Suppressive Effect of Soil Microbiomes Associated with Tropical Fruit Trees on Meloidogyne enterolobii
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling and Extraction of Microbial Communities from the Soil
2.2. Soil Chemical and Physical Properties Measurement
2.3. Preparation of the Experiments and Nematode Inoculation
2.4. Nematode Extraction and Evaluation
2.5. Statistical Analyses
2.6. DNA Extraction, Next-Generation Sequencing and Sequence Analysis
2.7. Sequencing Data Processing, OTU Cluster and Taxonomic Annotation
2.8. Data Quality Control, Operational Taxonomic Unit Identification and Alpha and Beta Diversity
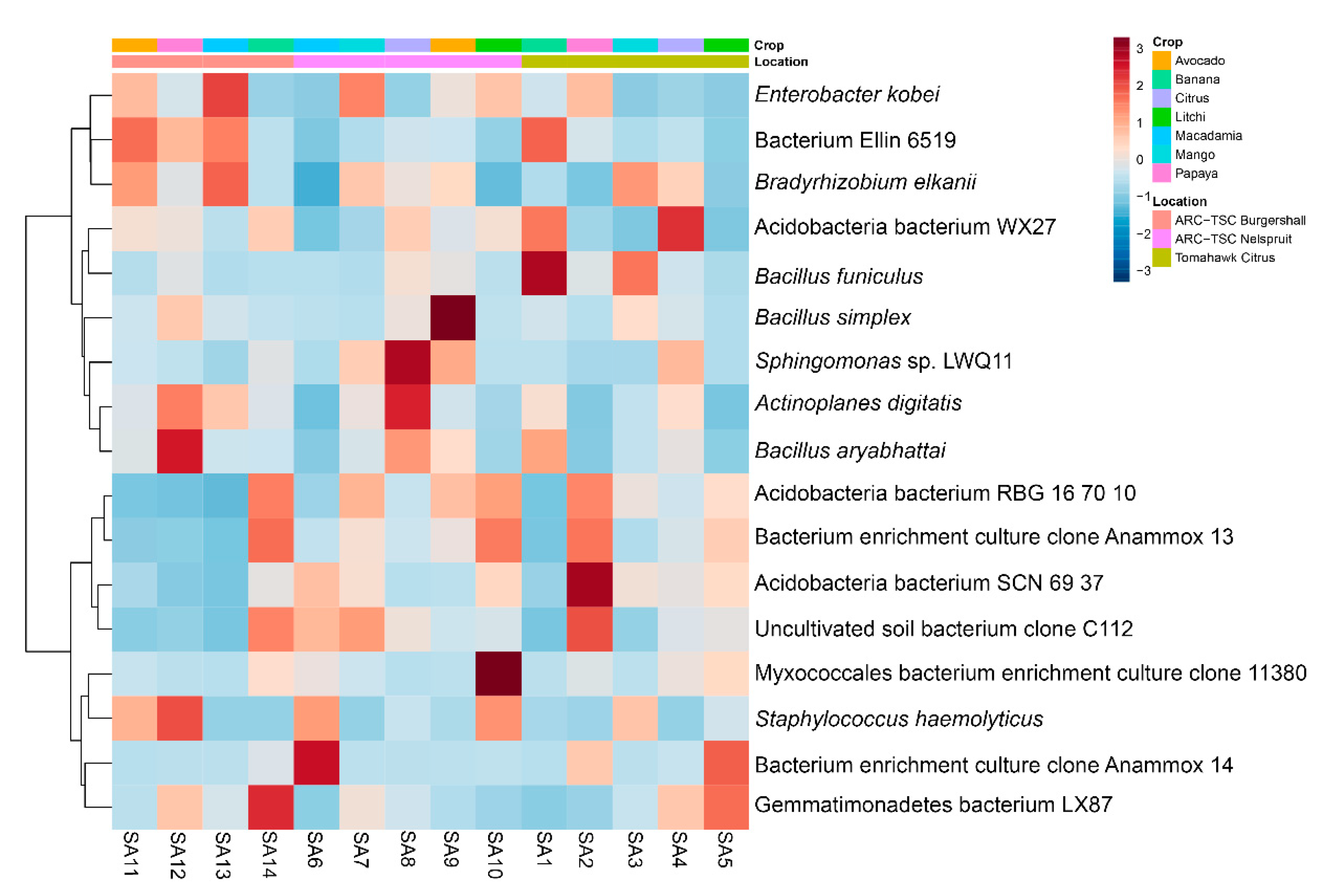
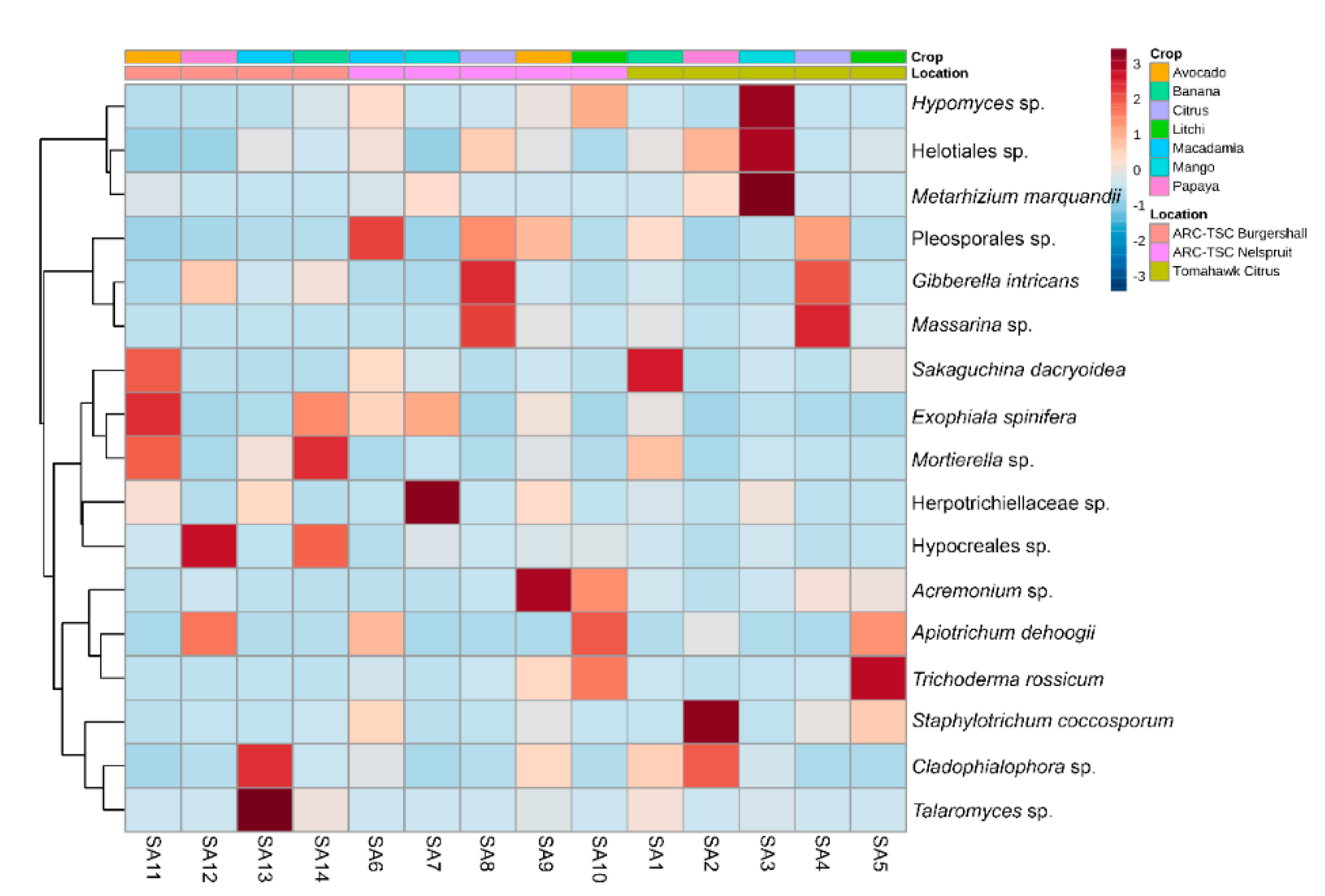
2.9. Linear Discriminant Analysis (LDA) Effect Size (LEfSe) and Heatmap Clustering
3. Results
3.1. Number of Eggs and J2 per Plant
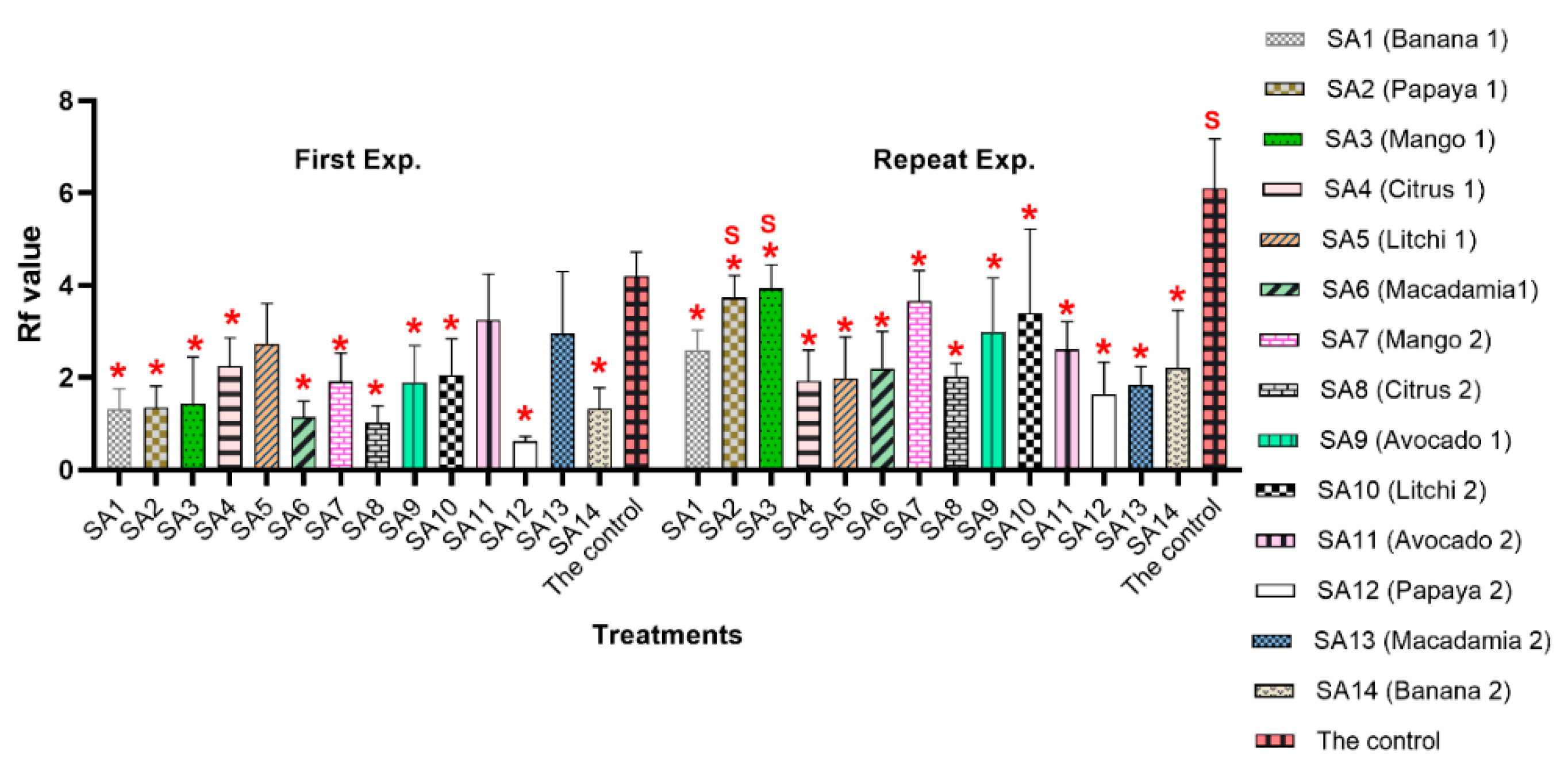
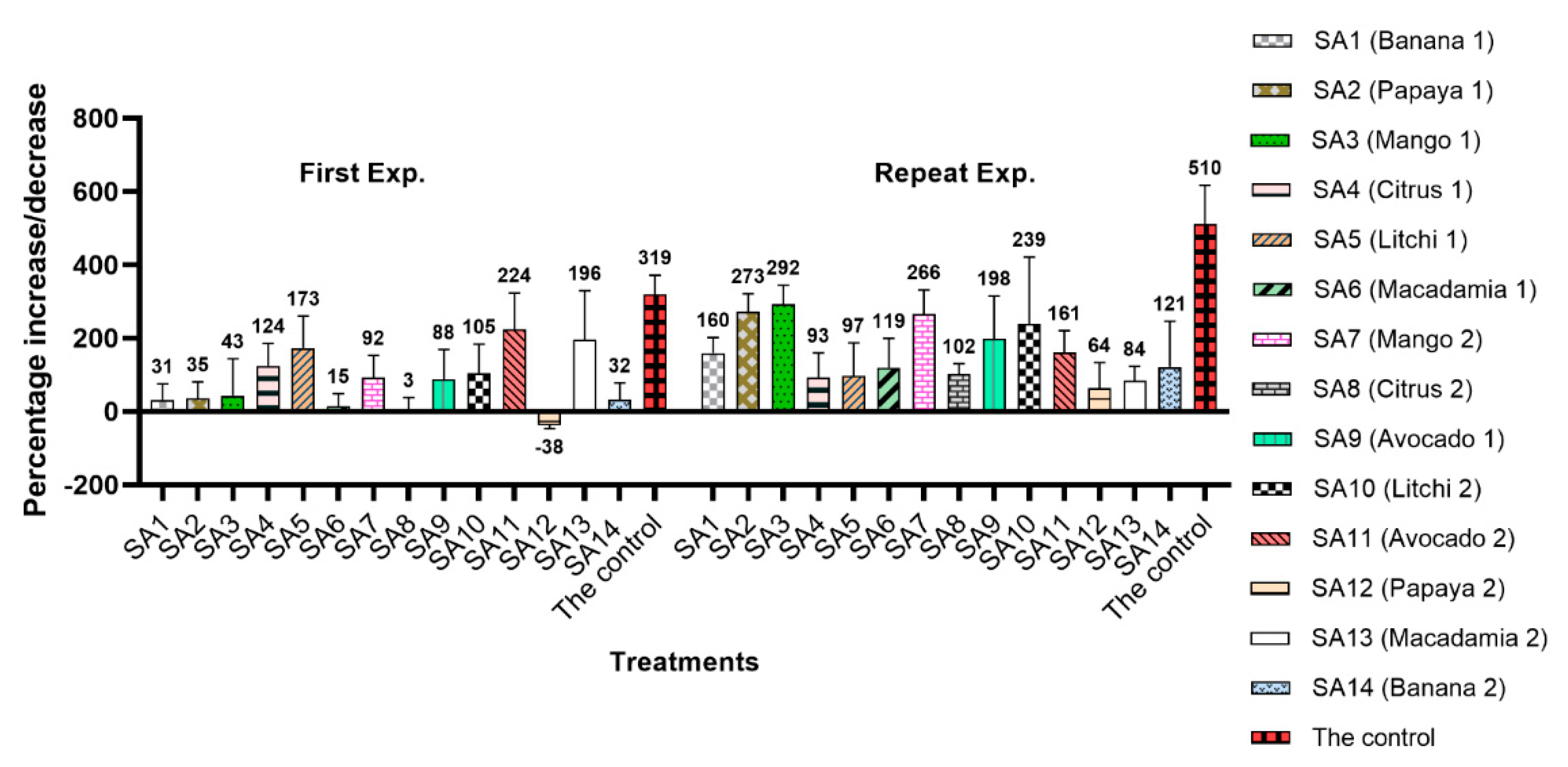
3.2. Reproduction Factor (Rf) Value
3.3. Suppression Effect of Microbial Communities on the Nematode Population
3.4. Bacterial and Fungal Datasets
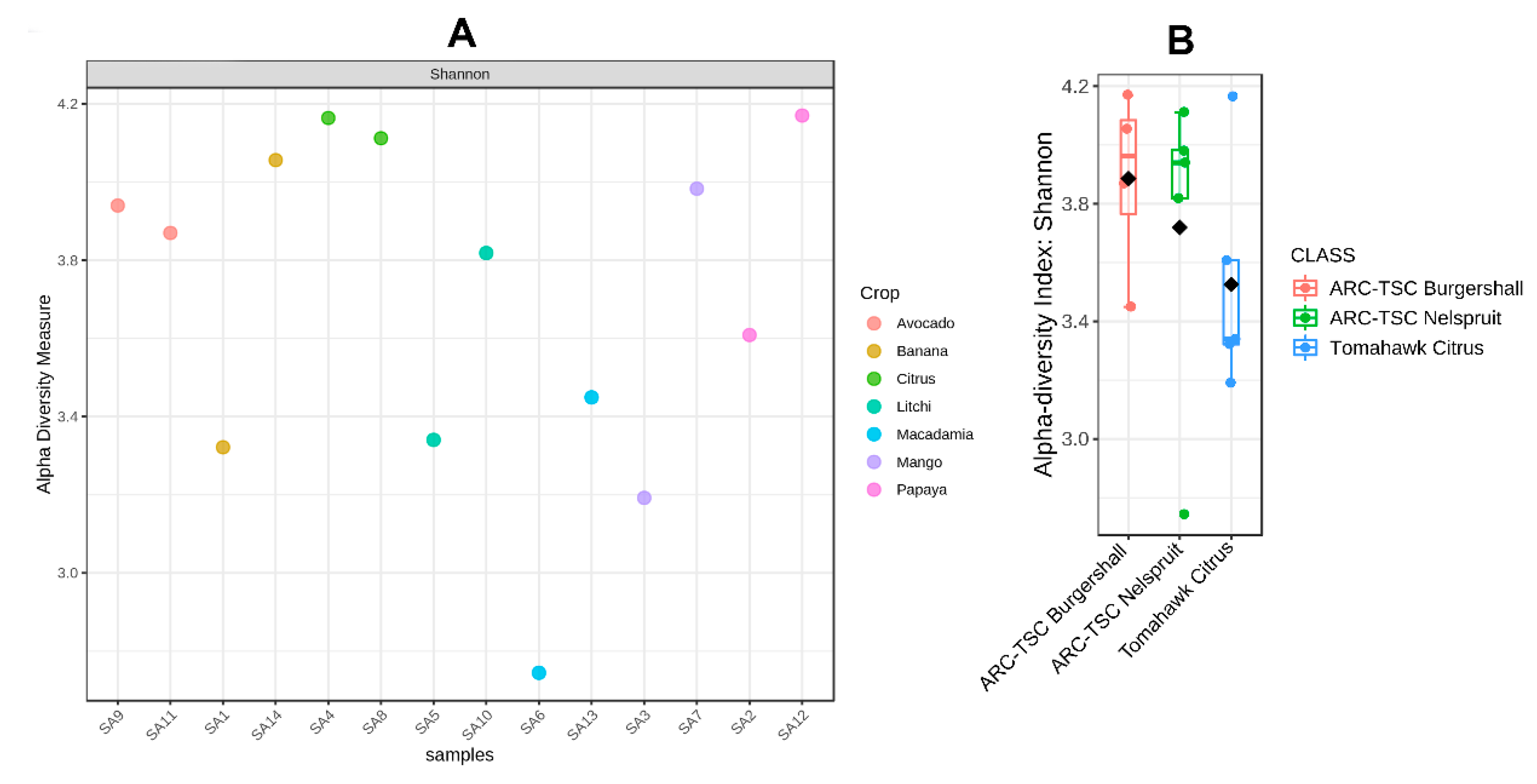
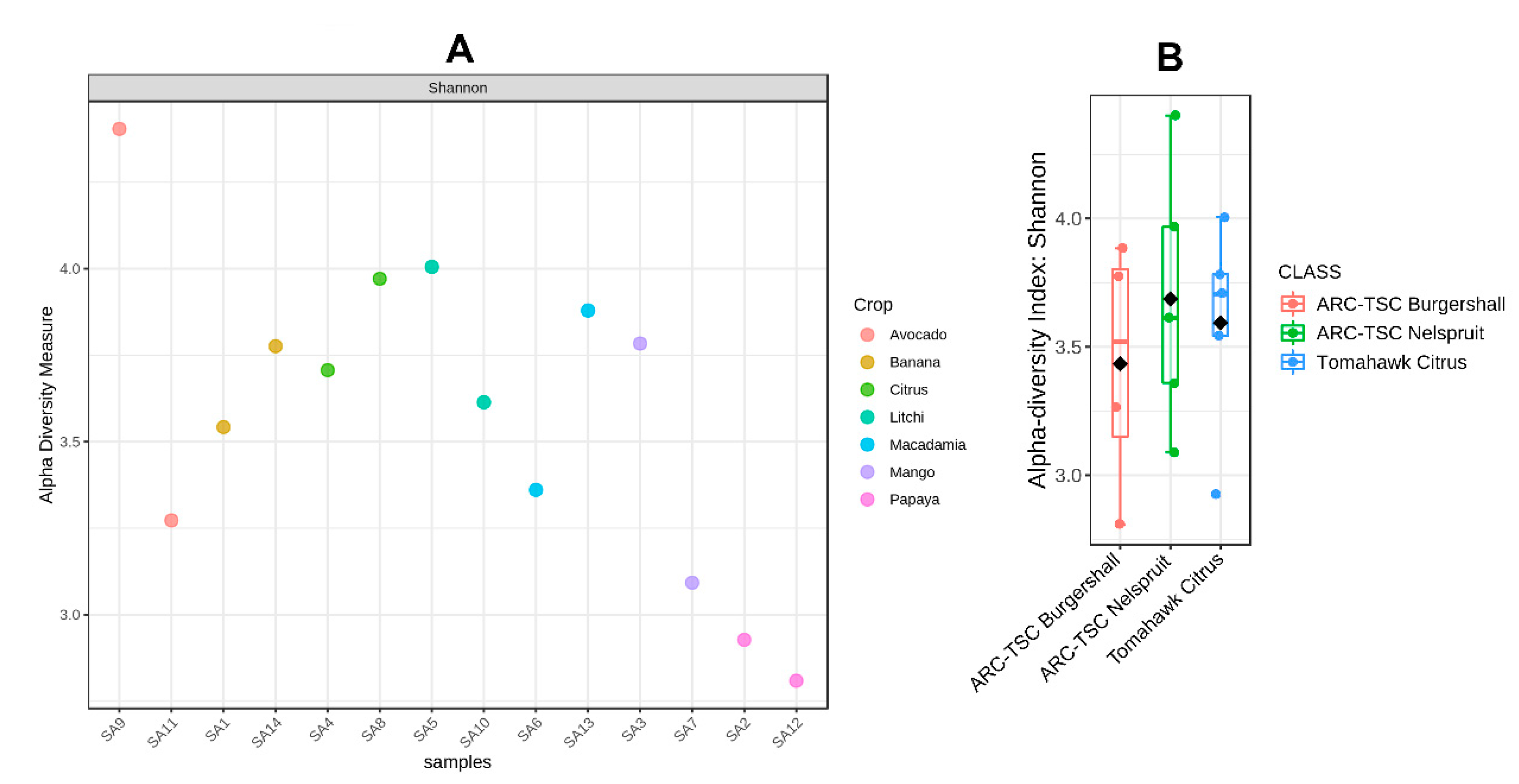
3.5. Alpha Diversity
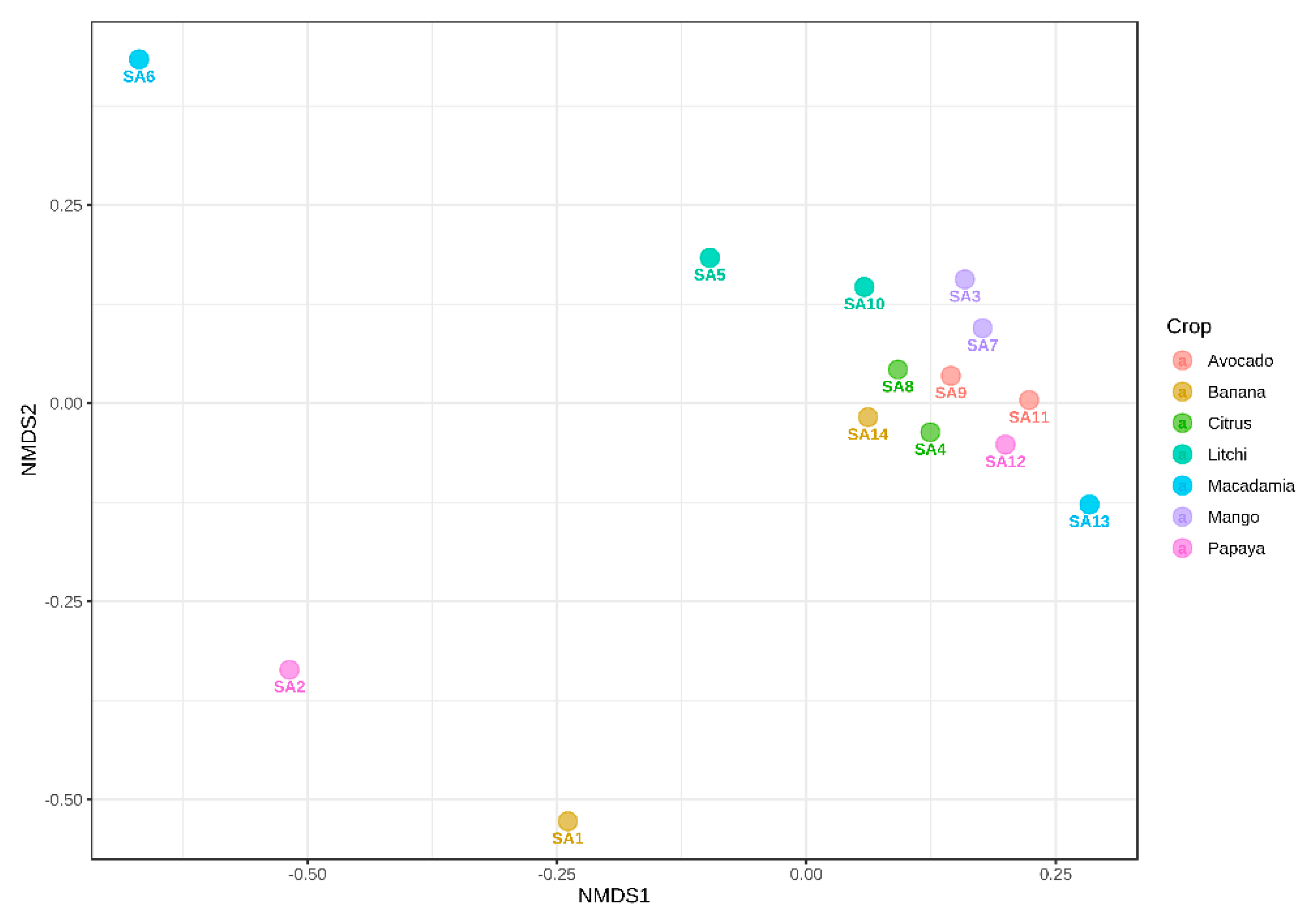
3.6. Beta Diversity
3.7. Microbial Diversity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cole, M.B.; Augustin, M.A.; Robertson, M.J.; Manners, J.M. The science of food security. NPJ Sci. Food 2018, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Daneel, M.S. Nematode Pests of Minor Tropical and Subtropical Crops. In Nematology in South Africa: A View from the 21st Century; Fourie, H., Spaull, V.W.S., Jones, R.K., Daneel, M.S., De Waele, D., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 373–393. [Google Scholar]
- Daneel, M.S.; De Waele, D. Nematode pests of banana. In Nematology in South Africa: A View from the 21st Century; Fourie, H., Spaull, V.W., Jones, R.K., Daneel, M.S., De Waele, D., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 359–371. [Google Scholar]
- Pretorius, M.C.; Le Roux, H.F. Nematode pests of citrus. In Nematology in South Africa: A View from the 21st Century; Fourie, H., Spaull, V., Jones, R., Daneel, M., De Waele, D., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 311–324. [Google Scholar]
