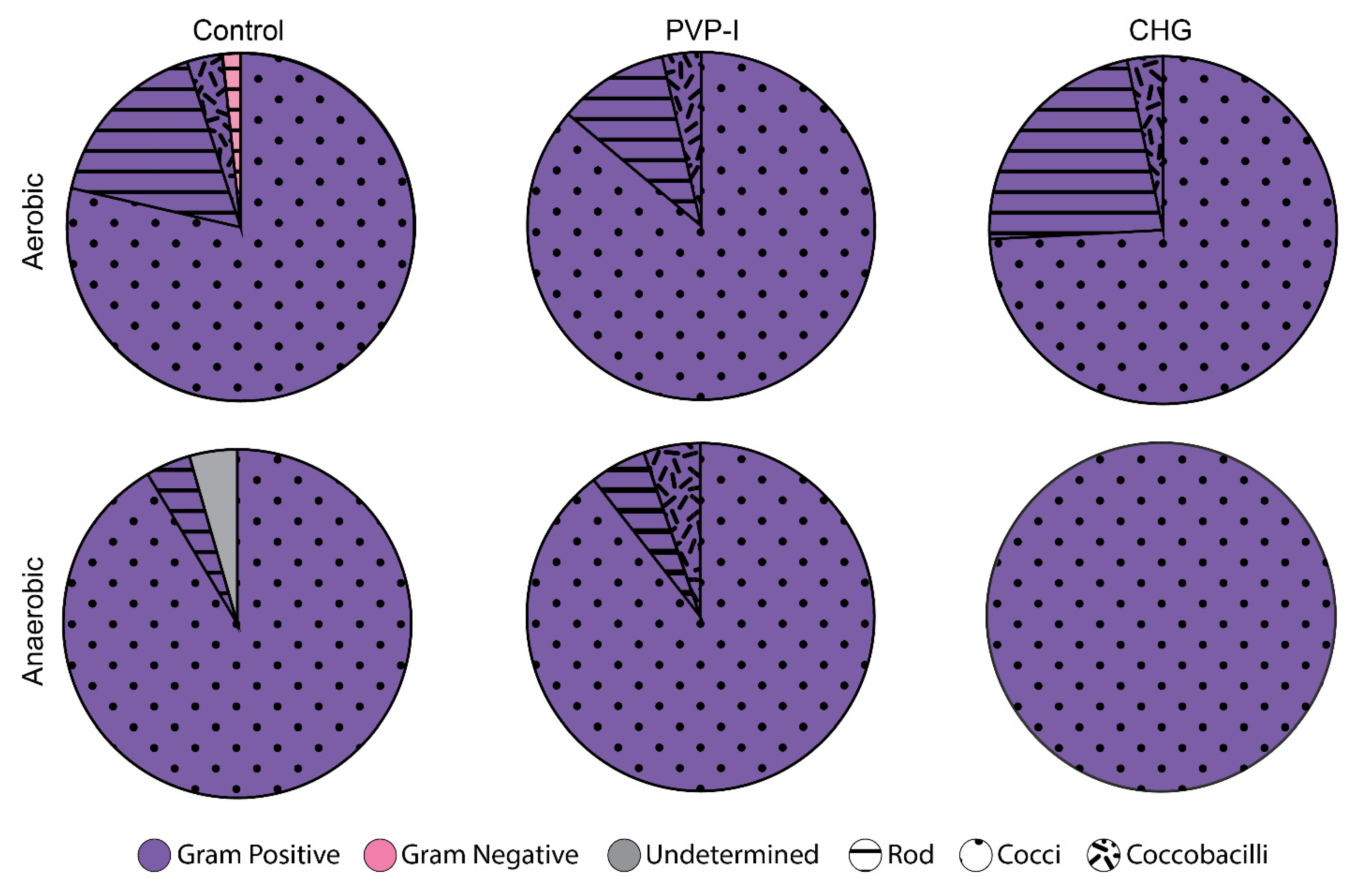
3.3. Bacterial Identification
3.4. Histology and Diffusion
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crosby, C.T.; Elliott, T.S.J.; Lambert, P.A.; Adams, D. Preoperative skin preparation: A historical perspective. Br. J. Hosp. Med. 2009, 70, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Lister, J. On a New Method of Treating Compound Fractures, Abscesses, ETC. Lancet 1867, 89, 326–329. [Google Scholar] [CrossRef]
- Digison, M.B. A review of anti-septic agents for pre-operative skin preparation. Plast. Surg. Nurs. 2007, 27, 185–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, K.A.; Minei, J.P.; Laronga, C.; Harbrecht, B.G.; Jensen, E.H.; Fry, D.E.; Itani, K.M.F.; Dellinger, E.P.; Ko, C.Y.; Duane, T.M. American College of Surgeons and Surgical Infection Society: Surgical Site Infection Guidelines, 2016 Update. J. Am. Coll. Surg. 2017, 224, 59–74. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Guidelines on Hand Hygine 2009; WHO: Geneva, Switzerland, 2009. [Google Scholar]
- WHO. WHO guidelines on hand hygiene in health care. In WHO Guidelines on Hand Hygiene in Health Care; WHO: Geneva, Switzerland, 2009; p. 270. [Google Scholar]
- Craig, C.P. Preparation of the skin for surgery. Infect. Control Hosp. Epidemiol. 1986, 8, 17–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumville, J.; Mcfarlane, E.; Edwards, P.; Lipp, A.; Holmes, A.; Liu, Z. Preoperative Skin Antiseptics for Preventing Surgical Wound Infections After Clean Surgery (Review). Cochrane Database Syst. Rev. 2015, 1–52. [Google Scholar] [CrossRef]
- Mackenzie, I. Preoperative skin preparation and surgical outcome. J. Hosp. Infect. 1988, 11, 27–32. [Google Scholar] [CrossRef]
- Mastrocola, M.; Matziolis, G.; Böhle, S.; Lindemann, C.; Schlattmann, P.; Eijer, H. Meta-analysis of the efficacy of preoperative skin preparation with alcoholic chlorhexidine compared to povidone iodine in orthopedic surgery. Sci. Rep. 2021, 11, 18634. [Google Scholar] [CrossRef]
- Swan, H.; Gonzalez, R.I.; Harris, A.; Couslon, C.; Hopwood, M.L. Use of Quaternary Ammonium Compounds For the Surgical Scrub. Am. J. Surg. 1949, 77, 24–37. [Google Scholar] [CrossRef]
- Williams, D.L.; Armbruster, D.A.; Summers, J.; Goeres, D.M.; Akers, K.S.; Wenke, J.C.; Murray, C.K.; Goswami, K.; Parvizi, J.; Myntti, M.; et al. Targeting Biofilms in Translational Research, Device Development, and Industrial Sectors; Williams, D.L., Ed.; Springer: Cham, Switzerland, 2019; ISBN 9783030306663. [Google Scholar]
- Nablo, B.J.; Rothrock, A.R.; Schoenfisch, M.H. Nitric oxide-releasing sol-gels as antibacterial coatings for orthopedic implants. J. Biomater. 2005, 26, 917–924. [Google Scholar] [CrossRef]
