The Microbiota–Gut–Brain Axis: Gut Microbiota Modulates Conspecific Aggression in Diversely Selected Laying Hens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Central Monoamines
2.3. Plasma Serotonin and Tryptophan
2.4. Plasma Cytokines and Immunoglobulin G
2.5. Plasma Corticosterone and Heterophil to Lymphocyte Ratio
2.6. DNA Extraction and Sequencing
2.7. Data Processing and Analysis
2.7.1. Physiological Data
2.7.2. 16S rRNA Sequencing Data
2.8. Correlation Analysis
3. Results
3.1. Physiological Parameters
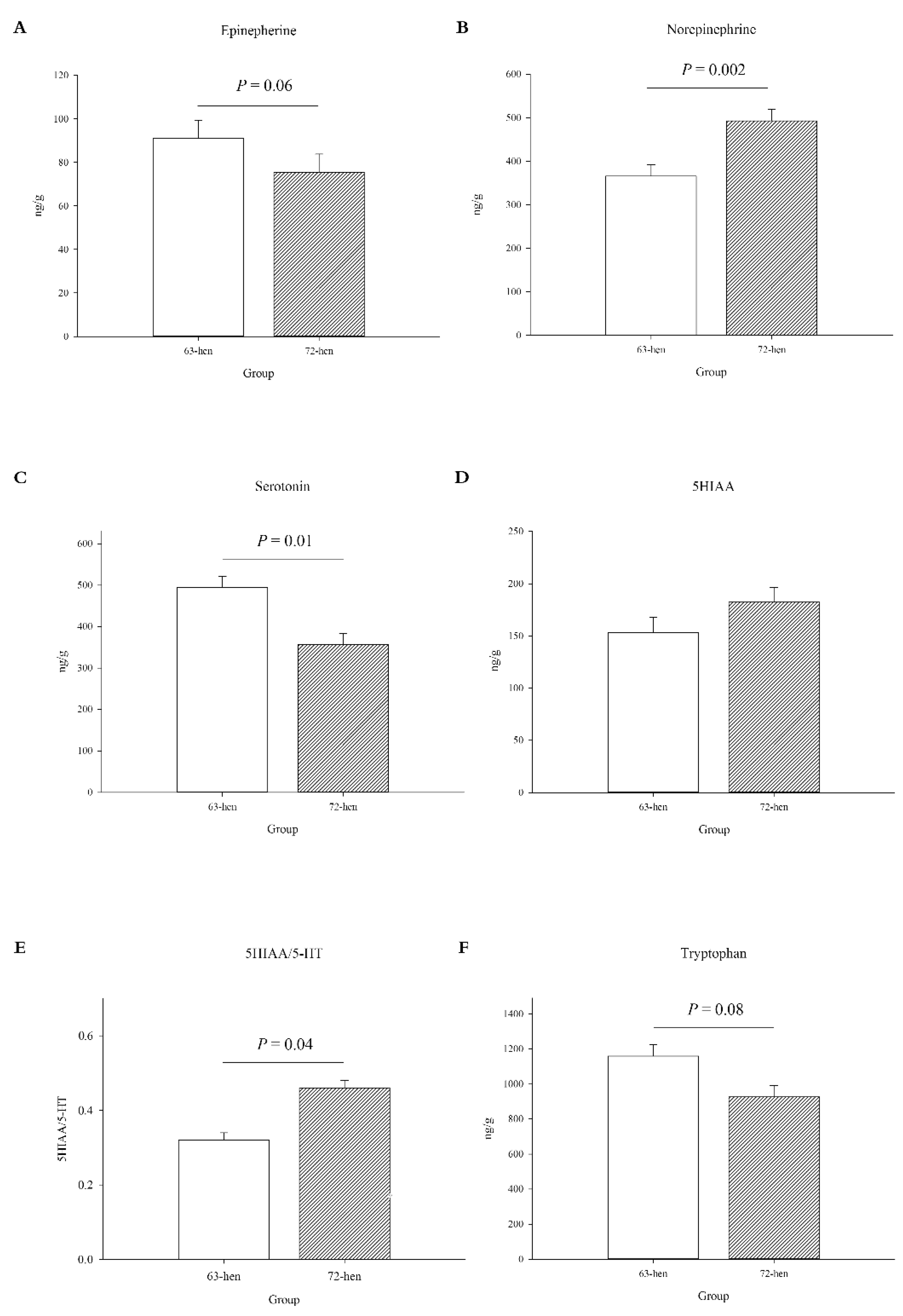
3.1.1. Central Monoamines Levels
3.1.2. Peripheral Serotonergic, Stress and Immune Parameters
3.2. Microbiota Profile
3.2.1. Alpha Diversity
3.2.2. Beta Diversity
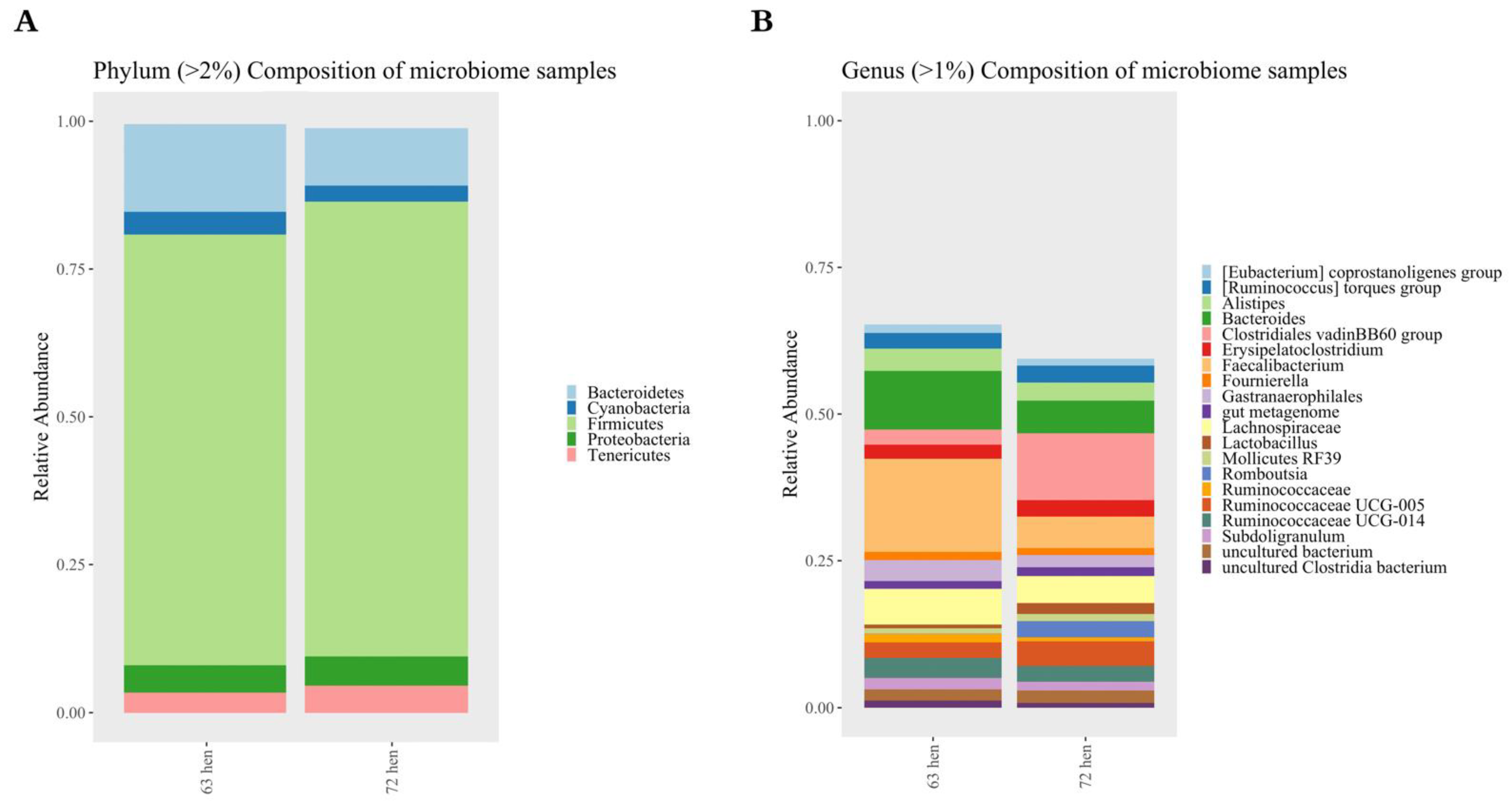
3.2.3. Taxonomic Composition
3.2.4. Metagenome Functions Prediction
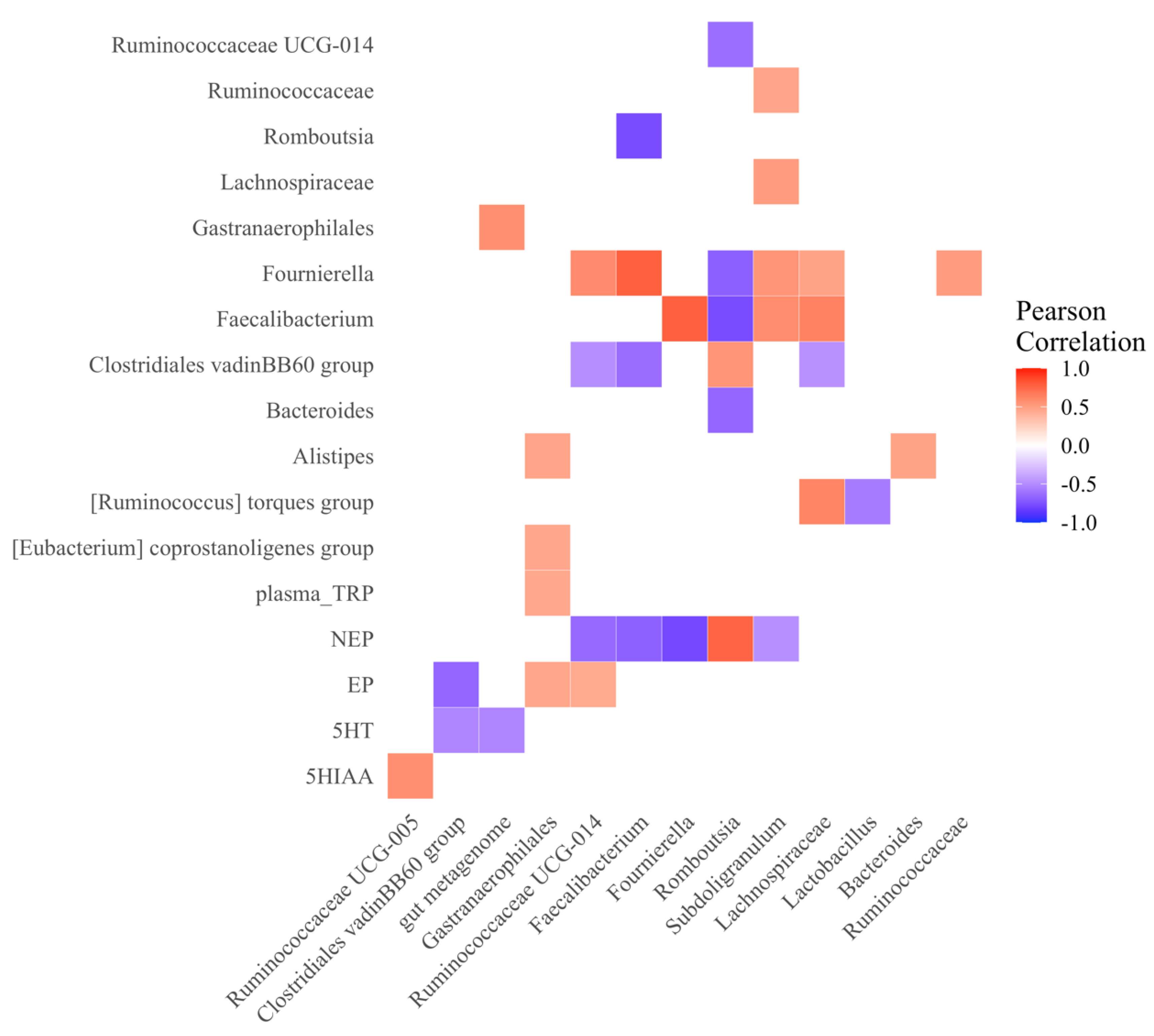
3.2.5. Microbial and Neurotransmitter Correlations
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cryan, J.F.; O’Mahony, S.M. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Łoniewski, I.; Misera, A.; Skonieczna-Żydecka, K.; Kaczmarczyk, M.; Kaźmierczak-Siedlecka, K.; Misiak, B.; Marlicz, W.; Samochowiec, J. Major Depressive Disorder and gut microbiota—Association not causation. A scoping review. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 106, 110111. [Google Scholar] [CrossRef] [PubMed]
- Bioque, M.; González-Rodríguez, A.; Garcia-Rizo, C.; Cobo, J.; Monreal, J.A.; Usall, J.; Soria, V.; Labad, J. Targeting the microbiome-gut-brain axis for improving cognition in schizophrenia and major mood disorders: A narrative review. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 105, 110130. [Google Scholar] [CrossRef]
