Comparative Evaluation of Three Immunoassays for the Simultaneous Detection of Clostridioides difficile Glutamate Dehydrogenase and Toxin A/B
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Three Immunoassays for the Detection of GDH Antigen and CD Toxin A/B
2.3. C. difficile Culture and Nucleic Acid Amplification Test for C. difficile Toxin A/B
2.4. Statistical Analysis
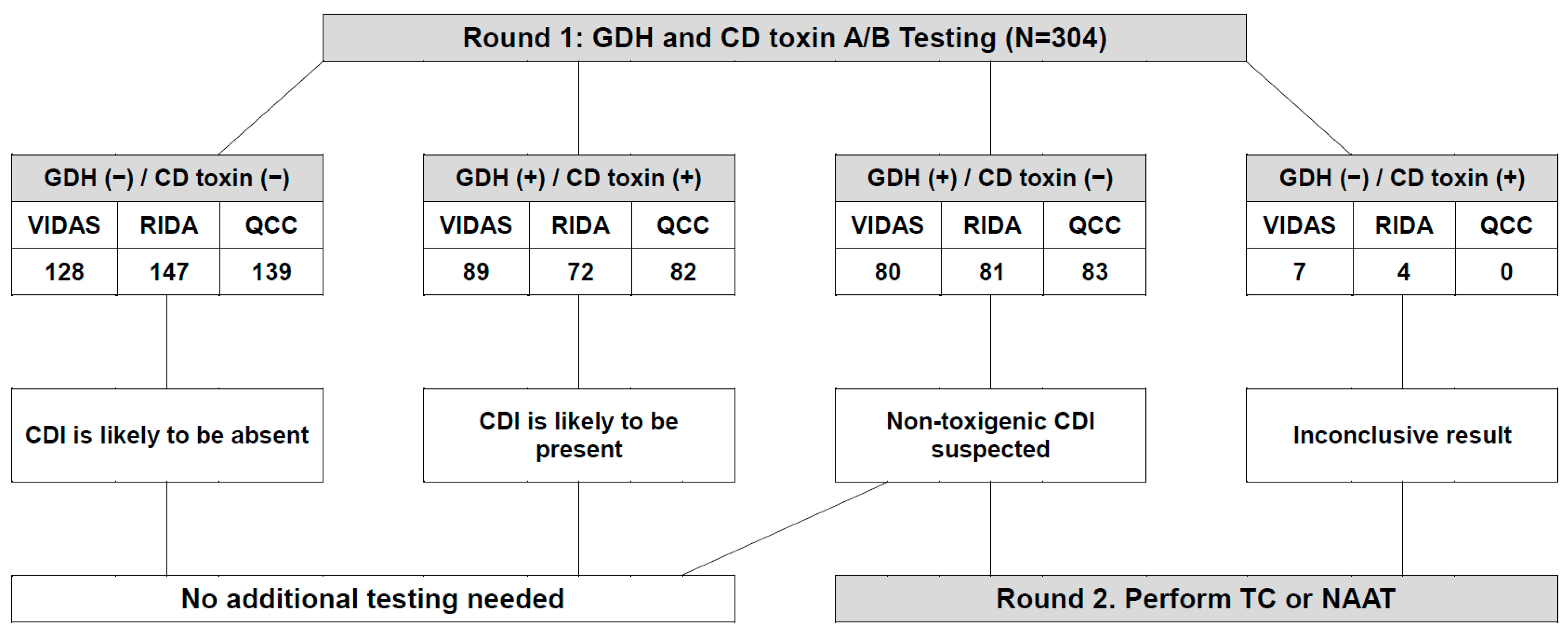
3. Results
3.1. Analytical Performance of the Three GDH Immunoassays as Compared to C. difficile Culture
3.2. Analytical Performance of the Three Toxin A/B Immunoassays as Compared to the Xpert C. difficile Assay
3.3. Analytical Performance of the Three GDH Immunoassays as Compared to the Consensus GDH Results
3.4. Analytical Performance of the Three Toxin A/B Immunoassays as Compared to the Consensus Toxin A/B Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kelly, C.P.; LaMont, J.T. Clostridium difficile-more difficult than ever. N. Engl. J. Med. 2008, 359, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Depestel, D.D.; Aronoff, D.M. Epidemiology of Clostridium difficile infection. J. Pharm. Pract. 2013, 26, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Vanpoucke, H.; De Baere, T.; Claeys, G.; Vaneechoutte, M.; Verschraegen, G. Evaluation of six commercial assays for the rapid detection of Clostridium difficile toxin and/or antigen in stool specimens. Clin. Microbiol. Infect. 2001, 7, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Kawada, M.; Annaka, M.; Kato, H.; Shibasaki, S.; Hikosaka, K.; Mizuno, H.; Masuda, Y.; Inamatsu, T. Evaluation of a simultaneous detection kit for the glutamate dehydrogenase antigen and toxin A/B in feces for diagnosis of Clostridium difficile infection. J. Infect. Chemother. 2011, 17, 807–811. [Google Scholar] [CrossRef]
- Kim, H.; Kim, W.H.; Kim, M.; Jeong, S.H.; Lee, K. Evaluation of a rapid membrane enzyme immunoassay for the simultaneous detection of glutamate dehydrogenase and toxin for the diagnosis of Clostridium difficile infection. Ann. Lab. Med. 2014, 34, 235–239. [Google Scholar] [CrossRef]
- Yoo, I.Y.; Song, D.J.; Huh, H.J.; Lee, N.Y. Simultaneous Detection of Clostridioides difficile Glutamate Dehydrogenase and Toxin A/B: Comparison of the C. DIFF QUIK CHEK COMPLETE and RIDASCREEN Assays. Ann. Lab. Med. 2019, 39, 214–217. [Google Scholar] [CrossRef]
- Schmidt, M.L.; Gilligan, P.H. Clostridium difficile testing algorithms: What is practical and feasible? Anaerobe 2009, 15, 270–273. [Google Scholar] [CrossRef]
- Aghajanian, S.; Hovsepyan, M.; Geoghegan, K.F.; Chrunyk, B.A.; Engel, P.C. A thermally sensitive loop in clostridial glutamate dehydrogenase detected by limited proteolysis. J. Biol. Chem. 2003, 278, 1067–1074. [Google Scholar] [CrossRef]