- Mc Donald, A.H.; De Waele, D.; Fourie, H. Nematode pests of maize and other cereal crops. In Nematology in South Africa: A View from the 21st Century; Fourie, H., Spaull, V.W., Jones, R.K., Daneel, M.K., De Waele, D., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 183–199. [Google Scholar]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Fourie, H.; Mc Donald, A.H.; Steenkamp, S.; De Waele, D. Nematode pests of leguminous and oilseed crops. In Nematology in South Africa: A View from the 21st Century; Fourie, H., Spaull, V.W., Jones, R.K., Daneel, M.S., De Waele, D., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 201–230. [Google Scholar]
- Rashidifard, M.; Fourie, H.; Daneel, M.S.; Marais, M. Morphological and morphometrical identification of Meloidogyne populations from various crop production areas in South Africa with emphasis on M. enterolobii. Zootaxa 2019, 4658, 251–274. [Google Scholar] [CrossRef] [PubMed]
- Rashidifard, M.; Marais, M.; Daneel, M.S.; Mienie, C.M.S.; Fourie, H. Molecular characterisation of Meloidogyne enterolobii and other Meloidogyne spp. from South Africa. Trop. Plant Pathol. 2019, 44, 213–224. [Google Scholar] [CrossRef]
- Visagie, M.; Mienie, C.M.; Marais, M.; Daneel, M.; Karssen, G.; Fourie, H. Identification of Meloidogyne spp. associated with agri-and horticultural crops in South Africa. Nematology 2018, 20, 397–401. [Google Scholar] [CrossRef]
- Seong, J.; Shin, J.; Kim, K.; Cho, B.-K. Microbial production of nematicidal agents for controlling plant-parasitic nematodes. Process Biochem. 2021, 108, 69–79. [Google Scholar] [CrossRef]
- Raymaekers, K.; Ponet, L.; Holtappels, D.; Berckmans, B.; Cammue, B.P. Screening for novel biocontrol agents applicable in plant disease management—A review. Biol. Control 2020, 144, 104240. [Google Scholar] [CrossRef]
- Engelbrecht, G.; Horak, I.; Jansen van Rensburg, P.J.; Claassens, S. Bacillus-based bionematicides: Development, modes of action and commercialisation. Biocontrol Sci. Technol. 2018, 28, 629–653. [Google Scholar] [CrossRef]
- Topalović, O.; Hussain, M.; Heuer, H. Plants and associated soil microbiota cooperatively suppress plant-parasitic nematodes. Front. Microbiol. 2020, 11, 313. [Google Scholar] [CrossRef]
