The Application of Mixed Organic and Inorganic Fertilizers Drives Soil Nutrient and Bacterial Community Changes in Teak Plantations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Experimental Design
2.3. Soil Sampling and Chemical Analysis
2.4. DNA Extraction and PCR Amplification
2.5. Illumina NovaSeq Sequencing and Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Changes in Soil Chemical Properties in Teak Plantations
3.2. Sequence Characteristics and Soil Bacterial Community Diversity
3.3. Impact of Fertilization on Bacterial Community Structure
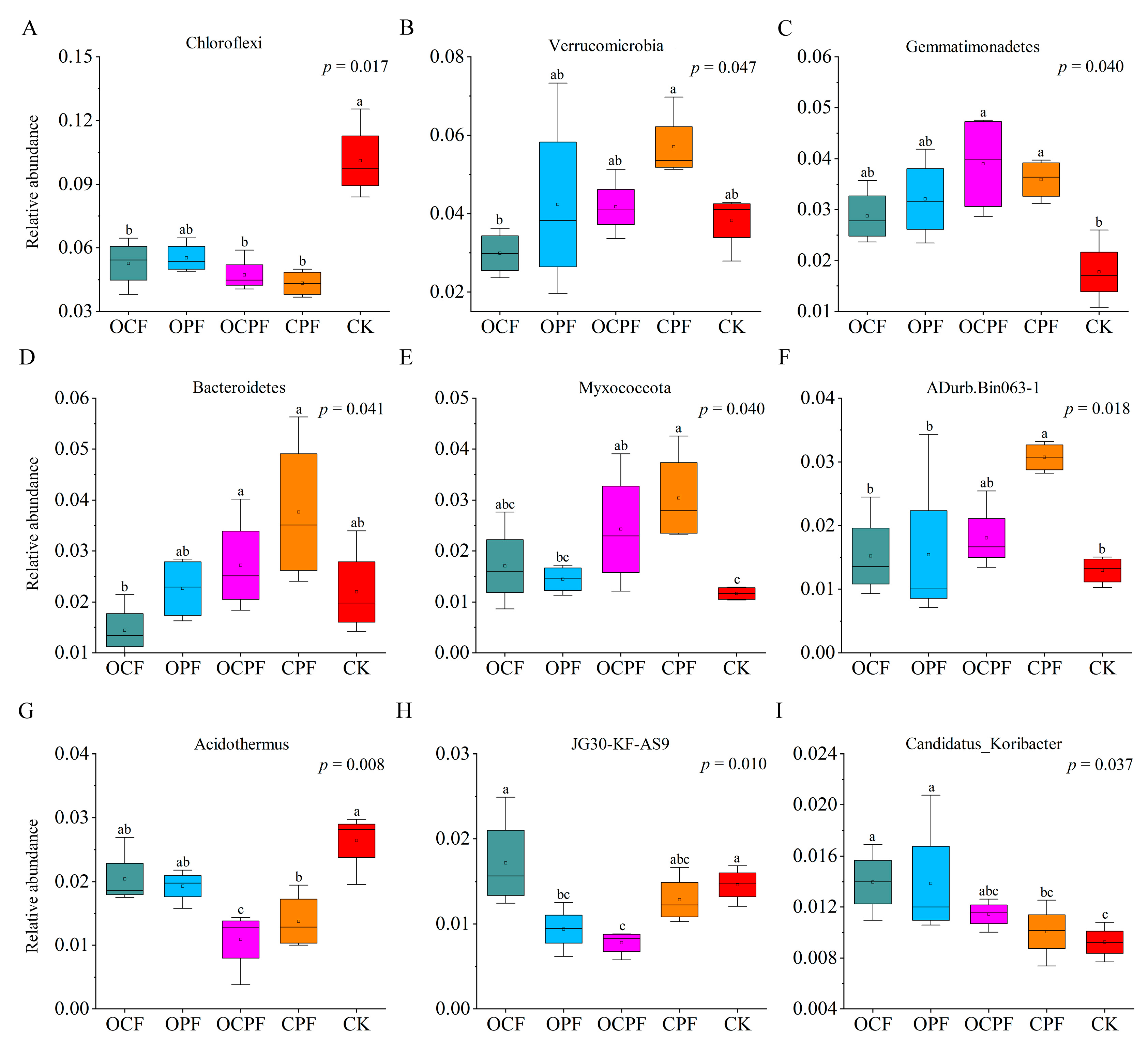
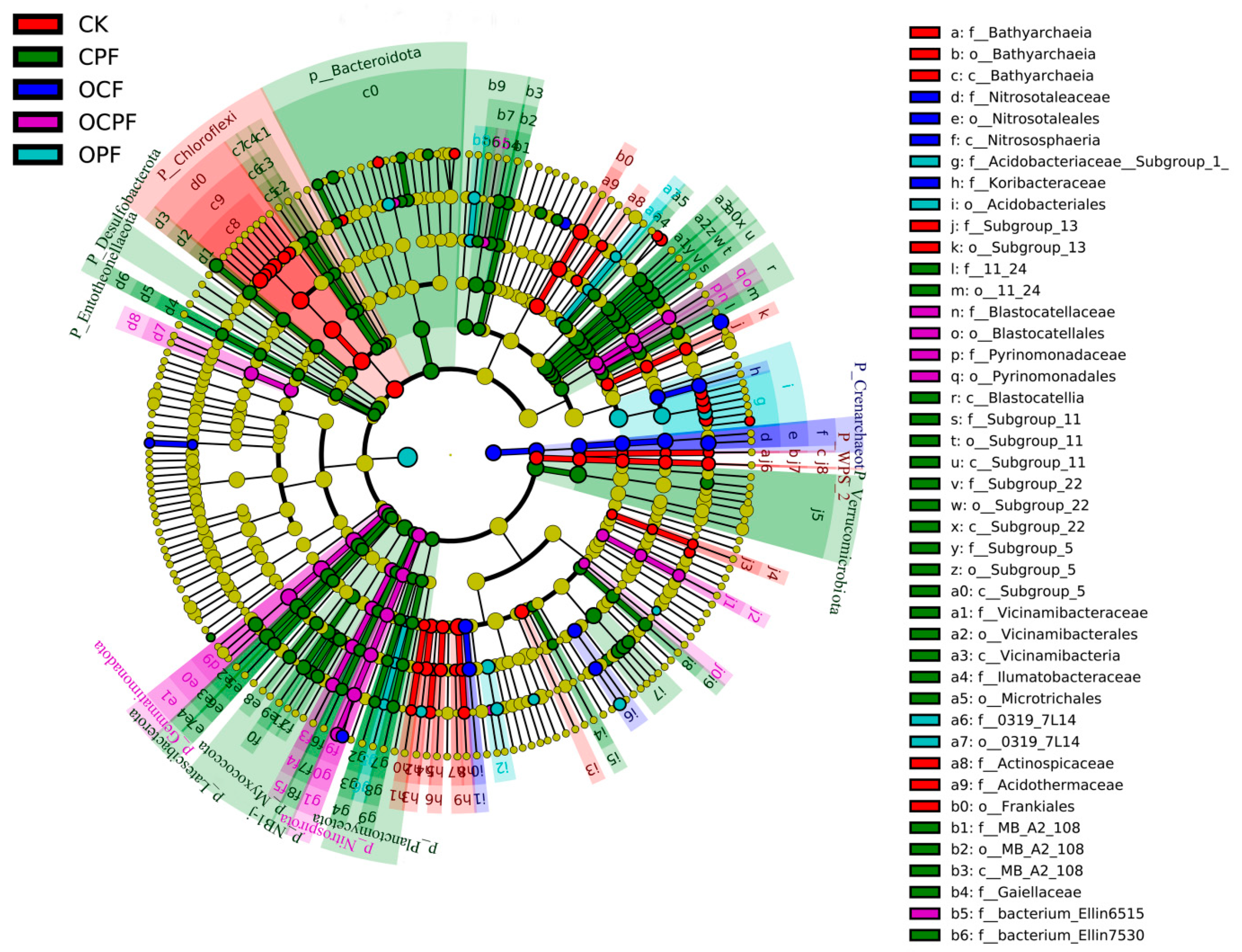
3.4. Biomarker Taxa of Soil Bacterial Communities
3.5. Co-Occurrence Network of Bacterial Communities
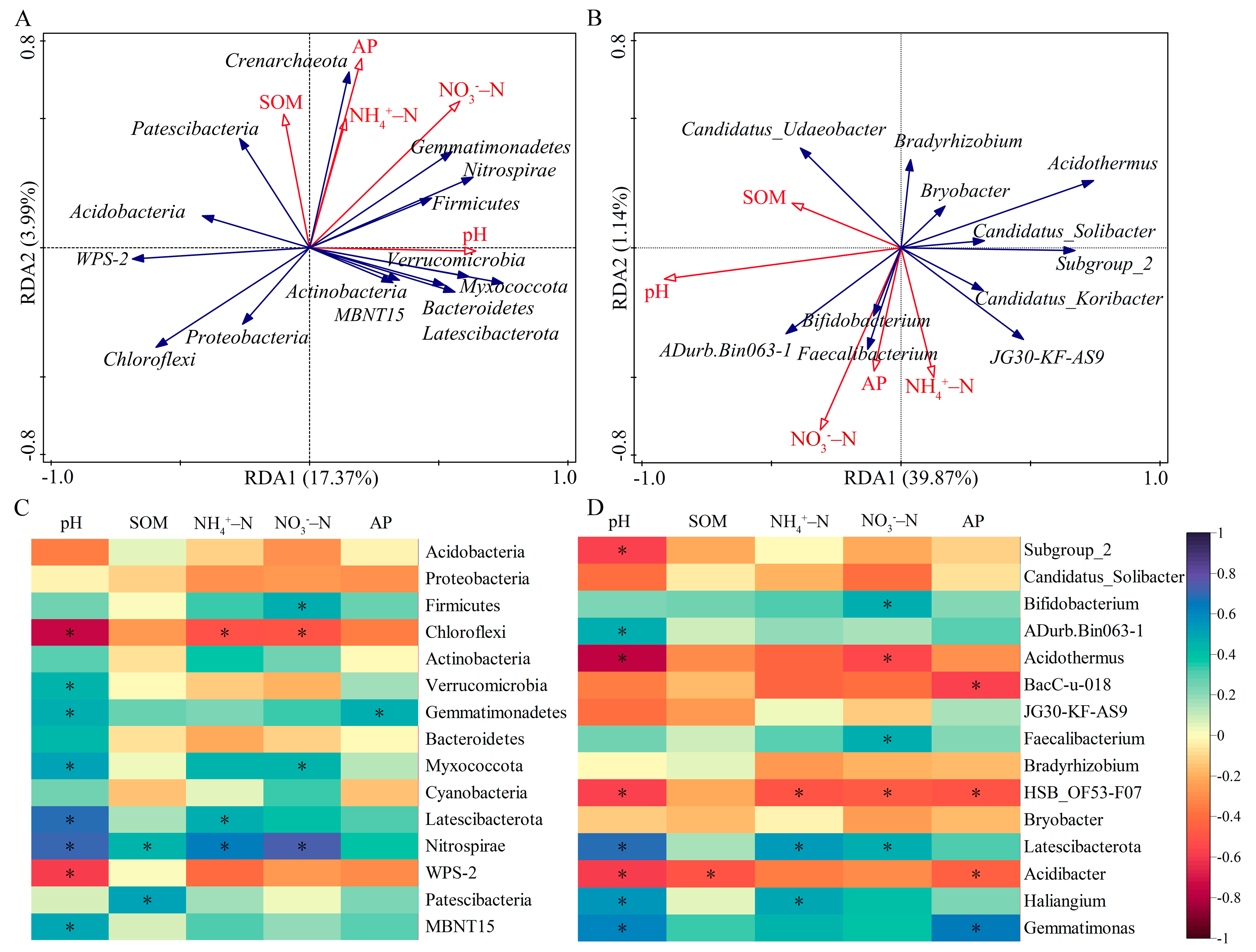
3.6. Relationships between Soil Factors and Bacterial Community Structure
4. Discussion
4.1. Variations in Soil Chemical Properties under Different Fertilization Treatments
4.2. Response of Bacterial Community to Soil Chemical Properties
4.3. Effects of Fertilization on Soil Bacterial Community Composition and Structure
4.4. Network Patterns of Soil Bacterial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eo, J.; Park, K.-C. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Han, S.; Delgado-Baquerizo, M.; Luo, X.; Liu, Y.; Van Nostrand, J.D.; Chen, W.; Zhou, J.; Huang, Q. Soil aggregate size-dependent relationships between microbial functional diversity and multifunctionality. Soil Biol. Biochem. 2021, 154, 108143. [Google Scholar] [CrossRef]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant Acidification in Major Chinese Croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.-C.; Chang, Y.-Y.; Hussain, M.; Lu, B.; Zhang, J.-P.; Song, X.-B.; Lei, X.-S.; Pei, D. Soil Chemical and Microbiological Properties Are Changed by Long-Term Chemical Fertilizers That Limit Ecosystem Functioning. Microorganisms 2020, 8, 694. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105 (Suppl. 1), 11512–11519. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, X.; Song, L.; Lin, X.; Zhang, H.; Shen, C.; Chu, H. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 2016, 92, 41–49. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, X.; Wei, D.; Zhao, B.S.; Ma, M.C.; Chen, S.F.; Cao, F.M.; Shen, D.L.; Guan, D.W.; Li, J. Consistent effects of nitrogen fertilization on soil bacterial communities in black soils for two crop seasons in China. Sci. Rep. 2017, 7, 3267. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, Y.; Ma, T.; Raza, W.; Li, J.; Howland, J.G.; Huang, Q.; Shen, Q. Impacts of inorganic and organic fertilization treatments on bacterial and fungal communities in a paddy soil. Appl. Soil Ecol. 2017, 112, 42–50. [Google Scholar] [CrossRef]
- Li, R.; Tao, R.; Ling, N.; Chu, G. Chemical, organic and bio-fertilizer management practices effect on soil physicochemical property and antagonistic bacteria abundance of a cotton field: Implications for soil biological quality. Soil Tillage Res. 2017, 167, 30–38. [Google Scholar] [CrossRef]