- Wren, M.W.; Kinson, R.; Sivapalan, M.; Shemko, M.; Shetty, N.R. Detection of Clostridium difficile infection: A suggested laboratory diagnostic algorithm. Br. J. Biomed. Sci. 2009, 66, 175–179. [Google Scholar] [CrossRef]
- Kvach, E.J.; Ferguson, D.; Riska, P.F.; Landry, M.L. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J. Clin. Microbiol. 2010, 48, 109–114. [Google Scholar] [CrossRef]
- Planche, T.; Wilcox, M. Reference assays for Clostridium difficile infection: One or two gold standards? J. Clin. Pathol. 2011, 64, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Fenner, L.; Widmer, A.F.; Goy, G.; Rudin, S.; Frei, R. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J. Clin. Microbiol. 2008, 46, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.; Planche, T.; Eckert, C.; Barbut, F.; Terveer, E.M.; Dekkers, O.M.; Wilcox, M.H.; Kuijper, E.J. European Society of Clinical Microbiology and Infectious Diseases: Update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2016, 22 (Suppl. 4), S63–S81. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Ki, C.S.; Lee, N.Y. Isolation and Identification of Clostridium difficile Using ChromID C. difficile Medium Combined with Gram Staining and PRO Disc Testing: A Proposal for a Simple Culture Process. Ann. Lab. Med. 2015, 35, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Lessa, F.C.; Gould, C.V.; McDonald, L.C. Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 2012, 55 (Suppl. 2), S65–S70. [Google Scholar] [CrossRef]
- Collins, D.A.; Sohn, K.M.; Wu, Y.; Ouchi, K.; Ishii, Y.; Elliott, B.; Riley, T.V.; Tateda, K. Clostridioides difficile infection in the Asia-Pacific region. Emerg. Microbes. Infect. 2020, 9, 42–52. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Testelin, S.; Mattrat, M.A.; Maillard, K.; Lemeland, J.F.; Pons, J.L. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 2004, 42, 5710–5714. [Google Scholar] [CrossRef]
- Paitan, Y.; Miller-Roll, T.; Adler, A. Comparative performance study of six commercial molecular assays for rapid detection of toxigenic Clostridium difficile. Clin. Microbiol. Infect. 2017, 23, 567–572. [Google Scholar] [CrossRef]
- Lai, H.; Huang, C.; Cai, J.; Ye, J.; She, J.; Zheng, Y.; Wang, L.; Wei, Y.; Fang, W.; Wang, X.; et al. Simultaneous detection and characterization of toxigenic Clostridium difficile directly from clinical stool specimens. Front. Med. 2018, 12, 196–205. [Google Scholar] [CrossRef]
