Bladder Microbiota Are Associated with Clinical Conditions That Extend beyond the Urinary Tract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population
2.2. Sample Handling
2.3. DNA Extraction and PCR
2.4. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; NIH HMP Working Group. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [PubMed] [Green Version]
- Loke, Y.L.; Chew, M.T.; Ngeow, Y.F.; Lim, W.W.D.; Peh, S.C. Colon Carcinogenesis: The Interplay Between Diet and Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 603086. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Łaniewski, P.; Ilhan, Z.E.; Herbst-Kralovetz, M.M. The microbiome and gynaecological cancer development, prevention and therapy. Nat. Rev. Urol. 2020, 17, 232–250. [Google Scholar] [CrossRef]
- Siddiqui, H.; Nederbragt, A.J.; Lagesen, K.; Jeansson, S.L.; Jakobsen, K.S. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol. 2011, 11, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilt, E.E.; McKinley, K.; Pearce, M.M.; Rosenfeld, A.B.; Zilliox, M.J.; Mueller, E.R.; Brubaker, L.; Gai, X.; Wolfe, A.J.; Schreckenberger, P.C. Urine Is Not Sterile: Use of Enhanced Urine Culture Techniques to Detect Resident Bacterial Flora in the Adult Female Bladder. J. Clin. Microbiol. 2014, 52, 871–876. [Google Scholar] [CrossRef] [Green Version]
- Fouts, D.E.; Pieper, R.; Szpakowski, S.; Pohl, H.; Knoblach, S.; Suh, M.-J.; Huang, S.-T.; Ljungberg, I.; Sprague, B.M.; Lucas, S.K.; et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J. Transl. Med. 2012, 10, 174. [Google Scholar] [CrossRef] [Green Version]
- Bajic, P.; Van Kuiken, M.E.; Burge, B.K.; Kirshenbaum, E.J.; Joyce, C.; Wolfe, A.J.; Branch, J.D.; Bresler, L.; Farooq, A.V. Male Bladder Microbiome Relates to Lower Urinary Tract Symptoms. Eur. Urol. Focus 2020, 15, 376–382. [Google Scholar] [CrossRef]
- Burnett, L.A.; Hochstedler, B.R.; Weldon, K.; Wolfe, A.J.; Brubaker, L. Recurrent urinary tract infection: Association of clinical profiles with urobiome composition in women. Neurourol. Urodyn. 2021, 40, 1479–1489. [Google Scholar] [CrossRef]
- Siddiqui, H.; Lagesen, K.; Nederbragt, A.J.; Jeansson, S.L.; Jakobsen, K.S. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol. 2012, 12, 205. [Google Scholar] [CrossRef] [Green Version]
- Shoskes, D.A.; Altemus, J.; Polackwich, A.S.; Tucky, B.; Wang, H.; Eng, C. The Urinary Microbiome Differs Significantly Between Patients with Chronic Prostatitis/Chronic Pelvic Pain Syndrome and Controls as Well as Between Patients with Different Clinical Phenotypes. Urology 2016, 92, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zhang, G.; Zhao, J.; Chen, J.; Chen, Y.; Huang, W.; Zeng, J. Profiling the Urinary Microbiota in Male Patients with Bladder Cancer in China. Front. Cell. Infect. Microbiol. 