- Syed Ab Rahman, S.F.; Singh, E.; Pieterse, C.M.J.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Wang, Y.-l.; Li, L.-f.; Li, D.-x.; Wang, B.; Zhang, K.; Niu, X. Yellow Pigment Aurovertins Mediate Interactions between the Pathogenic Fungus Pochonia chlamydosporia and Its Nematode Host. J. Agric. Food Chem. 2015, 63, 6577–6587. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, K.; Xu, J.; Dong, J.; Liu, Y. Nematicidal substances from fungi. Recent Pat. Biotechnol. 2007, 1, 212–233. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, S.; Stadler, M.; Dababat, A.A.; Richert-Pöggeler, K.R.; Finckh, M.R.; Maier, W. Monocillium gamsii sp. nov. and Monocillium bulbillosum: Two nematode-associated fungi parasitising the eggs of Heterodera filipjevi. MycoKeys 2017, 27, 21–38. [Google Scholar] [CrossRef]
- Giné, A.; Carrasquilla, M.; Martínez-Alonso, M.; Gaju, N.; Sorribas, F.J. Characterization of Soil Suppressiveness to Root-Knot Nematodes in Organic Horticulture in Plastic Greenhouse. Front. Plant Sci. 2016, 7, 164. [Google Scholar] [CrossRef]
- Castillo, J.D.; Vivanco, J.M.; Manter, D.K. Bacterial Microbiome and Nematode Occurrence in Different Potato Agricultural Soils. Microb. Ecol. 2017, 74, 888–900. [Google Scholar] [CrossRef]
- Hussain, M.; Hamid, M.I.; Tian, J.; Hu, J.; Zhang, X.; Chen, J.; Xiang, M.; Liu, X. Bacterial community assemblages in the rhizosphere soil, root endosphere and cyst of soybean cyst nematode-suppressive soil challenged with nematodes. FEMS Microbiol. Ecol. 2018, 94, 1–11. [Google Scholar] [CrossRef]
- Lopez-Llorca, L.V.; Olivares-Bernabeu, C.; Salinas, J.; Jansson, H.-B.; Kolattukudy, P.E. Pre-penetration events in fungal parasitism of nematode eggs. Mycol. Res. 2002, 106, 499–506. [Google Scholar] [CrossRef]
- Kara, Ö.; Bolat, İ. Influence of soil compaction on microfungal community structure in two soil types in Bartin Province, Turkey. J. Basic Microbiol. 2007, 47, 394–399. [Google Scholar] [CrossRef]
- Palma-Cano, L.E.; Piñon-Castillo, H.A.; Tarango-Rivero, S.H.; Carbon, A.; Salas-Leiva, J.; Muñoz-Castellanos, L.N.; Cravo-Laureau, C.; Duran, R.; Orrantia-Borunda, E. Effect of organic and conventional farming on soil bacterial diversity of pecan tree (Carya illinoensis K. Kosh) orchard across two phenological stages. Lett. Appl. Microbiol. 2021, 72, 556–569. [Google Scholar] [CrossRef]