- Hetrick, E.M.; Schoenfisch, M.H.; Hetrick, E.M.; Hetrick, E.M. Reducing implant-related infections: Active release strategies. Chem. Soc. Rev. 2006, 35, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Mclaughlin, J.S.; Hornick, R.B.; Ronica, M.; Frank, M. Sources of Contamination in Open Heart Surgery. JAMA 2019, 230, 1415–1418. [Google Scholar]
- Chu, N.S.; Chan-myers, H.; Aam, R.M.; Ghazanfari, N.; Antonoplos, P. Levels of naturally occurring microorganisms on surgical instruments after clinical use and after washing. Am. J. Infect. Control 1999, 27, 315–319. [Google Scholar] [CrossRef]
- Agarwal, A.; Lin, B.; Elgafy, H. Intraoperative Guard to Avoid Biofilm and Infection at Pedicle Screw & Bone Interface. In Proceedings of the Poster Presentation at the International Combined Orthopaedic Research Societies Annual Meeting, Montreal, QC, Canada, 19–22 June 2019. [Google Scholar]
- Achermann, Y.; Goldstein, E.J.C.; Coenye, T.; Shirtliff, M.E. Propionibacterium acnes: From Commensal to Opportunistic Biofilm- Associated Implant Pathogen. Clin. Microbiol. Rev. 2014, 27, 419–440. [Google Scholar] [CrossRef] [Green Version]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Comparative, N.; Program, S.; Bouffard, G.G.; Blakesley, R.W.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1193. [Google Scholar] [CrossRef] [Green Version]
- Portillo, M.E.; Corvec, S.; Borens, O.; Trampuz, A. Propionibacterium acnes: An Underestimated Pathogen in Implant-Associated Infections. Biomed Res. Int. 2013, 2013, 804391. [Google Scholar] [CrossRef] [Green Version]
- Sahin, Y.A.F.; Schwyzer, H.K. Characteristics and outcome of 16 periprosthetic shoulder joint infections. Infection 2013, 41, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Levy, P.Y.; Fenollar, F.; Stein, A.; Borrione, F.; Cohen, E.; Lebail, B.; Raoult, D. Propionibacterium acnes Postoperative Shoulder Arthritis: An Emerging Clinical Entity. Clin. Infect. Dis. 2008, 46, 1884–1886. [Google Scholar] [CrossRef] [Green Version]
- Contracture, C.; Del Pozo, J.L.; Tran, N.V.; Petty, P.M.; Johnson, C.H.; Walsh, M.F.; Bite, U.; Clay, R.P.; Mandrekar, J.N.; Piper, K.E.; et al. Pilot Study of Association of Bacteria on Breast Implants with. J. Clin. Microbiol. 2009, 47, 1333–1337. [Google Scholar] [CrossRef] [Green Version]
- Sampedro, M.F.; Huddleston, P.M.; Piper, K.E.; Karau, M.J.; Dekutoski, M.B.; Yaszemski, M.J.; Currier, B.L.; Mandrekar, J.N.; Osmon, D.R.; Mcdowell, A.; et al. A Biofilm Approach to Detect Bacteria on Removed Spinal Implants. Spine 2010, 35, 1218–1224. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev. Dermatol. 2010, 5, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, S.; Baru, K. Notes On the Vertical Distribution of the Human Sweat Glands. Jpn. J. Physiol. 1954, 4, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Moriarty, T.F.; Kuehl, R.; Coenye, T.; Metsemakers, W.; Morgenstern, M.; Schwarz, E.M.; Riool, M.; Khana, N.; Kates, S.L.; Richards, R.G. Orthopaedic device-related infection: Current and future interventions for improved prevention and treatment. EFORT Open Rev. 2016, 1, 13–17. [Google Scholar] [CrossRef] [PubMed]