- Sihi, D.; Dari, B.; Sharma, D.K.; Pathak, H.; Nain, L.; Sharma, O.P. Evaluation of soil health in organic vs. conventional farming of basmati rice in North India. J. Plant Nutr. Soil Sci. 2017, 180, 389–406. [Google Scholar] [CrossRef]
- Sheoran, H.S.; Kakar, R.; Kumar, N. Seema impact of organic and conventional farming practices on soil quality: A global review. Appl. Ecol. Environ. Res. 2019, 17, 951–968. [Google Scholar] [CrossRef]
- Muhammad, Q.; Huang, J.; Waqas, A.; Li, D.C.; Liu, S.J.; Zhang, L.; Cai, A.D.; Liu, L.S.; Xu, Y.M.; Gao, J.S.; et al. Yield sustainability, soil organic carbon sequestration and nutrients balance under long-term combined application of manure and inorganic fertilizers in acidic paddy soil. Soil Till. Res. 2020, 198, 1–6. [Google Scholar]
- Wei, M.; Hu, G.; Wang, H.; Bai, E.; Lou, Y.; Zhang, A.; Zhuge, Y. 35 years of manure and chemical fertilizer application alters soil microbial community composition in a Fluvo-aquic soil in Northern China. Eur. J. Soil Biol. 2017, 82, 27–34. [Google Scholar] [CrossRef]
- Mei, N.; Zhang, X.; Wang, X.; Peng, C.; Gao, H.; Zhu, P.; Gu, Y. Effects of 40 years applications of inorganic and organic fertilization on soil bacterial community in a maize agroecosystem in northeast China. Eur. J. Agron. 2021, 130, 126332. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The Microbial Engines That Drive Earth’s Biogeochemical Cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Z.; Huang, L.; Qi, Q.; Liu, L.; Zhao, Y.; Wang, Z.; Zhou, H.; Lv, X.; Mao, Z.; et al. Shifts in soil microbial community functional gene structure across a 61-year desert revegetation chronosequence. Geoderma 2019, 347, 126–134. [Google Scholar] [CrossRef]
- Grzyb, A.; Wolna-Maruwka, A.; Niewiadomska, A. Environmental Factors Affecting the Mineralization of Crop Residues. Agronomy 2020, 10, 1951. [Google Scholar] [CrossRef]
- Wu, H.; Cai, A.; Xing, T.; Huai, S.; Zhu, P.; Xu, M.; Lu, C. Fertilization enhances mineralization of soil carbon and nitrogen pools by regulating the bacterial community and biomass. J. Soils Sediments 2021, 21, 1633–1643. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, X.; Guan, D.; Wei, D.; Zhao, B.; Ma, M.; Chen, S.; Li, L.; Cao, F.; Li, J. Long-term fertilization changes bacterial diversity and bacterial communities in the maize rhizosphere of Chinese Mollisols. Appl. Soil Ecol. 2018, 125, 88–96. [Google Scholar] [CrossRef]
- Bissett, A.; Richardson, A.E.; Baker, G.; Thrall, P.H. Long-term land use effects on soil microbial community structure and function. Appl. Soil Ecol. 2011, 51, 66–78. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.; Wittwer, R.A.; Banerjee, S.; Walser, J.C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 1–14. [Google Scholar]