- Na, X.; Ma, C.; Ma, S.; Ma, X.; Zhu, X.; Xu, P.; Zhu, H.; Cao, X.; Liang, W. Monocropping decouples plant–bacteria interaction and strengthens phytopathogenic fungi colonization in the rhizosphere of a perennial plant species. Plant Soil 2019, 445, 549–564. [Google Scholar] [CrossRef]
- Elhady, A.; Giné, A.; Topalovic, O.; Jacquiod, S.; Sørensen, S.J.; Sorribas, F.J.; Heuer, H. Microbiomes associated with infective stages of root-knot and lesion nematodes in soil. PLoS ONE 2017, 12, e0177145. [Google Scholar] [CrossRef] [PubMed]
- Fourie, H.; Mothata, T.; Ntidi, K.N.; Mc Donald, A.H.; De Waele, D.G.M.A. Indications of variation in host suitability to root knot nematode populations in commercial tomato varieties. Afr. J. Agric. Res. 2012, 7, 2344–2355. [Google Scholar] [CrossRef]
- Riekert, H. A modified sodium hypochlorite technique for the extraction of root-knot nematode eggs and larvae from maize root samples. Afr. Plant Prot. 1995, 1, 41–43. [Google Scholar]
- De Grisse, A. A counting dish for nematodes excluding border effect. Nematologica 1963, 9, 162. [Google Scholar] [CrossRef]
- Windham, G.L.; Williams, W.P. Host suitability of commercial corn hybrids to Meloidogyne arenaria and M. incognita. J. Nematol. 1987, 19, 13–16. [Google Scholar] [PubMed]
- Rashidifard, M.; Ashrafi, S.; Claassens, S.; Thuenen, T.; Fourie, H. A pilot approach investigating the potential of crop rotation with sainfoin to reduce Meloidogyne enterolobii infection of maize under greenhouse conditions. Front. Plant Sci. 2021, 12, 659322. [Google Scholar] [CrossRef]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Fernandez-Gnecco, G.; Smalla, K.; Maccario, L.; Sørensen, S.J.; Barbieri, P.; Consolo, V.F.; Covacevich, F.; Babin, D. Microbial community analysis of soils under different soybean cropping regimes in the Argentinean south-eastern Humid Pampas. FEMS Microbiol. Ecol. 2021, 97, fiab007. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- McSorley, R.; Wang, K.; Kokalis-Burelle, N.; Church, G. Effects of soil type and steam on nematode biological control potential of the rhizosphere community. Nematropica 2006, 36, 197–214. [Google Scholar]
