Persulfide-Responsive Transcription Factor SqrR Regulates Gene Transfer and Biofilm Formation via the Metabolic Modulation of Cyclic di-GMP in Rhodobacter capsulatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
2.2. Cloning and Mutagenesis
2.3. GTA Transduction Assay
2.4. Western Blotting of Capsid Protein
2.5. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
2.6. Overexpression and Purification of SqrR
2.7. Gel Mobility Shift Analysis
2.8. Quantification of c-di-GMP
2.9. Quantification of Biofilm Formation
3. Results
3.1. SqrR Contributes to GTA Production and Release
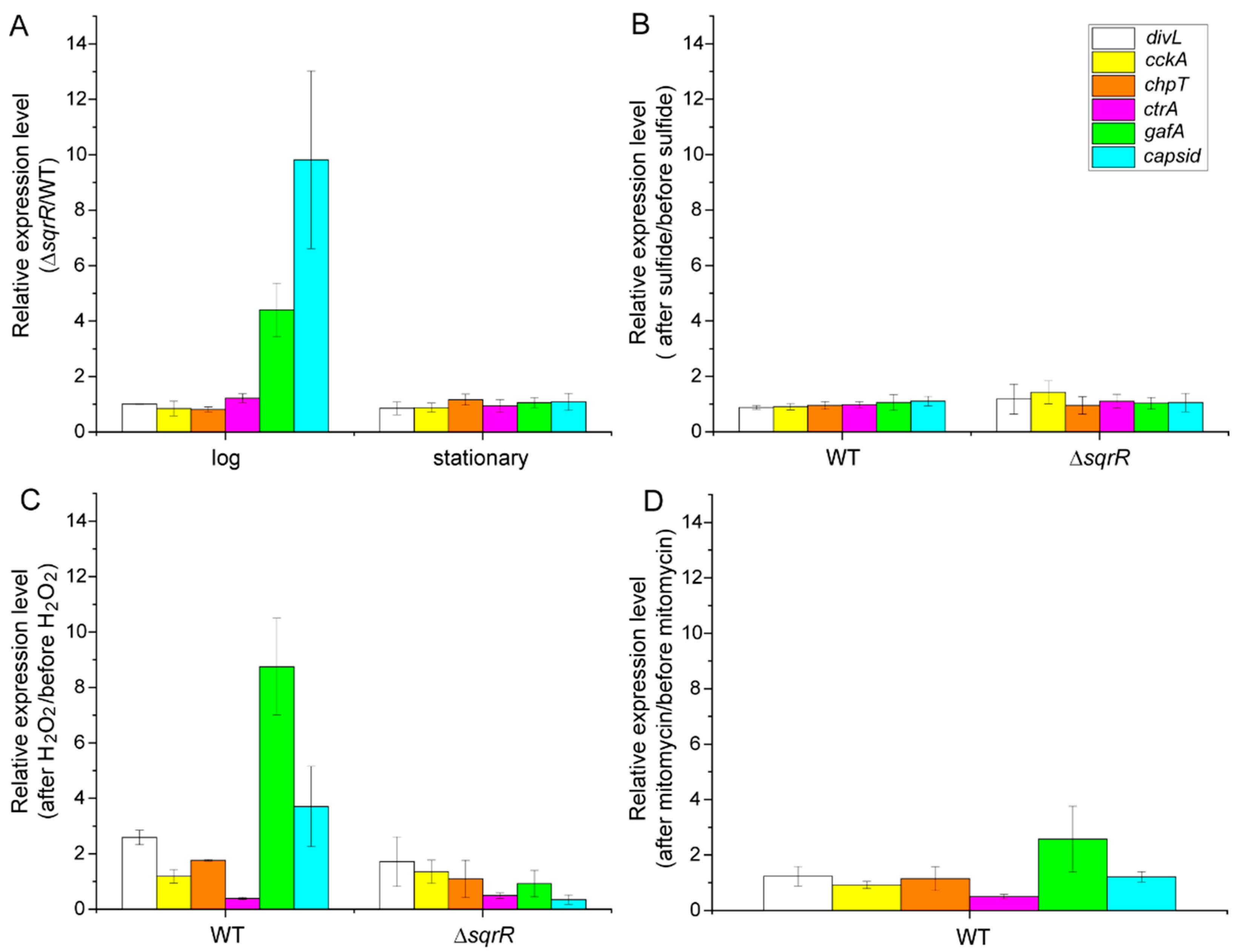
3.2. Molecular Mechanism of the SqrR-Related H2O2-Induced Transcription of GTA-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Boto, L. Horizontal gene transfer in evolution: Facts and challenges. Proc. R. Soc. B Biol. Sci. 2010, 277, 819–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanton, T.B. Prophage-like gene transfer agents-Novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 2007, 13, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.S.; Beatty, J.T. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 2007, 15, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Marrs, B. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 1974, 71, 971–973. [Google Scholar] [CrossRef] [Green Version]
- Lang, A.S.; Westbye, A.B.; Beatty, J.T. The Distribution, Evolution, and Roles of Gene Transfer Agents in Prokaryotic Genetic Exchange. Annu. Rev. Virol. 2017, 4, 87–104. [Google Scholar] [CrossRef]
- Lang, A.S.; Zhaxybayeva, O.; Beatty, J.T. Gene transfer agents: Phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012, 10, 472–482. [Google Scholar] [CrossRef] [Green Version]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; MacLeod, D.M.; Rivkin, R.B.; Chen, F.; Buchan, A.; Lang, A.S. High diversity of Rhodobacterales in the subarctic North Atlantic ocean and gene transfer agent protein expression in isolated strains. Aquat. Microb. Ecol. 2010, 59, 283–293. [Google Scholar] [CrossRef] [Green Version]
- McDaniel, L.D.; Young, E.; Delaney, J.; Ruhnau, F.; Ritchie, K.B.; Paul, J.H. High frequency of horizontal gene transfer in the oceans. Science 2012, 330, 50. [Google Scholar] [CrossRef] [Green Version]
- Fogg, P.C.M.; Westbye, A.B.; Beatty, J.T. One for all or all for one: Heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS ONE 2012, 7, e43772. [Google Scholar] [CrossRef] [Green Version]
- Westbye, A.B.; O’Neill, Z.; Schellenberg-Beaver, T.; Thomas Beatty, J. The Rhodobacter capsulatus gene transfer agent is induced by nutrient depletion and the RNAP omega subunit. Microbiology 2017, 163, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.L.; Taylor, T.A.; Beatty, J.T.; Greenberg, E.P. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J. Bacteriol. 2002, 184, 6515–6521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, M.M.; Brimacombe, C.A.; Spiegelman, G.B.; Beatty, J.T. The GtaR protein negatively regulates transcription of the gtaRI operon and modulates gene transfer agent (RcGTA) expression in Rhodobacter capsulatus. Mol. Microbiol. 2012, 83, 759–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, A.S.; Beatty, J.T. Genetic analysis of a bacterial genetic exchange element: The gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA 2000, 97, 859–864. [Google Scholar] [CrossRef] [Green Version]
