Hydrodynamic Effects on Biofilm Development and Recombinant Protein Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Flow Cell System and Experimental Conditions
2.3. Biofilm and Planktonic Monitoring
2.3.1. Assessment of eGFP-Expressing Cells and Specific Protein Concentrations
2.3.2. Plasmid Extraction and Quantification
2.4. Statistical Analysis
3. Results
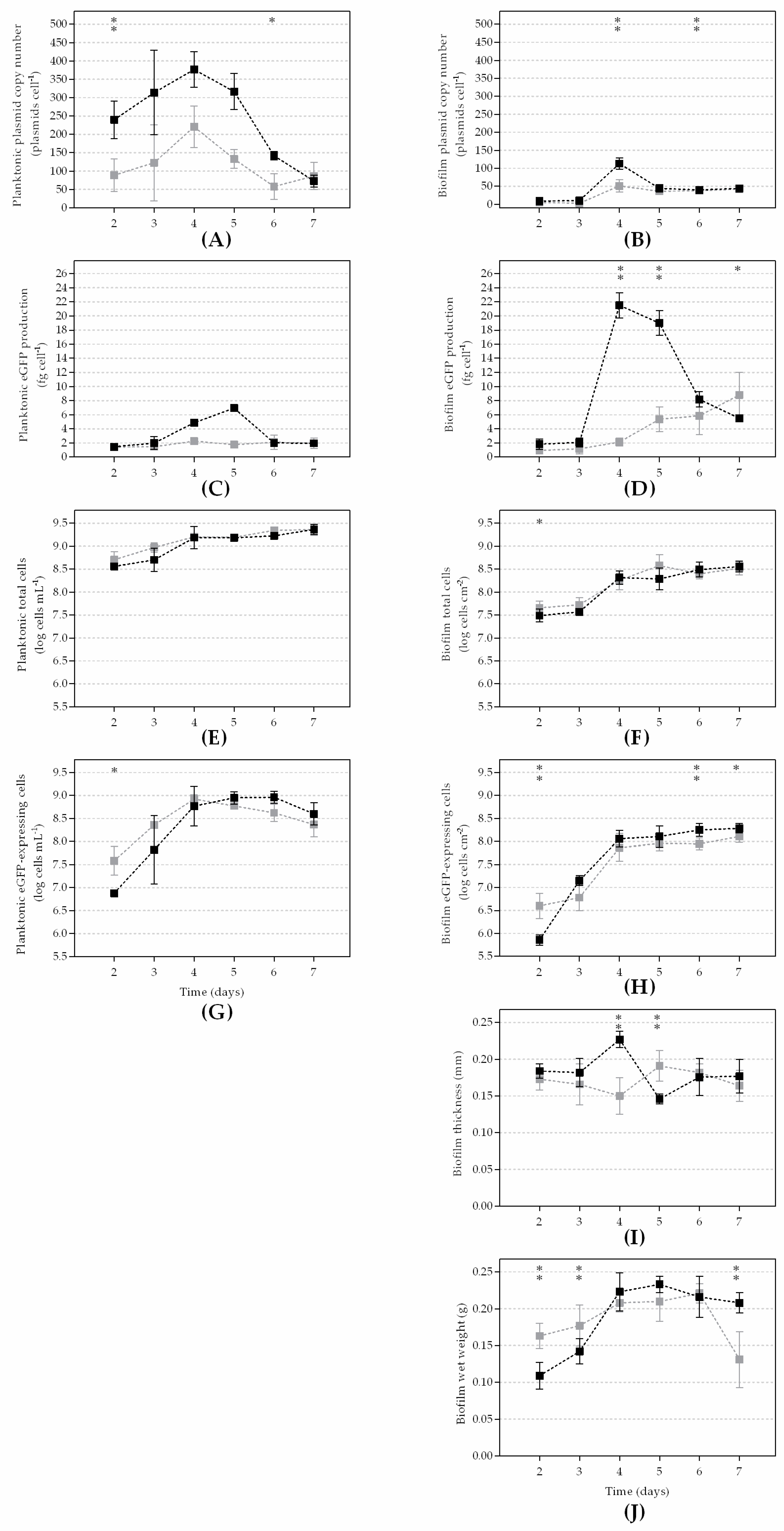
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Overton, T.W. Recombinant protein production in bacterial hosts. Drug Discov. Today 2014, 19, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.; Demain, A.L. Enzymes and bioconversions of industrial, pharmaceutical, and biotechnological significance. Org. Process Res. Dev. 2011, 15, 224–230. [Google Scholar] [CrossRef]
- Hoffmann, F.; Rinas, U. Stress induced by recombinant protein production in Escherichia coli. In Physiological Stress Responses in Bioprocesses; Springer: Berlin/Heidelberg, Germany, 2004; pp. 73–92. [Google Scholar]
- Cheng, K.C.; Demirci, A.; Catchmark, J.M. Advances in biofilm reactors for production of value-added products. Appl. Microbiol. Biotechnol. 2010, 87, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Azevedo, A.; Gomes, L.C.; Mergulhão, F.J. Recombinant protein expression in biofilms. AIMS Microbiol. 2019, 5, 232. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, H.A.; Niu, C.; Gilbert, E.S. Enhanced high copy number plasmid maintenance and heterologous protein production in an Escherichia coli biofilm. Biotechnol. Bioeng. 2007, 97, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Monteiro, G.A.; Gomes, L.C.; Mergulhão, F.J. The Influence of Nutrient Medium Composition on Escherichia coli Biofilm Development and Heterologous Protein Expression. Appl. Sci. 2021, 11, 8667. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881. [Google Scholar] [CrossRef]
- Rani, S.A.; Pitts, B.; Beyenal, H.; Veluchamy, R.A.; Lewandowski, Z.; Davison, W.M.; Buckingham, M.K.; Stewart, P.S. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol. 2007, 189, 4223–4233. [Google Scholar] [CrossRef] [Green Version]
- Bjergbæk, L.A.; Haagensen, J.A.J.; Reisner, A.; Molin, S.; Roslev, P. Effect of oxygen and growth medium on in vitro biofilm formation by Escherichia coli. Biofilms 2006, 3, 1–10. [Google Scholar] [CrossRef]
- Moreira, J.M.R.; Gomes, L.C.; Simões, M.; Melo, L.F.; Mergulhão, F.J. The impact of material properties, nutrient load and shear stress on biofouling in food industries. Food Bioprod. Process. 2015, 95, 228–236. [Google Scholar] [CrossRef] [Green Version]