- Wu, L.P.; Wang, Y.D.; Zhang, S.R.; Wei, W.L.; Kuzyakov, Y.; Ding, X.D. Fertilization effects on microbial community compo-sition and aggregate formation in saline-alkaline soil. Plant Soil 2021, 463, 523–535. [Google Scholar] [CrossRef]
- Li, X.; Jiao, X.; Wang, H.; Wang, G. Organic-inorganic combined fertilization alters reclaimed soil bacterial communities in an opencast coal mine area and improves soil quality. Arab. J. Geosci. 2021, 14, 1–13. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Huang, S.; Sha, C.; Wu, J.; Cui, C.; Su, J.; Ruan, J.; Tan, J.; Tang, H.; Xue, J. Changes of bacterial community in arable soil after short-term application of fresh manures and organic fertilizer. Environ. Technol. 2022, 43, 824–834. [Google Scholar] [CrossRef]
- Tao, R.; Liang, Y.; Wakelin, S.; Chu, G. Supplementing chemical fertilizer with an organic component increases soil biological function and quality. Appl. Soil Ecol. 2015, 96, 42–51. [Google Scholar] [CrossRef]
- Wolna-Maruwka, A.; Piechota, T.; Niewiadomska, A.; Kamiński, A.; Kayzer, D.; Grzyb, A.; Pilarska, A.A. The effect of bio-char-based organic amendments on the structure of soil bacterial community and yield of maize (Zea mays L.). Agronomy 2021, 11, 1286. [Google Scholar] [CrossRef]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; Van Der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Ma, B.; Lv, X.; Cai, Y.; Chang, S.X.; Dyck, M. Liming does not counteract the influence of long-term fertilization on soil bacterial community structure and its co-occurrence pattern. Soil Biol. Biochem. 2018, 123, 45–53. [Google Scholar] [CrossRef]
- Yao, T.; Chen, R.; Zhang, J.; Feng, Y.; Huang, M.; Lin, X. Divergent patterns of microbial community composition shift under two fertilization regimes revealed by responding species. Appl. Soil Ecol. 2020, 154, 103590. [Google Scholar] [CrossRef]
- Xu, Y.; Li, C.; Zhu, Y.; Wang, Z.; Zhu, W.; Wu, L.; Du, A. The shifts in soil microbial community and association network induced by successive planting of Eucalyptus plantations. For. Ecol. Manag. 2022, 505, 119877. [Google Scholar] [CrossRef]
- Muneer, M.A.; Hou, W.; Li, J.; Huang, X.; Kayani, M.U.R.; Cai, Y.; Yang, W.; Wu, L.; Ji, B.; Zheng, C. Soil pH: A key edaphic factor regulating distribution and functions of bacterial community along vertical soil profiles in red soil of pomelo orchard. BMC Microbiol. 2022, 22, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, H.; He, X.; Thomas, B.; Lupwayi, N.; Hao, X.; Thomas, M.C.; Shi, X. Fertilization Shapes Bacterial Community Structure by Alteration of Soil pH. Front. Microbiol. 2017, 8, 1325. [Google Scholar] [CrossRef] [PubMed]
- Brennan, E.B.; Acosta-Martinez, V. Cover cropping frequency is the main driver of soil microbial changes during six years of organic vegetable production. Soil Biol. Biochem. 2017, 109, 188–204. [Google Scholar] [CrossRef]
- Li, W.X.; Zhang, F.Y.; Cui, G.H.; Wang, Y.N.; Yang, J.G.; Cheng, H.C.; Liu, H.W.; Zhang, L.P. Effects of bio-organic fertilizer on soil fertility microbial community composition, and potato growth. Sci. Asia 2021, 47, 347–356. [Google Scholar] [CrossRef]