- Mercer, R.G.; Quinlan, M.; Rose, A.R.; Noll, S.; Beatty, J.T.; Lang, A.S. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol. Lett. 2012, 331, 53–62. [Google Scholar] [CrossRef]
- Fogg, P.C.M. Identification and characterization of a direct activator of a gene transfer agent. Nat. Commun. 2019, 10, 595. [Google Scholar] [CrossRef]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density- responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, C.; Domian, I.J.; Maddock, J.R.; Shapiro, L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 1999, 97, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Biondi, E.G.; Reisinger, S.J.; Skerker, J.M.; Arif, M.; Perchuk, B.S.; Ryan, K.R.; Laub, M.T. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 2006, 444, 899–904. [Google Scholar] [CrossRef]
- Westbye, A.B.; Kater, L.; Wiesmann, C.; Ding, H.; Yip, C.K.; Beatty, J.T. The protease ClpXP and the PAS domain protein DivL regulate CtrA and gene transfer agent production in Rhodobacter capsulatus. Appl. Environ. Microbiol. 2018, 84, e00275-18. [Google Scholar] [CrossRef] [Green Version]
- Pallegar, P.; Langille, E.; Gomelsky, M.; Lang, A.S. Cyclic di-GMP-Mediated Regulation of Gene Transfer and Motility in Rhodobacter capsulatus. J. Bacteriol. 2020, 202, e00554-19. [Google Scholar] [CrossRef]
- Shimizu, T.; Shen, J.; Fang, M.; Zhang, Y.; Hori, K.; Trinidad, J.C.; Bauer, C.E.; Giedroc, D.P.; Masuda, S. SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl. Acad. Sci. USA 2017, 114, 2355–2360. [Google Scholar] [CrossRef] [Green Version]
- Ida, T.; Sawa, T.; Ihara, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fujii, S.; Matsunaga, T.; et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef] [Green Version]
- Nishida, M.; Sawa, T.; Kitajima, N.; Ono, K.; Inoue, H.; Ihara, H.; Motohashi, H.; Yamamoto, M.; Suematsu, M.; Kurose, H.; et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012, 8, 714–724. [Google Scholar] [CrossRef]
- Cuevasanta, E.; Lange, M.; Bonanata, J.; Coitiño, E.L.; Ferrer-Sueta, G.; Filipovic, M.R.; Alvarez, B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 2015, 290, 26866–26880. [Google Scholar] [CrossRef] [Green Version]
- Yadav, P.K.; Martinov, M.; Vitvitsky, V.; Seravalli, J.; Wedmann, R.; Filipovic, M.R.; Banerjee, R. Biosynthesis and reactivity of cysteine persulfides in signaling. J. Am. Chem. Soc. 2016, 138, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Rauch, B.J.; Klimek, J.; David, L.; Perona, J.J. Persulfide Formation Mediates Cysteine and Homocysteine Biosynthesis in Methanosarcina acetivorans. Biochemistry 2017, 56, 1051–1061. [Google Scholar] [CrossRef]
- Olson, K.R. Hydrogen sulfide, reactive sulfur species and coping with reactive oxygen species. Free Radic. Biol. Med. 2019, 140, 74–83. [Google Scholar] [CrossRef]