- Elhady, A.; Adds, S.; Hallmann, J.; Heuer, H. Rhizosphere microbiomes modulated by pre-crop assisted plants in defence against plant-parasitic nematodes. Front. Microbiol. 2018, 9, 1133. [Google Scholar] [CrossRef] [PubMed]
- Adam, M.; Westphal, A.; Hallmann, J.; Heuer, H. Specific microbial attachment to root knot nematodes in suppressive soil. Appl. Environ. Microbiol. 2014, 80, 2679–2686. [Google Scholar] [CrossRef]
- Meisner, A.; Jacquiod, S.; Snoek, B.L.; ten Hooven, F.C.; van der Putten, W.H. Drought Legacy Effects on the Composition of Soil Fungal and Prokaryote Communities. Front. Microbiol. 2018, 9, 294. [Google Scholar] [CrossRef]
- Preece, C.; Verbruggen, E.; Liu, L.; Weedon, J.T.; Peñuelas, J. Effects of past and current drought on the composition and diversity of soil microbial communities. Soil Biol. Biochem. 2019, 131, 28–39. [Google Scholar] [CrossRef]
- Fernández-Calviño, D.; Bååth, E. Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol. Ecol. 2010, 73, 149–156. [Google Scholar] [CrossRef]
- Klimek, B.; Niklińska, M.; Jaźwa, M.; Tarasek, A.; Tekielak, I.; Musielok, Ł. Covariation of soil bacteria functional diversity and vegetation diversity along an altitudinal climatic gradient in the Western Carpathians. Pedobiologia 2015, 58, 105–112. [Google Scholar] [CrossRef]
- Zeng, J.; Zhao, D.; Li, H.; Huang, R.; Wang, J.; Wu, Q.L. A monotonically declining elevational pattern of bacterial diversity in freshwater lake sediments. Environ. Microbiol. 2016, 18, 5175–5186. [Google Scholar] [CrossRef]
- Moffett, B.F.; Nicholson, F.A.; Uwakwe, N.C.; Chambers, B.J.; Harris, J.A.; Hill, T.C.J. Zinc contamination decreases the bacterial diversity of agricultural soil. FEMS Microbiol. Ecol. 2003, 43, 13–19. [Google Scholar] [CrossRef]
- Liu, J.; Vipulanandan, C.; Cooper, T.F.; Vipulanandan, G. Effects of Fe nanoparticles on bacterial growth and biosurfactant production. J. Nanopart. Res. 2013, 15, 1405. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Yergeau, E.; Wong, L.C.; Pijl, A.S.; van Veen, J.A.; Kowalchuk, G.A. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 2012, 79, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Chodak, M.; Gołębiewski, M.; Morawska-Płoskonka, J.; Kuduk, K.; Niklińska, M. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl. Soil Ecol. 2013, 64, 7–14. [Google Scholar] [CrossRef]