- ASTM E1173-15; Standard Test Method for Evaluation of Preoperative, Precatheterization, or Preinjection Skin Preparations; Active Standard ASTM E1173|Developed by Subcommittee: E35.15 Book of Standards Volume: 11.08. ASTM International: West Conshohocken, PA, USA, 2015.
- Edmiston, C.E.; Seabrook, G.R.; Johnson, C.P.; Paulson, D.S.; Beausoleil, C.M. Comparative of a new and innovative 2% chlorhexidine gluconate-impregnated cloth with 4% chlorhexidine gluconate as topical antiseptic for preparation of the skin prior to surgery. Am. J. Infect. Control 2007, 35, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Paulson, D.S.; Topp, R.; Boykin, R.E.; Schultz, G.; Yang, Q. Efficacy and safety of a novel skin cleansing formulation versus chlorhexidine gluconate. Am. J. Infect. Control 2018, 46, 1262–1265. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.G.; Miles, A.A.; Niven, J.S.F. The enhancement of bacterial infections by adrenaline. Br. J. Exp. Pathol. 1948, 29, 20–39. [Google Scholar]
- Miles, A.A.; Niven, J.S.F. The Enhancement of Infection Durring Shock Produced by Bacterial Toxins and Other Agents. Br. J. Exp. Pathol. 1949, 31, 73. [Google Scholar]
- Kass, E.H. Diagnostic Urology Asymptomatic Infections of the Urinary Tract. Trans. Am. Physiol. 1956, 69, 56–64. [Google Scholar]
- Antiseptic, U. Bacteriuria and the Diagnosis of Infections of the Urinary Tract. AMA Arch. Intern. Med. 1957, 100, 709–714. [Google Scholar]
- Robson, M.C.; Heggers, J.P. Delayed wound closures based on bacterial counts. J. Surg. Oncol. 1970, 2, 379–383. [Google Scholar] [CrossRef]
- Liedberg, N.C.F.; Reiss, E.; Artz, C.P. The effect of bacteria on the take of split-thickness skin grafts in rabbits. Ann. Surg. 1955, 142, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Krizek, T.J.; Robson, M.C. Evolution of quantitative bacteriology in wound management. Am. J. Surg. 1975, 130, 579–584. [Google Scholar] [CrossRef]
- Bowler, P.G. The 10 (5) Bacterial growth guideline: Reassessing its clinical relevance in. Ostomy Wound Manag. 2003, 49, 44–53. [Google Scholar]
- Elek, S.D.; Conen, P.E. The virulence of Staphylococcus pyogenes for man; a study of the problems of wound infection. Br. J. Exp. Pathol. 1957, 38, 573–586. [Google Scholar] [PubMed]
- James, R.C.; MacLEOD, C.J. Induction of Staphyloccocal Infections in Mice With Small Inocula Introduced on Sutures. Br. J. Exp. Pathol. 1961, 42, 266–277. [Google Scholar]
- Ratner, B.D. Biomaterials Science: An Introduction to Materials in Medicine; Academic Press: Cambridge, MA, USA, 2004; ISBN 0125824637. [Google Scholar]
- Costerton, J.W.; Geesey, G.G.; Cheng, K.-J. How Bacteria Stick. Sci. Am. 1978, 238, 86–95. [Google Scholar] [CrossRef]