- van Rossen, T.M.; van Prehn, J.; Koek, A.; Jonges, M.; van Houdt, R.; van Mansfeld, R.; Kuijper, E.J.; Vandenbroucke-Grauls, C.; Budding, A.E. Simultaneous detection and ribotyping of Clostridioides difficile, and toxin gene detection directly on fecal samples. Antimicrob. Resist. Infect. Control 2021, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Cançado, G.G.L.; Silva, R.O.S.; Nader, A.P.; Lobato, F.C.F.; Vilela, E.G. Impact of simultaneous glutamate dehydrogenase and toxin A/B rapid immunoassay on Clostridium difficile diagnosis and treatment in hospitalized patients with antibiotic-associated diarrhea in a university hospital of Brazil. J. Gastroenterol. Hepatol. 2018, 33, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Polage, C.R.; Gyorke, C.E.; Kennedy, M.A.; Leslie, J.L.; Chin, D.L.; Wang, S.; Nguyen, H.H.; Huang, B.; Tang, Y.W.; Lee, L.W.; et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern. Med. 2015, 175, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- Dubberke, E.R.; Han, Z.; Bobo, L.; Hink, T.; Lawrence, B.; Copper, S.; Hoppe-Bauer, J.; Burnham, C.A.; Dunne, W.M., Jr. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J. Clin. Microbiol. 2011, 49, 2887–2893. [Google Scholar] [CrossRef]
- Cohen, S.H.; Gerding, D.N.; Johnson, S.; Kelly, C.P.; Loo, V.G.; McDonald, L.C.; Pepin, J.; Wilcox, M.H. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect. Control. Hosp. Epidemiol. 2010, 31, 431–455. [Google Scholar] [CrossRef]
- Swindells, J.; Brenwald, N.; Reading, N.; Oppenheim, B. Evaluation of diagnostic tests for Clostridium difficile infection. J. Clin. Microbiol. 2010, 48, 606–608. [Google Scholar] [CrossRef]
- Ota, K.V.; McGowan, K.L. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J. Clin. Microbiol. 2012, 50, 1185–1188. [Google Scholar] [CrossRef]
- Eckert, C.; Burghoffer, B.; Lalande, V.; Barbut, F. Evaluation of the chromogenic agar chromID C. difficile. J. Clin. Microbiol. 2013, 51, 1002–1004. [Google Scholar] [CrossRef]
- Han, S.B.; Chang, J.; Shin, S.H.; Park, K.G.; Lee, G.D.; Park, Y.G.; Park, Y.J. Performance of chromID Clostridium difficile agar compared with BBL C. difficile selective agar for detection of C. difficile in stool specimens. Ann. Lab. Med. 2014, 34, 376–379. [Google Scholar] [CrossRef][Green Version]
- Toltzis, P.; Nerandzic, M.M.; Saade, E.; O’Riordan, M.A.; Smathers, S.; Zaoutis, T.; Kim, J.; Donskey, C.J. High proportion of false-positive Clostridium difficile enzyme immunoassays for toxin A and B in pediatric patients. Infect. Control Hosp. Epidemiol. 2012, 33, 175–179. [Google Scholar] [CrossRef]
| Characteristic | VIDAS C. difficile GDH and Toxin A&B | RIDASCREEN C. difficile GDH and Toxin A/B | C. DIFF QUIK CHEK COMPLETE |
|---|---|---|---|
| Manufacturer | bioMérieux | R-Biopharm | TechLab |
| Immunoassay system | VIDAS | GEMINI | Not required |