- Huang, G.; Liang, K.; Zhou, Z.; Yang, G.; Muralidharan, E.M. Variation in Photosynthetic Traits and Correlation with Growth in Teak (Tectona grandis Linn.) Clones. Forests 2019, 10, 44. [Google Scholar] [CrossRef]
- Masilamani, P.; Albert, V.A.; Venkatesan, S.; Janaki, P. Effect of phosphorus application on growth and stump quality of teak (Tectona grandis L.f.) seedlings. Madras Agric. J. 2020, 107, 1–9. [Google Scholar]
- Balam-Che, M.; Gomez-Guerrero, A.; Vargas-Hernández, J.V.; Aldrete, A.; Obrador-Olán, J.J. Initial fertilization in commercial plantations of teak (Tectona grandis L.f.) in southeast México. Rev. Fitotec. Mex. 2015, 38, 205–212. [Google Scholar]
- Wehr, J.B.; Blamey, F.P.C.; Smith, T.E.; Menzies, N.W. Growth and physiological responses of teak (Tectona grandis L.f.) clones to Ca, H and Al stress in solution and acid soils. New For. 2017, 48, 137–152. [Google Scholar] [CrossRef]
- Wiratama, A. The Fertilization Giving Effect Basic Plant Growth of Teak (Tectona grandis) Age to 6 Months in the Village Um-Bulmartani, Ngemplak District, Sleman Regeny; Universitas Cadjah Mada: Yogyakarta, Indonesia, 2015. [Google Scholar]
- Dong, W.-Y.; Zhang, X.-Y.; Dai, X.-Q.; Fu, X.-L.; Yang, F.-T.; Liu, X.-Y.; Sun, X.-M.; Wen, X.-F.; Schaeffer, S. Changes in soil microbial community composition in response to fertilization of paddy soils in subtropical China. Appl. Soil Ecol. 2014, 84, 140–147. [Google Scholar] [CrossRef]
- Wu, L.; Li, Z.; Li, J.; Khan, M.A.; Huang, W.; Zhang, Z.; Lin, W. Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture. Appl. Soil Ecol. 2013, 67, 1–9. [Google Scholar] [CrossRef]
- Reijonen, I.; Metzler, M.; Hartikainen, H. Impact of soil pH and organic matter on the chemical bioavailability of vanadium species: The underlying basis for risk assessment. Environ. Pollut. 2016, 210, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, Y.; Kong, L.; Tong, L.; Cao, H.; Zhou, H.; Lv, Y. Long-Term Application of Bio-Compost Increased Soil Microbial Community Diversity and Altered Its Composition and Network. Microorganisms 2022, 10, 462. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Li, M.J.; Shao, D.T.; Zhou, J.C.; Gu, J.H.; Qin, J.J.; Chen, W.; Wei, W.Q. Signatures within esophageal microbiota with pro-gression of esophageal squamous cell carcinoma. Chin. J. Cancer. Res. 2020, 32, 755–767. [Google Scholar] [CrossRef]
- Li, S.; Wu, F. Diversity and Co-occurrence Patterns of Soil Bacterial and Fungal Communities in Seven Intercropping Systems. Front. Microbiol. 2018, 9, 1521. [Google Scholar] [CrossRef]
- Yu, Z.; Liang, K.; Huang, G.; Wang, X.; Lin, M.; Chen, Y.; Zhou, Z. Soil Bacterial Community Shifts Are Driven by Soil Nutrient Availability along a Teak Plantation Chronosequence in Tropical Forests in China. Biology 2021, 10, 1329. [Google Scholar] [CrossRef]