- Tarin, M.W.K.; Fan, L.; Xie, D.; Tayyab, M.; Rong, J.; Chen, L.; Muneer, M.A.; Zheng, Y. Response of soil fungal diversity and community composition to varying levels of bamboo biochar in red soils. Microorganisms 2021, 9, 1385. [Google Scholar] [CrossRef] [PubMed]
- Bachelot, B.; Uriarte, M.; Zimmerman, J.K.; Thompson, J.; Leff, J.W.; Asiaii, A.; Koshner, J.; McGuire, K. Long-lasting effects of land use history on soil fungal communities in second-growth tropical rain forests. Ecol. Appl. 2016, 26, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.J.; Silver, W.L. Iron oxidation stimulates organic matter decomposition in humid tropical forest soils. Glob. Chang. Biol. 2013, 19, 2804–2813. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, D.; Wang, Y.; Zhu, X.; Chen, L.; Duan, Y. Evaluation of Bacillus aryabhattai Sneb517 for control of Heterodera glycines in soybean. Biol. Control 2020, 142, 104147. [Google Scholar] [CrossRef]
- Antil, S.; Kumar, R.; Pathak, D.; Kumar, A.; Panwar, A.; Kumari, A.; Kumar, V. On the potential of Bacillus aryabhattai KMT-4 against Meloidogyne javanica. Egypt. J. Biol. Pest Control 2021, 31, 1–9. [Google Scholar] [CrossRef]
- Yao, Y.-R.; Tian, X.-L.; Shen, B.-M.; Mao, Z.-C.; Chen, G.-h.; Xie, B.-Y. Transformation of the endophytic fungus Acremonium implicatum with GFP and evaluation of its biocontrol effect against Meloidogyne incognita. World J. Microbiol. Biotechnol. 2015, 31, 549–556. [Google Scholar] [CrossRef]
- Thongkaewyuan, A.; Chairin, T. Biocontrol of Meloidogyne incognita by Metarhizium guizhouense and its protease. Biol. Control 2018, 126, 142–146. [Google Scholar] [CrossRef]
- Baron, N.C.; de Souza Pollo, A.; Rigobelo, E.C. Purpureocillium lilacinum and Metarhizium marquandii as plant growth-promoting fungi. PeerJ 2020, 8, e9005. [Google Scholar] [CrossRef] [PubMed]
- Daragó, Á.; Szabó, M.; Hrács, K.; Takács, A.; Nagy, P.I. In vitro investigations on the biological control of Xiphinema index with Trichoderma species. Helminthologia 2013, 50, 132–137. [Google Scholar] [CrossRef]