| Principle of operation | Enzyme-linked fluorescent assay | Enzyme-linked immunosorbent assay | Lateral flow membrane enzyme assay |
| Testing concurrency * | Separately | Separately | Simultaneously |
| Sample type | Stool | Stool | Stool |
| Minimum sample volume | 200 μL | 100 μL | 25 μL |
| Running time of process | 90 min | 120 min | 30 min |
| Immunoassay | Result | C. difficile Culture | Kappa (95% CI) | Sensitivity, % (95% CI) | Specificity, % (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|
| Positive (n = 124) | Negative (n = 180) | %Diff (95% CI) | |||||
| VIDAS GDH | Positive | 120 | 49 | 0.66 (0.58–0.74) | 96.8 (92.0–99.1) | 72.8 (65.7–79.1) | <0.0001 |
| Negative | 4 | 131 | −14.52 (−18.89 to −10.15) | ||||
| RIDA GDH | Positive | 111 | 42 | 0.64 (0.55–0.72) | 89.5 (82.7–94.3) | 76.7 (69.8–82.6) | 0.0001 |
| Negative | 13 | 138 | −9.54 (−14.2 to −4.88) | ||||
| QCC GDH | Positive | 118 | 47 | 0.66 (0.58–0.74) | 95.2 (89.8–98.2) | 73.9 (66.8–80.1) | <0.0001 |
| Negative | 6 | 133 | −13.49 (−17.93 to −9.04) | ||||
| Immunoassay | Result | Xpert C. difficile Assay | Kappa (95% CI) | Sensitivity, % (95% CI) | Specificity, % (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|
| GDH | %Diff (95% CI) | ||||||
| Positive (n = 152) | Negative (n = 152) | ||||||
| VIDAS GDH | Positive | 143 | 26 | 0.77 (0.70–0.84) | 94.1 (89.1–97.3) | 82.9 (76.0–88.5) | 0.0060 |
| Negative | 9 | 126 | −5.59 (−9.35 to −1.83) | ||||
| RIDA GDH | Positive | 139 | 14 | 0.82 (0.76–0.87) | 91.5 (85.8–95.4) | 90.8 (85.0–94.9) | 1.0000 |
| Negative | 13 | 138 | −0.33 (−3.68 to 3.02) | ||||
| QCC GDH | Positive | 141 | 24 | 0.77 (0.70–0.84) | 92.8 (87.4–96.3) | 84.2 (77.4–89.6) | 0.0410 |
| Negative | 11 | 128 | −4.28 (−8.06 to −0.49) | ||||
| CD toxin A/B | |||||||
| Positive (n = 152) | Negative (n = 152) | ||||||
| VIDAS CDAB | Positive | 86 | 10 | 0.5 (0.41–0.59) | 56.6 (48.3–64.6) | 93.4 (88.2–96.8) | <0.0001 |
| Negative | 66 | 142 | 18.42 (13.2 to 23.65) | ||||
| RIDA toxin A/B | Positive | 76 | 0 | 0.5 (0.42–0.58) | 50.0 (41.8–58.2) | 100 (97.6–100) | <0.0001 |
| Negative | 76 | 152 | 25 (20.13 to 29.87) | ||||
| QCC toxin A/B | Positive | 78 | 4 | 0.49 (0.40–0.57) | 51.3 (43.1–59.5) | 97.4 (93.4–99.3) | <0.0001 |
| Negative | 74 | 148 | 23.03 (17.95 to 28.1) | ||||
| Immunoassay | Result | Consensus Result | Kappa (95% CI) | PPA, % (95% CI) | NPA, % (95% CI) | p-Value | |
|---|---|---|---|---|---|---|---|
| GDH | %Diff (95% CI) | ||||||
| Positive (n = 161) | Negative (n = 143) | ||||||
| VIDAS GDH | Positive | 160 | 9 | 0.94 (0.89–0.97) | 99.4 (96.6–100) | 93.7 (88.4–97.1) | 0.0215 |
| Negative | 1 | 134 | −2.63 (−4.65 to −0.61) | ||||
| RIDA GDH | Positive | 142 | 11 | 0.80 (0.94–0.87) | 88.2 (82.2–92.7) | 92.3 (86.7–96.1) | 0.2005 |