- Fernández-Moya, J.; Alvarado, A.; Mata, R.; Thiele, H.; Segura, J.M.; Vaides, E.; Miguel-Ayanz, A.S.; Marchamalo-Sacristán, M. Soil fertility characterisation of teak (Tectona grandis L.f.) plantations in Central America. Soil Res. 2015, 53, 423–432. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, S.; Liang, K.; Ma, H.; Huang, G. Growth and mineral nutrient analysis of teak (Tectona grandis) grown on acidic soils in south China. J. For. Res. 2016, 28, 503–511. [Google Scholar] [CrossRef]
- Mkhonza, N.P.; Buthelezi-Dube, N.N.; Muchaonyerwa, P. Effects of lime application on nitrogen and phosphorus availability in humic soils. Sci. Rep. 2020, 10, 8634. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liu, J.; Wei, D.; Zhu, P.; Cui, X.; Zhou, B.; Chen, X.; Jin, J.; Liu, X.; Wang, G. Soil Bacterial Communities Under Different Long-Term Fertilization Regimes in Three Locations Across the Black Soil Region of Northeast China. Pedosphere 2018, 28, 751–763. [Google Scholar] [CrossRef]
- Muhammad, Q.; Huang, J.; Waqas, A.; Muhammad, A.; Li, D.C.; Zulqarnain, H.K.; Gao, J.S.; Liu, S.J.; Zhang, H.M. Linkages between ecoenzymatic stoichiometry and microbial community structure under long-term fertilization in paddy soil: A case study in China. Appl. Soil Ecol. 2021, 161, 103860. [Google Scholar]
- Frey, S.; Six, J.; Elliott, E. Reciprocal transfer of carbon and nitrogen by decomposer fungi at the soil–litter interface. Soil Biol. Biochem. 2003, 35, 1001–1004. [Google Scholar] [CrossRef]
- Terrazas, R.A.; Giles, C.; Paterson, E.; Robertson-Albertyn, S.; Cesco, S.; Mimmo, T.; Pii, Y.; Bulgarelli, D. Chapter one-plant-microbiota interactions as a driver of the mineral turnover in the rhizosphere. Adv. Appl. Microbiol. 2016, 95, 1–67. [Google Scholar]
- Mujakić, I.; Piwosz, K.; Koblek, M. Phylum Gemmatimonadetes and its role in the environment. Microorganisms 2022, 10, 151. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Strauss, S.L.; Gonzálea- López, J.; Bedmar, E.J. Changes in the diversity and predicted functional composition of the bulk and rhizosphere soil bacterial microbiomes of tomoto and common bean after inorganic N-fertilization. Rhizosphere 2021, 18, 100362. [Google Scholar] [CrossRef]
- Beales, N. Adaptation of Microorganisms to Cold Temperatures, Weak Acid Preservatives, Low pH, and Osmotic Stress: A Review. Compr. Rev. Food Sci. Food Saf. 2004, 3, 1–20. [Google Scholar] [CrossRef]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting Soil pH Effects on Fungal and Bacterial Growth Suggest Functional Redundancy in Carbon Mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef]
- Marcote, I.; Hernández, T.; García, C.; Polo, A. Influence of one or two successive annual applications of organic fertilisers on the enzyme activity of a soil under barley cultivation. Bioresour. Technol. 2001, 79, 147–154. [Google Scholar] [CrossRef]