- Ashraf, M.S.; Khan, T.A. Effect of opportunistic fungi on the life cycle of the root-knot nematode (Meloidogyne javanica) on brinjal. Arch. Phytopathol. Plant Prot. 2005, 38, 227–233. [Google Scholar] [CrossRef]
- Karliński, L.; Ravnskov, S.; Rudawska, M. Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions. Forests 2020, 11, 262. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Bakker, M.G.; Bradeen, J.M.; Kinkel, L.L. Plant community richness and microbial interactions structure bacterial communities in soil. Ecology 2015, 96, 134–142. [Google Scholar] [CrossRef]
- Cao, H.; Chen, R.; Wang, L.; Jiang, L.; Yang, F.; Zheng, S.; Wang, G.; Lin, X. Soil pH, total phosphorus, climate and distance are the major factors influencing microbial activity at a regional spatial scale. Sci. Rep. 2016, 6, 25815. [Google Scholar] [CrossRef]
- Collett, R.L.; Marais, M.; Daneel, M.S.; Rashidifard, M.; Fourie, H. Meloidogyne enterolobii, a threat to crop production with particular reference to sub-Saharan Africa: A critical and updated review. Nematology 2021, 23, 1–39. [Google Scholar] [CrossRef]



| Sample Code | Location | GPS Coordinates | Crop | Altitude (m) | Irrigation Type |
|---|---|---|---|---|---|
| SA 1 | Tomahawk Citrus, Malalane | S25 36 42.3 E031 36 00.9 | Banana (Musa sp.) | 292 | Microjet |
| SA 2 | Tomahawk Citrus, Malalane | S25 37 05.7 E031 35 43.5 | Papaya (Carica papaya) | 295 | Drip |
| SA 3 | Tomahawk Citrus, Malalane | S25 37 14.2 E031 35 54.5 | Mango (Mangifera indica) | 296 | Drip |
| SA 4 | Tomahawk Citrus, Malalane | S25 37 06.8 E031 36 07.7 | Citrus (Citrus sp.) | 297 | Microjet |
| SA 5 | Tomahawk Citrus, Malalane | S25 36 38.6 E031 37 12.4 | Litchi (Litchi chinensis) | 316 | Microjet |
| SA 6 | ARC-TSC Nelspruit | S25 27 21.0 E030 58 16.1 | Macadamia (Macadamia integrifolia) | 657 | Microjet |
| SA 7 | ARC-TSC Nelspruit | S25 27 22.0 E030 58 13.8 | Mango (Mangifera indica) | 658 | Microjet |
| SA 8 | ARC-TSC Nelspruit | S25 27 22.0 E030 58 13.8 | Citrus (Citrus sp.) | 658 | Microjet |
| SA 9 | ARC-TSC Nelspruit | S25 27 24.4 E030 58 10.7 | Avocado (Persea americanum) | 654 | Microjet |