- Costerton, J.W. Biofilm theory can guide the treatment of device-related orthopaedic infections. Clin. Orthop. Relat. Res. 2005, 437, 7–11. [Google Scholar] [CrossRef]
- Stewart, P.S. Biophysics of Biofilm Infection. Pathog. Dis. 2014, 70, 212–218. [Google Scholar] [CrossRef] [Green Version]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Nizet, V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J. Allergy Clin. Immunol. 2007, 120, 13–22. [Google Scholar] [CrossRef]
- Sarantis, H.; Grinstein, S. Subversion of phagocytosis for pathogen survival. Cell Host Microbe 2012, 12, 419–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leid, J.G. Bacterial Biofilms Resist Key Host Defenses Once in biofilms, bacterial pathogens resist antibiotics and withstand several host-defense measures, including phagocytosis. Microbe 2009, 4, 66–70. [Google Scholar]
- Herant, M. Mechanics of neutrophil phagocytosis: Experiments and quantitative models. J. Cell Sci. 2006, 119, 1903–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, D.; Hidalgo-Cantabrana, C.; Rodríguez, A.; García, P.; Ruas-Madiedo, P. Monitoring in real time the formation and removal of biofilms from clinical related pathogens using an impedance-based technology. PLoS ONE 2016, 11, e0163966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, P.S. Antimicrobial Tolerance in Biofilms. Microbiol. Spectr. 2015, 3, 1–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConoughey, S.J.; Howlin, R.; Granger, J.F.; Manring, M.M.; Calhoun, J.H.; Shirtliff, M.; Kathju, S.; Stoodley, P. Biofilms in periprosthetic orthopedic infections. Future Microbiol. 2014, 9, 987–1007. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Costerton, J.W. The Biofilm Primer; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Brandwein, M.; Steinberg, D.; Meshner, S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes 2016, 2, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Aarts, E.; Verhage, M.; Veenvliet, J.V.; Dolan, C.V.; Van Der Sluis, S. A solution to dependency: Using multilevel analysis to accommodate nested data. Nat. Neurosci. 2014, 17, 491–496. [Google Scholar] [CrossRef] [Green Version]
- Treybal, R.E. Mass Transfer Operations; McGraw-Hill Book Company: Kuala Lumpur, Malaysia, 1980; pp. 35–36. [Google Scholar]
- Cantrel, L.; Chaouche, R.; Chopin-dumas, J. Diffusion Coefficients of Molecular Iodine in Aqueous Solutions. J. Chem. Eng. Data 1997, 2, 216–220. [Google Scholar] [CrossRef]
- Klinov, A.; Anashkin, I. Diffusion in binary aqueous solutions of alcohols by molecular simulation. Processes 2019, 7, 947. [Google Scholar] [CrossRef] [Green Version]
- Howard, C. Berg Chapter 1, Diffusion: Microscopic Theory. In Random Walks in Biology; Princeton University Press: Princeton, NJ, USA, 1934; pp. 2–8. [Google Scholar]