- Dahlawi, S.; Naeem, A.; Rengel, Z.; Naidu, R. Biochar application for the remediation of salt-affected soils: Challenges and opportunities. Sci. Total Environ. 2018, 625, 320–335. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Wei, Y.; Meng, H.; Cao, Y.; Lead, J.; Hong, J. Effects of fertilization and reclamation time on soil bacterial communities in coal mining subsidence areas. Sci. Total Environ. 2020, 739, 139882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.N.; Sun, L.T.; Wang, Y.; Fan, K.; Xu, Q.S.; Li, Y.S.; Ma, Q.P.; Wang, J.G.; Ren, W.M.; Ding, Z.T. Cow manure appli-cation effectively regulates the soil bacterial community in tea plantation. BMC Microbiol. 2020, 20, 190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tian, N.; Yong, L.; Xiong, W.; Ran, W. Responses of Bacterial Communities in Arable Soils in a Rice-Wheat Cropping System to Different Fertilizer Regimes and Sampling Times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global Biogeography and Quantitative Seasonal Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Bansal, S.; Yin, X.H.; Sykes, V.; Lee, J.; Jagadamma, S. Soil aggregate-associated organic carbon and nitrogen response to long-term no-till crop rotation, cover crop, and manure application. Soil Sci. Soc. Am. J. 2021, 85, 2169–2184. [Google Scholar] [CrossRef]
- Dai, Z.; Su, W.; Chen, H.; Barberán, A.; Zhao, H.; Yu, M.; Yu, L.; Brookes, P.C.; Schadt, C.W.; Chang, S.X.; et al. Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Chang. Biol. 2018, 24, 3452–3461. [Google Scholar] [CrossRef]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational Improvements Reveal Great Bacterial Diversity and High Metal Toxicity in Soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef]
- Xue, L.; Ren, H.; Brodribb, T.J.; Wang, J.; Yao, X.; Li, S. Long term effects of management practice intensification on soil microbial community structure and co-occurrence network in a non-timber plantation. For. Ecol. Manag. 2021, 459, 117805. [Google Scholar] [CrossRef]
- Wang, F.; Chen, S.; Qin, S.; Sun, R.; Zhang, Y.; Wang, S.; Hu, C.; Hu, H.; Liu, B. Long-term nitrogen fertilization alters microbial community structure and denitrifier abundance in the deep vadose zone. J. Soils Sediments 2021, 21, 2394–2403. [Google Scholar] [CrossRef]
- Qiu, S.-L.; Wang, L.-M.; Huang, D.-F.; Lin, X.-J. Effects of fertilization regimes on tea yields, soil fertility, and soil microbial diversity. Chil. J. Agric. Res. 2014, 74, 333–339. [Google Scholar] [CrossRef]
- Hamm, A.C.; Tenuta, M.; Krause, D.O.; Ominski, K.H.; Tkachuk, V.L.; Flaten, D.N. Bacterial communities of an agricultural soil amended with solid pig and dairy manures, and urea fertilizer. Appl. Soil Ecol. 2016, 103, 61–71. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, T.; Zhao, J.; Wang, L.; Yang, D.; Li, G.; Xiu, W. Variation of Soil Bacterial and Fungal Communities from Fluvo-Aquic Soil Under Chemical Fertilizer Reduction Combined with Organic Materials in North China Plain. J. Soil Sci. Plant Nutr. 2020, 21, 349–363. [Google Scholar] [CrossRef]