| SA 10 | ARC-TSC Nelspruit | S25 27 02.8 E030 58 21.7 | Litchi (Litchi chinensis) | 656 | Microjet |
| SA 11 | ARC-TSC Burgershall | S25 06 59.8 E031 04 56.1 | Avocado (Persea americanum) | 774 | Microjet |
| SA 12 | ARC-TSC Burgershall | S25 06 56.1 E031 04 58.1 | Papaya (Carica papaya) | 779 | No irrigation |
| SA 13 | ARC-TSC Burgershall | S25 06 57.1 E031 05 02.5 | Macadamia (Macadamia integrifolia) | 768 | Microjet |
| SA 14 | ARC-TSC Burgershall | S25 06 49.7 E031 05 08.0 | Banana (Musa sp.) | 761 | Microjet |
| Sampling Region | pH | K mg/kg | Zn mg/kg | Fe mg/kg | Clay % | Al mg/kg | Altitude (m) |
|---|---|---|---|---|---|---|---|
| Tomahawk Citrus, Malelane | 6.13 a | 351.8 a | 17.7 b | 139.7 a | 19.6 a | 5.4 a | 299.2 a |
| ARC-TSC, Nelspruit | 7.98 b | 56.2 b | 13.09 ab | 135.5 a | 6.8 b | 3.2 b | 656.6 b |
| ARC-TSC Burgershall | 5.94 a | 348.7 a | 9.6 a | 80.3 b | 19 a | 6.7 a | 770.5 c |
| p value | 0.000 | 0.002 | 0.024 | 0.039 | 0.020 | 0.030 | 0.00 |
| F ratio | 34.36 | 10.93 | 5.337 | 4.415 | 5.653 | 4.912 | 5549 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashidifard, M.; Fourie, H.; Ashrafi, S.; Engelbrecht, G.; Elhady, A.; Daneel, M.; Claassens, S. Suppressive Effect of Soil Microbiomes Associated with Tropical Fruit Trees on Meloidogyne enterolobii. Microorganisms 2022, 10, 894. https://doi.org/10.3390/microorganisms10050894
Rashidifard M, Fourie H, Ashrafi S, Engelbrecht G, Elhady A, Daneel M, Claassens S. Suppressive Effect of Soil Microbiomes Associated with Tropical Fruit Trees on Meloidogyne enterolobii. Microorganisms. 2022; 10(5):894. https://doi.org/10.3390/microorganisms10050894
Chicago/Turabian StyleRashidifard, Milad, Hendrika Fourie, Samad Ashrafi, Gerhard Engelbrecht, Ahmed Elhady, Mieke Daneel, and Sarina Claassens. 2022. "Suppressive Effect of Soil Microbiomes Associated with Tropical Fruit Trees on Meloidogyne enterolobii" Microorganisms 10, no. 5: 894. https://doi.org/10.3390/microorganisms10050894
APA StyleRashidifard, M., Fourie, H., Ashrafi, S., Engelbrecht, G., Elhady, A., Daneel, M., & Claassens, S. (2022). Suppressive Effect of Soil Microbiomes Associated with Tropical Fruit Trees on Meloidogyne enterolobii. Microorganisms, 10(5), 894. https://doi.org/10.3390/microorganisms10050894