- Li, N.; Wang, Q.; Wang, Y.; Sun, A.; Lin, Y.; Jin, Y.; Li, X. Fecal microbiota transplantation from chronic unpredictable mild stress mice donors affects anxiety-like and depression-like behavior in recipient mice via the gut microbiota-inflammation-brain axis. Stress 2019, 22, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Lukic, I.; Getselter, D.; Ziv, O.; Oron, O.; Reuveni, E.; Koren, O.; Elliott, E. Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Transl. Psychiatry 2019, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.V.; Sherwin, E.; Schellekens, H.; Stanton, C.; Dinan, T.; Cryan, J.F. Feeding the microbiota-gut-brain axis: Diet, microbiome, and neuropsychiatry. Transl. Res. 2017, 179, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Dodiya, H.B.; Forsyth, C.B.; Voigt, R.M.; Engen, P.A.; Patel, J.; Shaikh, M.; Green, S.J.; Naqib, A.; Roy, A.; Kordower, J.H.; et al. Chronic stress-induced gut dysfunction exacerbates Parkinson’s disease phenotype and pathology in a rotenone-induced mouse model of Parkinson’s disease. Neurobiol. Dis. 2020, 135, 104352. [Google Scholar] [CrossRef]
- Zimmermann, M.; Grabemann, M.; Mette, C.; Abdel-Hamid, M.; Ueckermann, J.; Kraemer, M.; Wiltfang, J.; Kis, B.; Zepf, F.D. The Effects of Acute Tryptophan Depletion on Reactive Aggression in Adults with Attention-Deficit/Hyperactivity Disorder (ADHD) and Healthy Controls. PLoS ONE 2012, 7, e32023. [Google Scholar] [CrossRef]
- Coccaro, E.F.; Fanning, J.R.; Phan, K.L.; Lee, R. Serotonin and impulsive aggression. CNS Spectr. 2015, 20, 295–302. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietraszek, M.; Takada, Y.; Yan, D.; Urano, T.; Serizawa, K.; Takada, A. Relationship between serotonergic measures in periphery and the brain of mouse. Life Sci. 1992, 51, 75–82. [Google Scholar] [CrossRef]
- Dennis, R.; Zhang, H.M.; Cheng, H.W. Effect of Selection for Resistance and Susceptibility to Viral Diseases on Concentrations of Dopamine and Immunological Parameters in Six-Week-Old Chickens. Poult. Sci. 2006, 85, 2135–2140. [Google Scholar] [CrossRef]
- Dennis, R.L.; Cheng, H.-W. Differential serotonergic mediation of aggression in roosters bred for resistance and susceptibility to Marek’s disease. Br. Poult. Sci. 2014, 55, 13–20. [Google Scholar] [CrossRef]