- Liu, S.; Li, P.; Van Zwieten, L.; Tu, J.; Gan, W.; Lu, S.; Wang, H.; Wu, L. Edaphic variables influence soil bacterial structure under successive fertilization of Paulownia plantation substituting native vegetation. J. Soils Sediments 2021, 21, 2922–2937. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Genet. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Meng, F.; Yan, X.; Zhang, M.; Yao, X.; Kim, K.S.; Zhao, J.; Qiu, Q.; Xie, F.; Zhang, W. Changes in Soil Microbial Activity, Bacterial Community Composition and Function in a Long-Term Continuous Soybean Cropping System After Corn Insertion and Fertilization. Front. Microbiol. 2021, 12, 638326. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, M.; Bai, S.H.; Xu, Z.; Liu, Y.; Chen, F.; Guo, X.; Luo, H.; Wang, S.; Xie, J.; et al. Successive mineral nitrogen or phosphorus fertilization alone significantly altered bacterial community rather than bacterial biomass in plantation soil. Appl. Microbiol. Biotechnol. 2020, 104, 7213–7224. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Genet. 2011, 9, 119–130. [Google Scholar] [CrossRef]
- Reddy, P.P. Plant Growth Promoting Rhizobacteria for Horticultural Crop Protection; Springer India: New Delhi, India, 2014. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Z.; Wang, L.; Li, Z.; Fang, J.; Wei, X.; Wei, W.; Cao, J.; Wei, Y.; Xie, Z. Bulk and Active Sediment Prokaryotic Communities in the Mariana and Mussau Trenches. Front. Microbiol. 2020, 11, 1521. [Google Scholar] [CrossRef] [PubMed]
- Durand, A.; Maillard, F.; Alvarez-Lopez, V.; Guinchard, S.; Bertheau, C.; Valot, B.; Blaudez, D.; Chalot, M. Bacterial diversity associated with poplar trees grown on a Hg-contaminated site: Community characterization and isolation of Hg-resistant plant growth-promoting bacteria. Sci. Total Environ. 2018, 622–623, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Fournier, B.; Dos Santos, S.P.; Gustavsen, J.A.; Imfeld, G.; Lamy, F.; Mitchell, E.A.; Mota, M.; Noll, D.; Planchamp, C.; Heger, T.J. Impact of a synthetic fungicide (fosetyl-Al and propamocarb-hydrochloride) and a biopesticide (Clonostachys rosea) on soil bacterial, fungal, and protist communities. Sci. Total Environ. 2020, 738, 139635. [Google Scholar] [CrossRef] [PubMed]
- Kyei-Boahen, S.; Slinkard, A.; Walley, F. Isotopic fractionation during N2 fixation by chickpea. Soil Biol. Biochem. 2002, 34, 417–420. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Morriën, E.; Hannula, S.E.; Snoek, B.; Helmsing, N.R.; Zweers, H.; De Hollander, M.; Soto, R.L.; Bouffaud, M.-L.; Buée, M.; Dimmers, W.; et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 2017, 8, 14349. [Google Scholar] [CrossRef]
- Yu, J.; Deem, L.M.; Crow, S.E.; Deenik, J.L.; Penton, C.R. Biochar application influences microbial assemblage complexity and composition due to soil and bioenergy crop type interactions. Soil Biol. Biochem. 2018, 117, 97–107. [Google Scholar] [CrossRef]