- Rochex, A.; Godon, J.J.; Bernet, N.; Escudié, R. Role of shear stress on composition, diversity and dynamics of biofilm bacterial communities. Water Res. 2008, 42, 4915–4922. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Koo, H.; Ren, D. Effects of material properties on bacterial adhesion and biofilm formation. J. Dent. Res. 2015, 94, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Moreira, J.M.R.; Simões, M.; Melo, L.F.; Mergulhão, F.J. The combined effects of shear stress and mass transfer on the balance between biofilm and suspended cell dynamics. Desalin. Water Treat. 2015, 53, 3348–3354. [Google Scholar] [CrossRef] [Green Version]
- Moreira, J.M.R.; Teodósio, J.S.; Silva, F.C.; Simões, M.; Melo, L.F.; Mergulhão, F.J. Influence of flow rate variation on the development of Escherichia coli biofilms. Bioprocess Biosyst. Eng. 2013, 36, 1787–1796. [Google Scholar] [CrossRef]
- Teodósio, J.S.; Silva, F.C.; Moreira, J.M.; Simões, M.; Melo, L.F.; Alves, M.A.; Mergulhão, F.J. Flow cells as quasi-ideal systems for biofouling simulation of industrial piping systems. Biofouling 2013, 29, 953–966. [Google Scholar] [CrossRef]
- Busscher, H.J.; van der Mei, H.C. Microbial adhesion in flow displacement systems. Clin. Microbiol. Rev. 2006, 19, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tay, J.H. The essential role of hydrodynamic shear force in the formation of biofilm and granular sludge. Water Res. 2002, 36, 1653–1665. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Muffler, K.; Lakatos, M.; Schlegel, C.; Strieth, D.; Kuhne, S.; Ulber, R. Application of biofilm bioreactors in white biotechnology. Product. Biofilms 2014, 146, 123–161. [Google Scholar]
- Stewart, P.S. Diffusion in biofilms. J. Bacteriol. 2003, 185, 1485–1491. [Google Scholar] [CrossRef] [Green Version]
- Gomes, L.C.; Carvalho, D.; Briandet, R.; Mergulhão, F.J. Temporal variation of recombinant protein expression in Escherichia coli biofilms analysed at single-cell level. Process Biochem. 2016, 51, 1155–1161. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.T.; Peretti, S.W.; Bryers, J.D. Plasmid retention and gene expression in suspended and biofilm cultures of recombinant Escherichia coli DH5α (pMJR1750). Biotechnol. Bioeng. 1993, 41, 211–220. [Google Scholar] [CrossRef]
- Rahman, M.S.; Ano, T.; Shoda, M. Biofilm fermentation of iturin A by a recombinant strain of Bacillus subtilis 168. J. Biotechnol. 2007, 127, 503–507. [Google Scholar] [CrossRef]
- Vogt, C.M.; Schraner, E.M.; Aguilar, C.; Eichwald, C. Heterologous expression of antigenic peptides in Bacillus subtilis biofilms. Microb. Cell Factories 2016, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Dhulster, P.; Thomas, D.; Barbotin, J.N. Agitation rate effects on plasmid stability in immobilized and free-cell continuous cultures of recombinant E. coli. Enzyme Microb. Technol. 1990, 12, 933–939. [Google Scholar] [CrossRef]
- Singh, R.S.; Yadav, M. Enhanced production of recombinant aspartase of Aeromonas media NFB-5 in a stirred tank reactor. Bioresour. Technol. 2013, 145, 217–223. [Google Scholar] [CrossRef]