- Bienenstock, J.; Kunze, W.; Forsythe, P. Microbiota and the gut–brain axis. Nutr. Rev. 2015, 73 (Suppl. 1), 28–31. [Google Scholar] [CrossRef]
- Caramaschi, D.; de Boer, S.F.; Koolhaas, J.M. Differential role of the 5-HT1A receptor in aggressive and non-aggressive mice: An across-strain comparison. Physiol. Behav. 2007, 90, 590–601. [Google Scholar] [CrossRef] [Green Version]
- Homberg, J.R.; Pattij, T.; Janssen, M.C.W.; Ronken, E.; De Boer, S.F.; Schoffelmeer, A.N.M.; Cuppen, E. Serotonin transporter deficiency in rats improves inhibitory control but not behavioural flexibility. Eur. J. Neurosci. 2007, 26, 2066–2073. [Google Scholar] [CrossRef] [Green Version]
- Olivier, B.; van Oorschot, R. 5-HT1B receptors and aggression: A review. Eur. J. Pharmacol. 2005, 526, 207–217. [Google Scholar] [CrossRef]
- Albert, P.R.; Le François, B.; Millar, A.M. Transcriptional dysregulation of 5-HT1A autoreceptors in mental illness. Mol. Brain 2011, 4, 21. [Google Scholar] [CrossRef] [Green Version]
- Albert, P.R.; Vahid-Ansari, F. The 5-HT1A receptor: Signaling to behavior. Biochimie 2019, 161, 34–45. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Mu, C.-L.; Farzi, A.; Zhu, W.-Y. Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef] [Green Version]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Fanning, J.R.; Lee, R.; Gozal, D.; Coussons-Read, M.; Coccaro, E.F. Childhood trauma and parental style: Relationship with markers of inflammation, oxidative stress, and aggression in healthy and personality disordered subjects. Biol. Psychol. 2015, 112, 56–65. [Google Scholar] [CrossRef]
- Kraus, M.R.; Schäfer, A.; Faller, H.; Csef, H.; Scheurlen, M. Psychiatric symptoms in patients with chronic hepatitis C receiving interferon alfa-2b therapy. J. Clin. Psychiatry 2003, 64, 708–714. [Google Scholar] [CrossRef]