- Ge, Z.; Li, S.Y.; Bol, R.; Zhu, P.; Peng, C.; An, T.T.; Cheng, N.; Liu, X.; Li, T.Y.; Xu, Z.Q.; et al. Differential long-term fertilization alters residue-derived labile organic carbon fractions and microbial community during straw residue decomposition. Soil Till. Res. 2021, 213, 105120. [Google Scholar] [CrossRef]
- Ratzke, C.; Barrere, J.; Gore, J. Strength of species interactions determines biodiversity and stability in microbial communities. Nat. Ecol. Evol. 2020, 4, 376–383. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, Z.; Gong, Q.; Zhai, B.; Li, Z. Responses of fungal–bacterial community and network to organic inputs vary among different spatial habitats in soil. Soil Biol. Biochem. 2018, 125, 54–63. [Google Scholar] [CrossRef]



| Treatment | pH | SOM (g·kg−1) | NH4+–N (mg·kg−1) | NO3−–N (mg·kg−1) | AP (mg·kg−1) |
|---|---|---|---|---|---|
| OCF | 5.04 ± 0.09 a,b | 42.21 ± 0.94 a,b | 78.21 ± 3.01 a | 11.95 ± 0.33 a | 2.52 ± 0.19 a |
| OPF | 5.03 ± 0.08 a,b | 40.86 ± 2.12 a,b | 33.75 ± 2.66 c,d | 6.68 ± 0.05 c | 2.32 ± 0.24 a,b |
| OCPF | 5.30 ± 0.08 a | 43.99 ± 1.39 a | 48.21 ± 3.96 b,c | 11.97 ± 0.41 a | 2.13 ± 0.06 a,b |
| CPF | 5.20 ± 0.08 a,b | 37.43 ± 0.97 b | 51.35 ± 2.05 b | 10.22 ± 0.26 b | 2.35 ± 0.19 a,b |
| CK | 4.90 ± 0.08 b | 36.54 ± 1.17 b | 30.92 ± 2.27 d | 6.27 ± 0.44 c | 1.70 ± 0.11 b |
| p-value | 0.028 | 0.008 | <0.001 | <0.001 | 0.035 |
| Topological Properties | OCF | OPF | OCPF | CPF | CK |
|---|---|---|---|---|---|
| Nodes | 97 | 98 | 99 | 96 | 98 |
| Edges | 285 | 431 | 500 | 426 | 273 |
| Positive edge proportion | 59.30% | 66.82% | 62.80% | 72.77% | 73.26% |
| Negative edge proportion | 40.70% | 33.18% | 32.70% | 27.23% | 26.74% |
| Network density | 0.061 | 0.091 | 0.103 | 0.091 | 0.057 |
| Average degree | 5.876 | 8.796 | 10.101 | 8.784 | 5.315 |
| Average clustering coefficient | 0.621 | 0.637 | 0.600 | 0.648 | 0.541 |
| Network diameter | 14 | 11 | 11 | 14 | 15 |
| Modularity | 0.735 | 0.608 | 0.527 | 0.550 | 0.653 |
| Average path length | 6.030 | 3.954 | 4.165 | 5.788 | 5.315 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Zhao, W.; Zhou, Z.; Huang, G.; Wang, X.; Han, Q.; Liu, G. The Application of Mixed Organic and Inorganic Fertilizers Drives Soil Nutrient and Bacterial Community Changes in Teak Plantations. Microorganisms 2022, 10, 958. https://doi.org/10.3390/microorganisms10050958
Zhang Q, Zhao W, Zhou Z, Huang G, Wang X, Han Q, Liu G. The Application of Mixed Organic and Inorganic Fertilizers Drives Soil Nutrient and Bacterial Community Changes in Teak Plantations. Microorganisms. 2022; 10(5):958. https://doi.org/10.3390/microorganisms10050958
Chicago/Turabian StyleZhang, Qingqing, Weiwei Zhao, Zaizhi Zhou, Guihua Huang, Xianbang Wang, Qiang Han, and Gaofeng Liu. 2022. "The Application of Mixed Organic and Inorganic Fertilizers Drives Soil Nutrient and Bacterial Community Changes in Teak Plantations" Microorganisms 10, no. 5: 958. https://doi.org/10.3390/microorganisms10050958