- Weaver, P.F.; Wall, J.D.; Gest, H. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 1975, 105, 207–216. [Google Scholar] [CrossRef]
- Yen, H.C.; Hu, N.T.; Marrs, B.L. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 1979, 131, 157–168. [Google Scholar] [CrossRef]
- Sganga, M.W.; Bauer, C.E. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell 1992, 68, 945–954. [Google Scholar] [CrossRef]
- Wall, J.D.; Weaver, P.F.; Gest, H. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch. Microbiol. 1975, 105, 217–224. [Google Scholar] [CrossRef]
- Masuda, S.; Bauer, C.E. Null Mutation of HvrA Compensates for Loss of an Essential relA/spoT-Like Gene in Rhodobacter capsulatus. J. Bacteriol. 2004, 186, 235–239. [Google Scholar] [CrossRef] [Green Version]
- Solioz, M.; Yen, H.C.; Marrs, B. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J. Bacteriol. 1975, 123, 651–657. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Cheng, Z.; Matsuura, K.; Masuda, S.; Bauer, C.E. Evidence that altered cis element spacing affects PpsR mediated redox control of photosynthesis gene expression in Rubrivivax gelatinosus. PLoS ONE 2015, 10, e0128446. [Google Scholar] [CrossRef]
- Hynes, A.P.; Mercer, R.G.; Watton, D.E.; Buckley, C.B.; Lang, A.S. DNA packaging bias and differential expression of gene transfer agent genes within a population during production and release of the Rhodobacter capsulatus gene transfer agent, RcGTA. Mol. Microbiol. 2012, 85, 314–325. [Google Scholar] [CrossRef]
- Ding, H.; Grüll, M.P.; Mulligan, M.E.; Lang, A.S.; Thomas Beatty, J. Induction of Rhodobacter capsulatus gene transfer agent gene expression is a bistable stochastic process repressed by an extracellular calcium-binding rtx protein homologue. J. Bacteriol. 2019, 201, e00430-19. [Google Scholar] [CrossRef]
- Barbé, J.; Vericat, J.A.; Cairó, J.; Guerrero, R. Further characterization of SOS system induction in recBC mutants of Escherichia coli. Mutat. Res. DNA Repair Rep. 1985, 146, 23–32. [Google Scholar] [CrossRef]
- Goerlich, O.; Quillardet, P.; Hofnung, M. Induction of the SOS response by hydrogen peroxide in various Escherichia coli mutants with altered protection against oxidative DNA damage. J. Bacteriol. 1989, 171, 6141–6147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuchinski, K.S.; Brimacombe, C.A.; Westbye, A.B.; Ding, H.; Beatty, T.J. The SOS response master regulator LexA regulates the gene transfer agent of Rhodobacter capsulatus and represses transcription of the signal transduction protein CckA. J. Bacteriol. 2016, 198, 1137–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christman, M.F.; Morgan, R.W.; Jacobson, F.S.; Ames, B.N. Oxidative Stress and Some Heat-Shock Proteins in Salmonella typhimurium. Cell 1985, 41, 753–762. [Google Scholar] [CrossRef]