- Hassanain, M.; Bhatt, S.; Zalcman, S.; Siegel, A. Potentiating role of interleukin-1β (IL-1β) and IL-1β type 1 receptors in the medial hypothalamus in defensive rage behavior in the cat. Brain Res. 2005, 1048, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Van De Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef] [PubMed]
- Halverson, T.; Alagiakrishnan, K. Gut microbes in neurocognitive and mental health disorders. Ann. Med. 2020, 52, 423–443. [Google Scholar] [CrossRef] [PubMed]
- Obrenovich, M.E.M. Leaky Gut, Leaky Brain? Microorganisms 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuehne, S.A.; Cartman, S.T.; Heap, J.T.; Kelly, M.L.; Cockayne, A.; Minton, N.P. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010, 467, 711–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.-N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558 Pt 1, 263–275. [Google Scholar] [CrossRef]
- Bailey, M.T.; Dowd, S.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Crumeyrolle-Arias, M.; Jaglin, M.; Bruneau, A.; Vancassel, S.; Cardona, A.; Daugé, V.; Naudon, L.; Rabot, S. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology 2014, 42, 207–217. [Google Scholar] [CrossRef]
- Dallman, M.F.; Akana, S.F.; Levin, N.; Walker, C.-D.; Bradbury, M.J.; Suemaru, S.; Scribner, K.S. Corticosteroids and the Control of Function in the Hypothalamo-Pituitary-Adrenal (HPA) Axisa. Ann. N. Y. Acad. Sci. 1994, 746, 22–31. [Google Scholar] [CrossRef]
- Pollard, C.M.; Booth, S. Addressing Food and Nutrition Security in Developed Countries. Int. J. Environ. Res. Public Health 2019, 16, 2370. [Google Scholar] [CrossRef] [Green Version]
- Pollard, C.M.; Booth, S. Food Insecurity and Hunger in Rich Countries—It Is Time for Action against Inequality. Int. J. Environ. Res. Public Health 2019, 16, 1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lay, D.C., Jr.; Fulton, R.M.; Hester, P.Y.; Karcher, D.M.; Kjaer, J.B.; Mench, J.A.; Mullens, B.A.; Newberry, R.C.; Nicol, C.J.; O’Sullivan, N.P.; et al. Hen welfare in different housing systems. Poult. Sci. 2011, 90, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Janczak, A.M.; Riber, A.B. Review of rearing-related factors affecting the welfare of laying hens. Poult. Sci. 2015, 94, 1454–1469. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H. Morphopathological changes and pain in beak trimmed laying hens. World Poult. Sci. J. 2006, 62, 41–52. [Google Scholar] [CrossRef]
- Barbosa, R.S.D.; Vieira-Coelho, M.A. Probiotics and prebiotics: Focus on psychiatric disorders—A systematic review. Nutr. Rev. 2020, 78, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Generoso, J.S.; Giridharan, V.V.; Lee, J.; Macedo, D.; Barichello, T. The role of the microbiota-gut-brain axis in neuropsychiatric disorders. Braz. J. Psychiatr. 2021, 43, 293–305. [Google Scholar] [CrossRef]
- Noonan, S.; Zaveri, M.; Macaninch, E.; Martyn, K. Food & mood: A review of supplementary prebiotic and probiotic interventions in the treatment of anxiety and depression in adults. BMJ Nutr. Prev. Health 2020, 3, 351–362. [Google Scholar] [CrossRef]
- Evrensel, A.; Tarhan, K.N. Emerging role of Gut-microbiota-brain axis in depression and therapeutic implication. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2020, 106, 110138. [Google Scholar] [CrossRef]
- Tremblay, A.; Lingrand, L.; Maillard, M.; Feuz, B.; Tompkins, T.A. The effects of psychobiotics on the microbiota-gut-brain axis in early-life stress and neuropsychiatric disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 105, 110142. [Google Scholar] [CrossRef]
- Van Der Eijk, J.A.J.; Rodenburg, T.B.; De Vries, H.; Kjaer, J.B.; Smidt, H.; Naguib, M.; Kemp, B.; Lammers, A. Early-life microbiota transplantation affects behavioural responses, serotonin and immune characteristics in chicken lines divergently selected on feather pecking. Sci. Rep. 2020, 10, 2750. [Google Scholar] [CrossRef]
- Van Staaveren, N.; Krumma, J.; Forsythe, P.; Kjaer, J.B.; Kwon, I.Y.; Mao, Y.-K.; West, C.; Kunze, W.; Harlander-Matauschek, A. Cecal motility and the impact of Lactobacillus in feather pecking laying hens. Sci. Rep. 2020, 10, 12978. [Google Scholar] [CrossRef] [PubMed]