2018, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Littman, D.R. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proin fl ammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4615–4622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlin, M.; Prast-Nielsen, S. EBioMedicine The gut microbiome and epilepsy. EBioMedicine 2019, 44, 741–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barandouzi, Z.A.; Starkweather, A.R.; Henderson, W.; Gyamfi, A.; Cong, X.S. Altered Composition of Gut Microbiota in Depression: A Systematic Review. Front. Psychiatry 2020, 11, 541. [Google Scholar] [CrossRef]
- Wu, P.; Chen, Y.; Zhao, J.; Zhang, G.; Chen, J.; Wang, J.; Zhang, H. Urinary Microbiome and Psychological Factors in Women with Overactive Bladder. Front. Cell. Infect. Microbiol. 2017, 7, 488. [Google Scholar] [CrossRef]
- Lewis, D.A.; Brown, R.; Williams, J.; White, P.; Jacobson, S.K.; Marchesi, J.R.; Drake, M.J. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell. Infect. Microbiol. 2013, 3, 41. [Google Scholar] [CrossRef] [Green Version]
- Kramer, H.; Kuffel, G.; Thomas-White, K.; Wolfe, A.J.; Vellanki, K.; Leehey, D.J.; Bansal, V.K.; Brubaker, L.; Flanigan, R.; Koval, J.; et al. Diversity of the midstream urine microbiome in adults with chronic kidney disease. Int. Urol. Nephrol. 2018, 50, 1123–1130. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Větrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platfomr for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef] [PubMed]
- Aronesty, E. Comparison of Sequencing Utility Programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P. Analysis of soil fungal communities by amplicon pyrosequencing: Current approaches to data analysis and the introduction of the pipeline SEED. Biol. Fertil. Soils 2013, 49, 1027–1037. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [Green Version]
- Willis, A. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [Green Version]
- Su, X. Elucidating the Beta-Diversity of the Microbiome: From Global Alignment to Local Alignment. mSystems 2021, 6. [Google Scholar] [CrossRef]
- Bylemans, J.; Gleeson, D.M.; Lintermans, M.; Hardy, C.M.; Beitzel, M.; Gilligan, D.M.; Furlan, E.M. Package ‘vegan. Community Ecol. Packag. 2018, 5, 2017. [Google Scholar]
- McMurdie, P.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. 2021. Available online: http://www.r-project.org/ (accessed on 22 September 2021).