- Storz, G.; Tartaglia, L.A.; Ames, B.N. Transcriptional regulator of oxidative stress-inducible genes: Direct activation by oxidation. Science 1990, 248, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Sanjuán, J. Exploiting the commons: Cyclic diguanylate regulation of bacterial exopolysaccharide production. Curr. Opin. Microbiol. 2016, 30, 36–43. [Google Scholar] [CrossRef]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Farrera-Calderon, R.G.; Pallegar, P.; Westbye, A.B.; Wiesmann, C.; Lang, A.S.; Beatty, J.T. The CckA-ChpT-CtrA phosphorelay controlling Rhodobacter capsulatus gene transfer agent production is bidirectional and regulated by cyclic di-GMP. J. Bacteriol. 2020, 203, e00525-20. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.L.; Hinton, D.M.; Waters, C.M. VpsR and cyclic di-GMP together drive transcription initiation to activate biofilm formation in Vibrio cholerae. Nucleic Acids Res. 2018, 46, 8876–8887. [Google Scholar] [CrossRef] [PubMed]
- Pursley, B.R.; Maiden, M.M.; Hsieh, M.L.; Fernandez, N.L.; Severin, G.B.; Waters, C.M. Cyclic di-GMP regulates TfoY in Vibrio cholerae to control motility by both transcriptional and posttranscriptional mechanisms. J. Bacteriol. 2018, 200, e00578-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasteva, P.V.; Jiunn, J.C.; Shikuma, N.J.; Beyhan, S.; Navarro, M.V.A.S.; Yildiz, F.H.; Sondermann, H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 2010, 327, 866–868. [Google Scholar] [CrossRef] [Green Version]
- Pallegar, P.; Canuti, M.; Langille, E.; Peña-Castillo, L.; Lang, A.S. A Two-Component System Acquired by Horizontal Gene Transfer Modulates Gene Transfer and Motility via Cyclic Dimeric GMP. J. Mol. Biol. 2020, 432, 4840–4855. [Google Scholar] [CrossRef]
- Capdevila, D.A.; Walsh, B.J.C.; Zhang, Y.; Dietrich, C.; Gonzalez-gutierrez, G.; Giedroc, D.P. Structural basis for persulfide-sensing specificity in a transcriptional regulator. Nat. Chem. Biol. 2020, 17, 65–70. [Google Scholar] [CrossRef]
- Hou, N.; Yan, Z.; Fan, K.; Li, H.; Zhao, R.; Xia, Y.; Xun, L.; Liu, H. OxyR senses sulfane sulfur and activates the genes for its removal in Escherichia coli. Redox Biol. 2019, 26, 101293. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Hayashi, Y.; Arai, M.; McGlynn, S.E.; Masuda, T.; Masuda, S. Repressor Activity of SqrR, a Master Regulator of Persulfide-Responsive Genes, Is Regulated by Heme Coordination. Plant Cell Physiol. 2021, 62, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Bunn, H.F.; Jandl, J.H. Exchange of heme among hemoglobins and between hemoglobin and albumin. J. Biol. Chem. 1968, 243, 465–475. [Google Scholar] [CrossRef]
- Kvam, E.; Noel, A.; Basu-Modak, S.; Tyrrell, R.M. Cyclooxygenase dependent release of heme from microsomal hemeproteins correlates with induction of heme oxygenase 1 transcription in human fibroblasts. Free Radic. Biol. Med. 1999, 26, 511–517. [Google Scholar] [CrossRef]




| Gene | Annotation | Transcript Fold Change in ΔsqrR | p-Value |
|---|---|---|---|
| rcc00042 | divL | 9.276648 | 4.94 × 10−64 |
| rcc01687 | capsid | 3.318318 | 2.28 × 10−7 |
| rcc03000 | chpT | 2.984249 | 2.6 × 10−8 |
| rcc00645 | c-di-GMP metabolic enzyme | 118.6299 | 3.7 × 10−167 |
| rcc02857 | c-di-GMP metabolic enzyme | 0.497602 | 0.002762 |
| Name | Sequence 5′–3′ | Purpose |