- Zeeuwen, P.L.J.M.; Boekhorst, J.; van den Bogaard, E.H.; de Koning, H.D.; van de Kerkhof, P.M.C.; Saulnier, D.M.; van Swam, I.I.; van Hijum, S.A.F.T.; Kleerebezem, M.; Schalkwijk, J.; et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012, 13, R101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovell, D.L. Skin Bacteria -Their Location with Reference to Skin Sterilization. Surg. Genecol. Obstet. Int. Abstracys Surg. 1945, 80, 174. [Google Scholar]
- Turnbull, P.C.B.; Kramer, J.M.; Melling, J. Chapter 15, Bacillus. In Medical Microbiology; The University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; pp. 1–7. [Google Scholar]
- Gillespie, S.H. Medical Microbiology Illustrated; Butterworth-Heinemann: Oxford, UK, 2014; ISBN 1483193632. [Google Scholar]
- Bernard, K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J. Clin. Microbiol. 2012, 50, 3152–3158. [Google Scholar] [CrossRef] [Green Version]
- Management, R.W. Lasers and Pulsed-Light Devices: Hair Removal. In Pfenninger and Fowler’s Procedures for Primary Care; Elsevier: Amsterdam, The Netherlands, 2010; pp. 153–160. [Google Scholar]
- Anderson, R.L.; Cassidy, J.M. Variations in physical dimensions and chemical composition of human stratum corneum. J. Investig. Dermatol. 1973, 61, 30–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stylianopoulos, T.; Poh, M.Z.; Insin, N.; Bawendi, M.G.; Fukumura, D.; Munn, L.L.; Jain, R.K. Diffusion of particles in the extracellular matrix: The effect of repulsive electrostatic interactions. Biophys. J. 2010, 99, 1342–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Depth (mm) | PVP-I | I- | CHG | Chlorhexidine | IPA | |
|---|---|---|---|---|---|---|
| Diffusion Coefficient | 6 × 10−10 m2/s [56] | 2 × 10−9 m2/s [57] | 4 × 10−10 m2/s [56] | 5 × 10−10 m2/s [56] | 1 × 10−9 m2/s [58] | |
| Follicle | 3.2 ± 0.7 | 145 ± 67 min | 44 ± 20 min | 218 ± 101 min | 175 ± 80 min | 87 ± 40 min |
| Stratum Corneum | 0.056 ± 0.029 | 3.2 ± 3.2 s | 1.0 ± 1.0 s | 4.8 ± 4.8 s | 3.9 ± 3.9 s | 1.9 ± 1.9 s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duffy, H.R.; Godfrey, R.W.; Williams, D.L.; Ashton, N.N. A Porcine Model for the Development and Testing of Preoperative Skin Preparations. Microorganisms 2022, 10, 837. https://doi.org/10.3390/microorganisms10050837
Duffy HR, Godfrey RW, Williams DL, Ashton NN. A Porcine Model for the Development and Testing of Preoperative Skin Preparations. Microorganisms. 2022; 10(5):837. https://doi.org/10.3390/microorganisms10050837
Chicago/Turabian StyleDuffy, Hannah R., Rose W. Godfrey, Dustin L. Williams, and Nicholas N. Ashton. 2022. "A Porcine Model for the Development and Testing of Preoperative Skin Preparations" Microorganisms 10, no. 5: 837. https://doi.org/10.3390/microorganisms10050837
APA StyleDuffy, H. R., Godfrey, R. W., Williams, D. L., & Ashton, N. N. (2022). A Porcine Model for the Development and Testing of Preoperative Skin Preparations. Microorganisms, 10(5), 837. https://doi.org/10.3390/microorganisms10050837