- Burt, D.W. Emergence of the Chicken as a Model Organism: Implications for Agriculture and Biology. Poult. Sci. 2007, 86, 1460–1471. [Google Scholar] [CrossRef] [PubMed]
- Caspi, A.; McClay, J.; Moffitt, T.E.; Mill, J.; Martin-Moreno, L.; Craig, I.W.; Taylor, A.; Poulton, R. Role of Genotype in the Cycle of Violence in Maltreated Children. Science 2002, 297, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Dennis, R.L.; Chen, Z.Q.; Cheng, H.W. Serotonergic Mediation of Aggression in High and Low Aggressive Chicken Strains. Poult. Sci. 2008, 87, 612–620. [Google Scholar] [CrossRef]
- Tiihonen, J.; Rautiainen, M.-R.; Ollila, H.; Repotiihonen, E.; Virkkunen, M.; Palotie, A.; Pietilainen, O.; Kristiansson, K.; Joukamaa, M.; Lauerma, H.; et al. Genetic background of extreme violent behavior. Mol. Psychiatry 2015, 20, 786–792. [Google Scholar] [CrossRef] [Green Version]
- Bacon, L.; Palmquist, D. Chicken lines differ in production of interferon-like activity by peripheral white blood cells stimulated with phytohemagglutinin. Poult. Sci. 2002, 81, 1629–1636. [Google Scholar] [CrossRef]
- Bacon, L.D.; Hunt, H.D.; Cheng, H.H. A Review of the Development of Chicken Lines to Resolve Genes Determining Resistance to Diseases. Poult. Sci. 2000, 79, 1082–1093. [Google Scholar] [CrossRef]
- Bacon, L.D.; Hunt, H.D.; Cheng, H.H. Genetic Resistance to Marek’s Disease. In Marek’s Disease; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2001; Volume 255, pp. 121–141. [Google Scholar] [CrossRef]
- Xu, M.; Xu, C.; Liu, F.; Shen, X.; Meng, J.; Chen, H.; Yang, J.; Zhou, P.; Gao, R.; Gan, S. Effects of active immunization with newly modified GnRH peptides on spermatogenesis and production performance of Holstein bulls. Biol. Reprod. 2018, 99, 461–472. [Google Scholar] [CrossRef]