|---|---|---|
| oxyR F1 | CGACTCTAGAGGATCATTGCCGTATTTCTTCTTGATCGGC | Cloning for gene disruption |
| oxyR R1 | CGAGAGGTTTATCATAATGAAAAACTATCGCAGGC | |
| oxyR F2 | ATGATAAACCTCTCGGCGCGGGAGGCGTGAGGTCGGCGGGTTCGG | |
| oxyR R2 | CGGTACCCGGGGATCGCGCATCCGCTGGCGCCCGAGACGC | |
| divL-F | CGGTACCCGGGGATCAGAATGCGCCGGTGCCGCGGCGCTC | Cloning for DNA probes of the gel shift assay |
| divL-R | CGACTCTAGAGGATCGACTGCAGCCCTCTCGCCTGTCCCG | |
| cckA-F | CGGTACCCGGGGATCCGCCGCAGCTATTCCCCGCGCGACG | |
| cckA-R | CGACTCTAGAGGATCGGGCTGATCGGGATGTACCACTGGC | |
| chpT-F | CGGTACCCGGGGATCAAGCTGCACCCGTCGCCCGTCGATC | |
| chpT-R | CGACTCTAGAGGATCGGGTCATGGTGGATCTCCCTTTCGG | |
| gafA-F | CGGTACCCGGGGATCGTAATCGCGCTGCCCGAAGCGTGCG | |
| gafA-R | CGACTCTAGAGGATCCTCCGGTCTCCCATCGACAGGCTGG | |
| capsid-F | CGGTACCCGGGGATCACCGGCGGGCATGCTTTTGCCGAGA | |
| capsid-R | CGACTCTAGAGGATCGTCTTGCGTGACCCGCCTCTCATGC | |
| ctrA-F | CGGTACCCGGGGATCGCCGCCGAAAGAAACGCGTCGTTGG | |
| ctrA-R | CGACTCTAGAGGATCCCTGGGTTCTCCGCATTAATCCCTC | |
| 02857-F | CGGTACCCGGGGATCTGGTGCCCCAGCCTAACCGCGGGAT | |
| 02857-R | CGACTCTAGAGGATCCGGGAACGGACCCCTTCGAGTGGAT | |
| 02630-F | CGGTACCCGGGGATCGTGCCCGGACCGGAGGCGGTTTTCC | |
| 02630-R | CGACTCTAGAGGATCGGACCCTCCTCGCGCGGACCATAGC | |
| 00620-F | CGGTACCCGGGGATCCTTGTCGGGGGGGATGACGCCGCTG | |
| uvrD qF | CAGAAGGAACACACGGTCAA | For qRT-PCR |
| uvrD qR | AAAGTGTCAGGCGGAATCTC | |
| divL qF | CCGACGCTTTATGCCTTTCT | |
| divL qR | CCTGTTCCAGTTCCGTCATCT | |
| cckA qF | GCGCATGATTTCAACAACTT | |
| cckA qR | TTCTGGCTGATCTGGTCAAG | |
| chpT qF | ACGGGGTGGAGTTGCTGAA | |
| chpT qR | AAAGGCGATGCGGAAGAA | |
| ctrA qF | TTTGGCGCCGATGATTAC | |
| ctrA qR | GGATGATCGACTGCGAATG | |
| gafA qF | GCTGAACGGCTGGATCTT | |
| gafA qR | TTCCAACAGCCGCTTCAA | |
| capsid qF | CGGTTGCCGAGGTGAAA | |
| capsid qR | CACACGCTCTTCCTGTTGTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimizu, T.; Aritoshi, T.; Beatty, J.T.; Masuda, T. Persulfide-Responsive Transcription Factor SqrR Regulates Gene Transfer and Biofilm Formation via the Metabolic Modulation of Cyclic di-GMP in Rhodobacter capsulatus. Microorganisms 2022, 10, 908. https://doi.org/10.3390/microorganisms10050908
Shimizu T, Aritoshi T, Beatty JT, Masuda T. Persulfide-Responsive Transcription Factor SqrR Regulates Gene Transfer and Biofilm Formation via the Metabolic Modulation of Cyclic di-GMP in Rhodobacter capsulatus. Microorganisms. 2022; 10(5):908. https://doi.org/10.3390/microorganisms10050908
Chicago/Turabian StyleShimizu, Takayuki, Toma Aritoshi, J. Thomas Beatty, and Tatsuru Masuda. 2022. "Persulfide-Responsive Transcription Factor SqrR Regulates Gene Transfer and Biofilm Formation via the Metabolic Modulation of Cyclic di-GMP in Rhodobacter capsulatus" Microorganisms 10, no. 5: 908. https://doi.org/10.3390/microorganisms10050908
APA StyleShimizu, T., Aritoshi, T., Beatty, J. T., & Masuda, T. (2022). Persulfide-Responsive Transcription Factor SqrR Regulates Gene Transfer and Biofilm Formation via the Metabolic Modulation of Cyclic di-GMP in Rhodobacter capsulatus. Microorganisms, 10(5), 908. https://doi.org/10.3390/microorganisms10050908