- Man, R.C.; Ismail, A.F.; Fuzi, S.F.Z.M.; Ghazali, N.F.; Illias, R.M. Effects of culture conditions of immobilized recombinant Escherichia coli on cyclodextrin glucanotransferase (CGTase) excretion and cell stability. Process Biochem. 2016, 51, 474–483. [Google Scholar] [CrossRef] [Green Version]
- Teodósio, J.S.; Simões, M.; Melo, L.F.; Mergulhão, F.J. Flow cell hydrodynamics and their effects on E. coli biofilm formation under different nutrient conditions and turbulent flow. Biofouling 2011, 27, 1–11. [Google Scholar] [CrossRef]
- Teodósio, J.S.; Simoes, M.; Alves, M.A.; Melo, L.F.; Mergulhao, F.J. Setup and validation of flow cell systems for biofouling simulation in industrial settings. Sci. World J. 2012, 2012, 361496. [Google Scholar] [CrossRef]
- Gomes, L.C.; Mergulhão, F.J. Heterologous protein production in Escherichia coli biofilms: A non-conventional form of high cell density cultivation. Process Biochem. 2017, 57, 1–8. [Google Scholar] [CrossRef]
- Mergulhão, F.J.; Monteiro, G.A. Analysis of factors affecting the periplasmic production of recombinant proteins in Escherichia coli. J. Microbiol. Biotechnol. 2007, 17, 1236–1241. [Google Scholar]
- Whelan, J.A.; Russell, N.B.; Whelan, M.A. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 2003, 278, 261–269. [Google Scholar] [CrossRef]
- Araújo, P.A.; Malheiro, J.; Machado, I.; Mergulhão, F.J.; Melo, L.F.; Simões, M. Influence of flow velocity on the characteristics of Pseudomonas fluorescens biofilms. J. Environ. Eng. 2016, 142, 04016031. [Google Scholar] [CrossRef]
- Cunningham, D.S.; Koepsel, R.; Ataai, M.; Domach, M. Factors affecting plasmid production in Escherichia coli from a resource allocation standpoint. Microb. Cell Factories 2009, 8, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Landini, P. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res. Microbiol. 2009, 160, 259–266. [Google Scholar] [CrossRef]
- Lowder, M.; Unge, A.; Maraha, N.; Jansson, J.K.; Swiggett, J.; Oliver, J.D. Effect of starvation and the viable-but-nonculturable state on green fluorescent protein (GFP) fluorescence in GFP-tagged Pseudomonas fluorescens A506. Appl. Environ. Microbiol. 2000, 66, 3160–3165. [Google Scholar] [CrossRef] [Green Version]
- Bentley, W.E.; Mirjalili, N.; Andersen, D.C.; Davis, R.H.; Kompala, D.S. Plasmid-encoded protein: The principal factor in the“metabolic burden”associated with recombinant bacteria. Biotechnol. Bioeng. 1990, 35, 668–681. [Google Scholar] [CrossRef]
- Kurland, C.; Dong, H. Bacterial growth inhibition by overproduction of protein. Mol. Microbiol. 1996, 21, 1–4. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 2005, 115, 113–128. [Google Scholar] [CrossRef]
- Helianti, I.; Ulfah, M.; Nurhayati, N.; Suhendar, D.; Finalissari, A.K.; Wardani, A.K. Production of xylanase by recombinant Bacillus subtilis DB104 cultivated in agroindustrial waste medium. HAYATI J. Biosci. 2016, 23, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Khamduang, M.; Packdibamrung, K.; Chutmanop, J.; Chisti, Y.; Srinophakun, P. Production of L-phenylalanine from glycerol by a recombinant Escherichia coli. J. Ind. Microbiol. Biotechnol. 2009, 36, 1267–1274. [Google Scholar] [CrossRef]