- Chiu, C.-H.; Wang, Y.-T.; Walther, B.A.; Chao, A. An improved nonparametric lower bound of species richness via a modified good-turing frequency formula. Biometrics 2014, 70, 671–682. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, R.B.; O’Hara, R. Species richness estimators: How many species can dance on the head of a pin? J. Anim. Ecol. 2005, 74, 375–386. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Dobbler, P.T.; Pylro, V.S.; Kolaczkowski, B.; Drew, J.C.; Triplett, E.W. Pime: A package for discovery of novel differences among microbial communities. Mol. Ecol. Resour. 2020, 20, 415–428. [Google Scholar] [CrossRef]
- Whiteside, S.A.; Razvi, H.; Dave, S.; Reid, G.; Burton, J. The microbiome of the urinary tract—A role beyond infection. Nat. Rev. Urol. 2015, 12, 81–90. [Google Scholar] [CrossRef]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.-Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [Green Version]
- Thomas-White, K.J.; Kliethermes, S.; Rickey, L.; Lukacz, E.S.; Richter, H.E.; Moalli, P.; Zimmern, P.; Norton, P.; Kusek, J.W.; Wolfe, A.J.; et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am. J. Obstet. Gynecol. 2017, 216, 55.e1–55.e16. [Google Scholar] [CrossRef] [Green Version]
- Harman, S.M.; Metter, E.J.; Tobin, J.D.; Pearson, J.; Blackman, M.R. Longitudinal Effects of Aging on Serum Total and Free Testosterone Levels in Healthy Men. J. Clin. Endocrinol. Metab. 2001, 86, 724–731. [Google Scholar] [CrossRef]
- Grishina, I.; Fenton, A.; Sankaran-Walters, S. Gender differences, aging and hormonal status in mucosal injury and repair. Aging Dis. 2014, 5, 160–169. [Google Scholar] [CrossRef]
- Ragonnaud, E.; Biragyn, A. Gut microbiota as the key controllers of ‘healthy’ aging of elderly people. Immun. Ageing 2021, 18, 2. [Google Scholar] [CrossRef]
- Yang, T.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. The gut microbiota and the brain–gut–kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Santisteban, M.M.; Rodriguez, V.; Li, E.; Ahmari, N.; Carvajal, J.M.; Zadeh, M.; Gong, M.; Qi, Y.; Zubcevic, J.; et al. Gut Dysbiosis Is Linked to Hypertension. Hypertension 2015, 65, 1331–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, N.; Vogensen, F.K.; Berg, F.W.J.V.D.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Abu Al-Soud, W.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ling, Z.; Xiao, Y.; Lv, L.; Yang, Q.; Wang, B.; Lu, H.; Zheng, L.; Jiang, P.; Wang, W.; et al. Dysbiosis of urinary microbiota is positively correlated with Type 2 diabetes mellitus. Oncotarget 2017, 8, 3798–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.B.; Lu, J.; Chen, P.P.; Lu, C.C.; Zhang, J.X.; Li, X.Q.; Yuan, B.Y.; Huang, S.J.; Ruan, X.Z.; Liu, B.C.; et al. Dysbiosis of intestinal microbiota mediates tubulointerstitial injury in diabetic nephropathy via the disruption of cholesterol homeostasis. Theranostics 2020, 10, 2803–2816. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, W.; Shen, L.; Liu, J.; Yang, F.; Maskey, N.; Wang, H.; Zhang, J.; Yan, Y.; Yao, X. Can Smoking Cause Differences in Urine Microbiome in Male Patients with Bladder Cancer? A Retrospective Study. Front. Oncol. 2021, 11, 677605. [Google Scholar] [CrossRef]