- Burgess, S.C.; Basaran, B.H.; Davison, T.F. Resistance to Marek’s Disease Herpesvirus-induced Lymphoma is Multiphasic and Dependent on Host Genotype. Vet. Pathol. 2001, 38, 129–142. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Babiuk, L.A.; Hurk, S.; van Drunen, L.-V.D. Strategies for loading dendritic cells with hepatitis C NS5a antigen and inducing protective immunity. J. Viral Hepat. 2008, 15, 459–470. [Google Scholar] [CrossRef]
- Kaiser, P.; Underwood, G.; Davison, F. Differential Cytokine Responses following Marek’s Disease Virus Infection of Chickens Differing in Resistance to Marek’s Disease. J. Virol. 2003, 77, 762–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, R.; Zhang, H.M.; Bacon, L.D.; Estevez, I.; Cheng, H.W. Behavioral and physiological features of chickens diversely selected for resistance to Avian Disease. 1. Selected inbred lines differ in behavioral and physical responses to social stress. Poult. Sci. 2004, 83, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.Y. Thermal Perches as Cooling Devices for Reducing Heat Stress in Caged Laying Hens. Ph.D. Dissertation, Purdue University, West Lafayette, IN, USA, 2017. [Google Scholar]
- Hu, J.; Hester, P.; Makagon, M.; Xiong, Y.; Gates, R.; Cheng, H. Effect of cooled perches on physiological parameters of caged White Leghorn hens exposed to cyclic heat. Poult. Sci. 2019, 98, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.W.; Dillworth, G.; Singleton, P.; Chen, Y.; Muirt, W.M. Effects of Group Selection for Productivity and Longevity on Blood Concentrations of Serotonin, Catecholamines, and Corticosterone of Laying Hens. Poult. Sci. 2001, 80, 1278–1285. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Caliński, T.; Steel, R.G.D.; Torrie, J.H. Principles and Procedures of Statistics: A Biometrical Approach. Biometrics 1981, 37, 859–860. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Oksanen, F.J. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 1 December 2020).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 1 December 2020).