- Kumar, R.; Banoth, L.; Banerjee, U.C.; Kaur, J. Enantiomeric separation of pharmaceutically important drug intermediates using a Metagenomic lipase and optimization of its large scale production. Int. J. Biol. Macromol. 2017, 95, 995–1003. [Google Scholar] [CrossRef]
- Talabardon, M.; Yang, S.T. Production of GFP and glucoamylase by recombinant Aspergillus niger: Effects of fermentation conditions on fungal morphology and protein secretion. Biotechnol. Prog. 2005, 21, 1389–1400. [Google Scholar] [CrossRef]
- Villena, G.; Fujikawa, T.; Tsuyumu, S.; Gutiérrez-Correa, M. Structural analysis of biofilms and pellets of Aspergillus niger by confocal laser scanning microscopy and cryo scanning electron microscopy. Bioresour. Technol. 2010, 101, 1920–1926. [Google Scholar] [CrossRef]
- Lenz, A.P.; Williamson, K.S.; Pitts, B.; Stewart, P.S.; Franklin, M.J. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2008, 74, 4463–4471. [Google Scholar] [CrossRef] [Green Version]
- Werner, E.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Heydorn, A.; Molin, S.; Pitts, B.; Stewart, P.S. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2004, 70, 6188–6196. [Google Scholar] [CrossRef] [Green Version]
- Beyenal, H.; Lewandowski, Z. Internal and external mass transfer in biofilms grown at various flow velocities. Biotechnol. Prog. 2002, 18, 55–61. [Google Scholar] [CrossRef]
- Singha, T.K.; Gulati, P.; Mohanty, A.; Khasa, Y.P.; Kapoor, R.K.; Kumar, S. Efficient genetic approaches for improvement of plasmid based expression of recombinant protein in Escherichia coli: A review. Process Biochem. 2017, 55, 17–31. [Google Scholar] [CrossRef]
- Gomes, L.C.; Monteiro, G.A.; Mergulhão, F.J. The impact of IPTG induction on plasmid stability and heterologous protein expression by Escherichia coli biofilms. Int. J. Mol. Sci. 2020, 21, 576. [Google Scholar] [CrossRef] [Green Version]
- Diaz Ricci, J.C.; Hernández, M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000, 20, 79–108. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, A.; Gomes, L.C.; Monteiro, G.A.; Mergulhão, F.J. Hydrodynamic Effects on Biofilm Development and Recombinant Protein Expression. Microorganisms 2022, 10, 931. https://doi.org/10.3390/microorganisms10050931
Soares A, Gomes LC, Monteiro GA, Mergulhão FJ. Hydrodynamic Effects on Biofilm Development and Recombinant Protein Expression. Microorganisms. 2022; 10(5):931. https://doi.org/10.3390/microorganisms10050931
Chicago/Turabian StyleSoares, Alexandra, Luciana C. Gomes, Gabriel A. Monteiro, and Filipe J. Mergulhão. 2022. "Hydrodynamic Effects on Biofilm Development and Recombinant Protein Expression" Microorganisms 10, no. 5: 931. https://doi.org/10.3390/microorganisms10050931