| Negative | 19 | 132 | 2.63 (0.89 to 6.12) | ||||
| QCC GDH | Positive | 159 | 6 | 0.95 (0.91–0.98) | 98.7 (95.6–100) | 95.8 (91.1–98.4) | 0.2891 |
| Negative | 2 | 137 | −1.32 (−3.13 to 0.5) | ||||
| CD toxin A/B | |||||||
| Positive (n = 86) | Negative (n = 218) | ||||||
| VIDAS CDAB | Positive | 83 | 13 | 0.88 (0.82–0.94) | 96.5 (90.1–99.3) | 94.0 (90.0–96.8) | 0.0213 |
| Negative | 3 | 205 | −3.29 (−5.84 to −0.74) | ||||
| RIDA toxin A/B | Positive | 72 | 4 | 0.85 (0.78–0.92) | 83.7 (74.2–90.8) | 98.2 (95.4–99.5) | 0.0309 |
| Negative | 14 | 214 | 3.29 (0.58 to 6) | ||||
| QCC toxin A/B | Positive | 79 | 3 | 0.92 (0.87–0.97) | 91.9 (84.0–96.7) | 98.6 (96.0–99.7) | 0.3438 |
| Negative | 7 | 215 | 1.32 (−0.72 to 3.35) | ||||
| Case (N = 39) | CD Culture | GDH | Xpert Assay | CD Toxin A/B | ||||
|---|---|---|---|---|---|---|---|---|
| VIDAS | RIDA | QCC | VIDAS | RIDA | QCC | |||
| A (N = 5) | Neg | Pos | Pos | Pos | Pos | Pos | Pos | Pos |
| B (N = 1) | Neg | Pos | Pos | Pos | Pos | Pos | Pos | Neg |
| C (N = 2) | Neg | Pos | Pos | Pos | Pos | Pos | Neg | Pos |
| D (N = 1) | Neg | Pos | Pos | Pos | Pos | Neg | Pos | Pos |
| E (N = 1) | Neg | Pos | Pos | Pos | Pos | Neg | Pos | Neg |
| F (N = 17) | Neg | Pos | Pos | Pos | Pos | Neg | Neg | Neg |
| G (N = 3) | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg |
| H (N = 5) | Neg | Neg | Pos | Neg | Pos | Neg | Neg | Neg |
| I (N = 1) | Neg | Neg | Neg | Neg | Pos | Neg | Neg | Neg |
| J (N = 3) | Neg | Pos | Pos | Pos | Neg | Neg | Neg | Neg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Lee, S.Y.; Park, J.; Lee, J. Comparative Evaluation of Three Immunoassays for the Simultaneous Detection of Clostridioides difficile Glutamate Dehydrogenase and Toxin A/B. Microorganisms 2022, 10, 947. https://doi.org/10.3390/microorganisms10050947
Kim N, Lee SY, Park J, Lee J. Comparative Evaluation of Three Immunoassays for the Simultaneous Detection of Clostridioides difficile Glutamate Dehydrogenase and Toxin A/B. Microorganisms. 2022; 10(5):947. https://doi.org/10.3390/microorganisms10050947
Chicago/Turabian StyleKim, Namsu, Seung Yeob Lee, Joonhong Park, and Jaehyeon Lee. 2022. "Comparative Evaluation of Three Immunoassays for the Simultaneous Detection of Clostridioides difficile Glutamate Dehydrogenase and Toxin A/B" Microorganisms 10, no. 5: 947. https://doi.org/10.3390/microorganisms10050947
APA StyleKim, N., Lee, S. Y., Park, J., & Lee, J. (2022). Comparative Evaluation of Three Immunoassays for the Simultaneous Detection of Clostridioides difficile Glutamate Dehydrogenase and Toxin A/B. Microorganisms, 10(5), 947. https://doi.org/10.3390/microorganisms10050947