- Moynihan, M.J.; Sullivan, T.; Provenzano, K.; Rieger-Christ, K. Urinary Microbiome Evaluation in Patients Presenting with Hematuria with a Focus on Exposure to Tobacco Smoke. Res. Rep. Urol. 2019, 11, 359–367. [Google Scholar] [CrossRef]
- Reid, G.; Denstedt, J.D.; Kang, Y.S.; Lam, D.; Nause, C. Microbial Adhesion and Biofilm Formation on Ureteral Stents in Vitro and in Vivo. J. Urol. 1992, 148, 1592–1594. [Google Scholar] [CrossRef]

| n | OTUs | ACE | iChao2 | Shannon | Simpson | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | Yes | No | Yes | p | No | Yes | p | No | Yes | p | No | Yes | p | No | Yes | p | |
| Age (≥75 years) | 34 | 16 | 89 | 34 | 0.057 | 316.5 | 221.9 | 0.052 | 344.4 | 191.5 | 0.035 | 2.59 | 2.52 | 0.507 | 0.79 | 0.80 | 0.602 |
| Hypertension | 19 | 31 | 92 | 49 | 0.100 | 347.1 | 244.0 | 0.132 | 348.6 | 250.0 | 0.090 | 3.00 | 2.39 | 0.054 | 0.86 | 0.78 | 0.105 |
| Diabetes mellitus | 36 | 14 | 87 | 19 | 0.021 | 307.0 | 173.6 | 0.028 | 317.2 | 178.0 | 0.029 | 2.84 | 2.15 | 0.033 | 0.84 | 0.72 | 0.017 |
| Chronic kidney disease 1 | 36 | 10 | 64 | 35 | 0.390 | 306.2 | 302.4 | 0.485 | 315.7 | 301.6 | 0.466 | 2.53 | 2.39 | 0.976 | 0.79 | 0.81 | 0.585 |
| Dyslipidemia | 35 | 15 | 80 | 48 | 0.132 | 307.8 | 193.0 | 0.012 | 344.4 | 185.9 | 0.013 | 2.59 | 2.51 | 0.426 | 0.79 | 0.81 | 0.939 |
| Current smoker | 29 | 11 | 62 | 95 | 0.557 | 370.4 | 299.4 | 0.970 | 369.7 | 312.1 | 0.910 | 2.40 | 3.02 | 0.104 | 0.77 | 0.85 | 0.038 |
| Ureteric stent | 50 | 8 | 62 | 63 | 0.761 | 294.6 | 170.2 | 0.324 | 296.5 | 168.3 | 0.333 | 2.58 | 2.39 | 0.609 | 0.79 | 0.75 | 0.511 |
| Post-void residual urine 2 | 30 | 4 | 81 | 80 | 0.953 | 356.8 | 392.4 | 0.814 | 348.4 | 382.4 | 0.953 | 2.67 | 2.40 | 0.289 | 0.84 | 0.78 | 0.239 |
| Insignificant growth 3 | 38 | 12 | 80 | 19 | 0.101 | 312.1 | 218.7 | 0.090 | 327.2 | 209.5 | 0.196 | 2.67 | 2.31 | 0.544 | 0.83 | 0.78 | 1.000 |
| Preoperative antibiotics | 28 | 22 | 97 | 19 | 0.001 | 388.0 | 238.7 | 0.057 | 388.1 | 252.2 | 0.037 | 2.80 | 2.40 | 0.144 | 0.79 | 0.81 | 0.550 |
| n | OTUs | p | ACE | p | iChao2 | p | Shannon | p | Simpson | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI | |||||||||||
| <25.0 | 12 | 92.0 ± 54.4 | 0.402 | 397.5 ± 282.0 | 0.550 | 399.8 ± 230.2 | 0.586 | 2.5 ± 0.9 | 0.795 | 0.8 ± 0.2 | 0.753 |
| 25.0–29.9 | 23 | 39.5 ± 51.7 | 252.6 ± 342.3 | 266.5 ± 325.7 | 2.5 ± 0.6 | 0.8 ± 0.1 | |||||
| ≥30.0 | 13 | 113.0 ± 92.8 | 273.4 ± 283.7 | 284.4 ± 277.9 | 3.1 ± 1.5 | 0.9 ± 0.3 | |||||
| IPSS | |||||||||||
| 1 | 17 | 103.0 ± 51.1 | 0.009 | 392.3 ± 311.0 | 0.205 | 357.1 ± 278.5 | 0.246 | 3.2 ± 0.9 | 0.034 | 0.9 ± 0.2 | 0.066 |
| 2 | 14 | 16.0 ± 55.5 | 241.8 ± 378.5 | 218.6 ± 361.7 | 1.8 ± 1.0 | 0.7 ± 0.3 | |||||
| 3 | 8 | 68.0 ± 34.6 | 365.0 ± 149.3 | 392.3 ± 176.2 | 2.9 ± 0.4 | 0.8 ± 0.1 | |||||
| IIEF-5 | |||||||||||
| 1 | 12 | 84.0 ± 59.7 | 0.190 | 308.4 ± 295.4 | 0.298 | 318.3 ± 279.3 | 0.294 | 2.9 ± 1.1 | 0.778 | 0.9 ± 0.3 | 0.857 |
| 2 | 8 | 92.5 ± 39.6 | 517.3 ± 200.4 | 493.3 ± 233.9 | 2.7 ± 1.1 | 0.8 ± 0.3 | |||||
| 3 | 3 | 34.5 ± 27.6 | 180.3 ± 83.2 | 172.1 ± 70.6 | 2.6 ± 0.7 | 0.8 ± 0.1 | |||||
| 4 | 5 | 70.0 ± 23.2 | 364.5 ± 82.3 | 398.0 ± 91.4 | 3.1 ± 0.5 | 0.9 ± 0.1 | |||||
| 5 | 7 | 9.0 ± 55.6 | 250.3 ± 102.3 | 213.0 ± 110.5 | 1.7 ± 1.2 | 0.7 ± 0.2 |
| n | PIME OTUs | R2 | PERMANOVA p | |
|---|---|---|---|---|
| Age (≥75 years) | 50 | 29 | 0.015 | 0.727 |
| Hypertension | 50 | 29 | 0.017 | 0.574 |
| Diabetes mellitus | 50 | 9 | 0.027 | 0.203 |
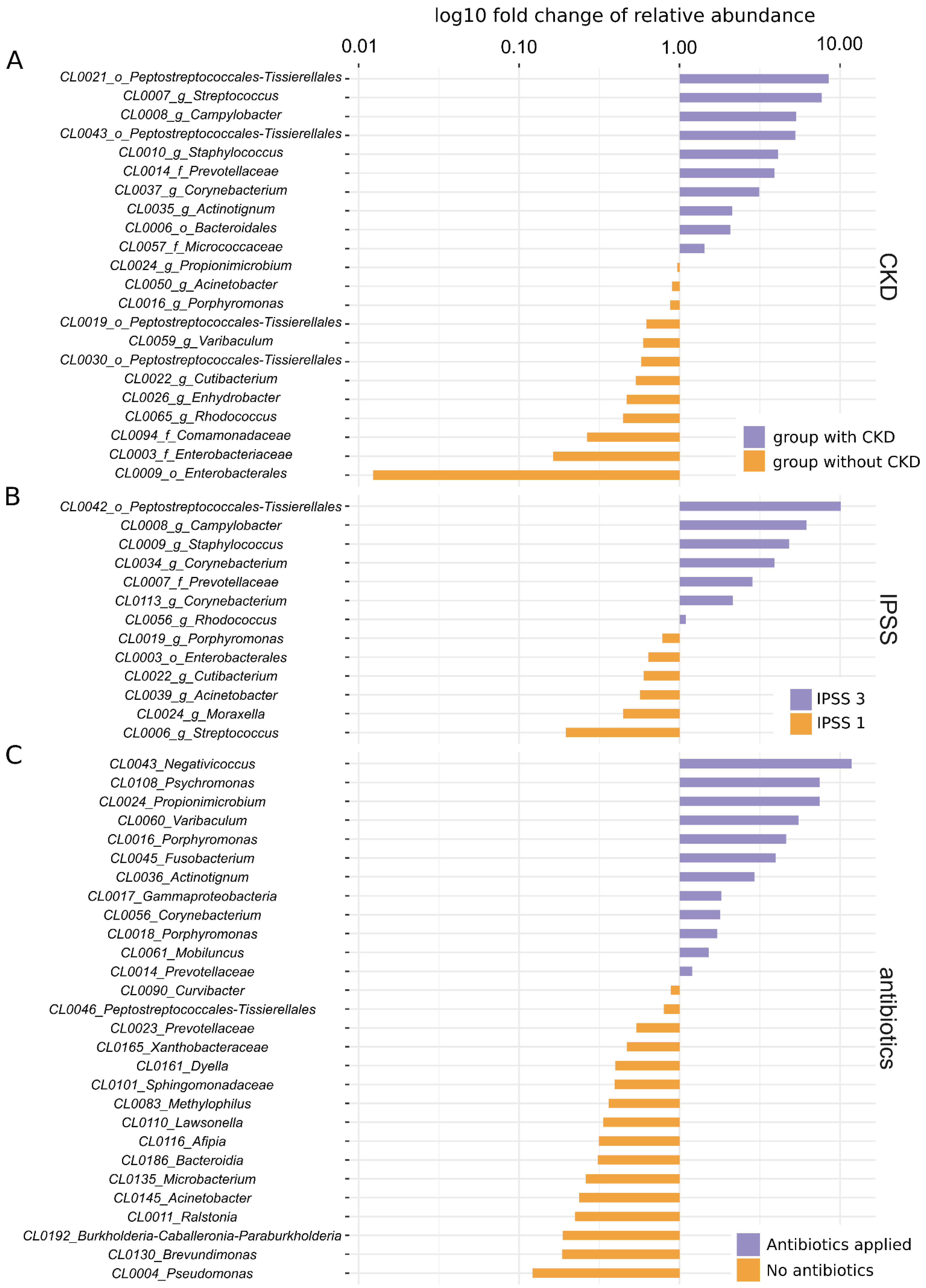
| Chronic kidney disease | 46 | 22 | 0.054 | 0.007 |
| Dyslipidemia | 50 | 29 | 0.021 | 0.364 |
| Obesity | 48 | 155 | 0.016 | 0.724 |
| Current smoker | 40 | 21 | 0.021 | 0.630 |
| Ureteric stent | 58 | 14 | 0.017 | 0.448 |
| Post-void residual urine | 34 | 29 | 0.018 | 0.806 |
| Insignificant growth | 50 | 29 | 0.019 | 0.547 |
| IPSS (mild vs. severe) | 39 | 13 | 0.083 | 0.041 |
| IIEF-5 | 35 | 14 | 0.033 | 0.331 |
| Preoperative antibiotics | 50 | 29 | 0.062 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hrbacek, J.; Tlaskal, V.; Cermak, P.; Hanacek, V.; Zachoval, R. Bladder Microbiota Are Associated with Clinical Conditions That Extend beyond the Urinary Tract. Microorganisms 2022, 10, 874. https://doi.org/10.3390/microorganisms10050874
Hrbacek J, Tlaskal V, Cermak P, Hanacek V, Zachoval R. Bladder Microbiota Are Associated with Clinical Conditions That Extend beyond the Urinary Tract. Microorganisms. 2022; 10(5):874. https://doi.org/10.3390/microorganisms10050874
Chicago/Turabian StyleHrbacek, Jan, Vojtech Tlaskal, Pavel Cermak, Vitezslav Hanacek, and Roman Zachoval. 2022. "Bladder Microbiota Are Associated with Clinical Conditions That Extend beyond the Urinary Tract" Microorganisms 10, no. 5: 874. https://doi.org/10.3390/microorganisms10050874
APA StyleHrbacek, J., Tlaskal, V., Cermak, P., Hanacek, V., & Zachoval, R. (2022). Bladder Microbiota Are Associated with Clinical Conditions That Extend beyond the Urinary Tract. Microorganisms, 10(5), 874. https://doi.org/10.3390/microorganisms10050874