- Revelle, W. Package ‘Psych’. Version 2.1.3. 2021. Available online: https://cran.r-project.org/web/packages/psych/psych.pdf (accessed on 1 December 2020).
- Palumbo, S.; Mariotti, V.; Iofrida, C.; Pellegrini, S. Genes and Aggressive Behavior: Epigenetic Mechanisms Underlying Individual Susceptibility to Aversive Environments. Front. Behav. Neurosci. 2018, 12, 117. [Google Scholar] [CrossRef] [PubMed]
- Runions, K.C.; Morandini, H.; Rao, P.; Wong, J.W.Y.; Kolla, N.J.; Pace, G.; Mahfouda, S.; Hildebrandt, C.S.; Stewart, R.; Zepf, F.D. Serotonin and aggressive behaviour in children and adolescents: A systematic review. Acta Psychiatr. Scand. 2019, 139, 117–144. [Google Scholar] [CrossRef] [PubMed]
- Browne, C.; Clarke, G.; Dinan, T.G.; Cryan, J.F. An effective dietary method for chronic tryptophan depletion in two mouse strains illuminates a role for 5-HT in nesting behaviour. Neuropharmacology 2012, 62, 1903–1915. [Google Scholar] [CrossRef]
- Lu, F.; Zhang, Y.; Trivedi, A.; Jiang, X.; Chandra, D.; Zheng, J.; Nakano, Y.; Uyghurturk, D.A.; Jalai, R.; Onur, S.G.; et al. Fluoride related changes in behavioral outcomes may relate to increased serotonin. Physiol. Behav. 2019, 206, 76–83. [Google Scholar] [CrossRef]
- Lindseth, G.; Helland, B.; Caspers, J. The Effects of Dietary Tryptophan on Affective Disorders. Arch. Psychiatr. Nurs. 2015, 29, 102–107. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, T.A.; Nguyen, J.C.D.; Polglaze, K.E.; Bertrand, P.P. Influence of Tryptophan and Serotonin on Mood and Cognition with a Possible Role of the Gut-Brain Axis. Nutrients 2016, 8, 56. [Google Scholar] [CrossRef]
- Jacobsen, J.P.R.; Medvedev, I.O.; Caron, M.G. The 5-HT deficiency theory of depression: Perspectives from a naturalistic 5-HT deficiency model, the tryptophan hydroxylase 2 Arg 439 His knockin mouse. Philos. Trans. R. Soc. Lond B Biol. Sci. 2012, 367, 2444–2459. [Google Scholar] [CrossRef] [Green Version]
- Manchia, M.; Carpiniello, B.; Valtorta, F.; Comai, S. Serotonin Dysfunction, Aggressive Behavior, and Mental Illness: Exploring the Link Using a Dimensional Approach. ACS Chem. Neurosci. 2017, 8, 961–972. [Google Scholar] [CrossRef]
- Rosado, B.; García-Belenguer, S.; Palacio, J.; Chacón, G.; Villegas, A.; Alcalde, A.I. Serotonin transporter activity in platelets and canine aggression. Vet. J. 2010, 186, 104–105. [Google Scholar] [CrossRef]
- Cheng, H.; Muir, W.M. The effects of genetic selection for survivability and productivity on chicken physiological homeostasis. World Poult. Sci. J. 2005, 61, 15. [Google Scholar] [CrossRef]
- Luykx, J.; Bakker, S.C.; Lentjes, E.; Neeleman, M.; Strengman, E.; Mentink, L.; Deyoung, J.; De Jong, S.; Sul, J.H.; Eskin, E.; et al. Genome-wide association study of monoamine metabolite levels in human cerebrospinal fluid. Mol. Psychiatry 2014, 19, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Costedio, M.M.; Hyman, N.; Mawe, G.M. Serotonin and Its Role in Colonic Function and in Gastrointestinal Disorders. Dis. Colon Rectum 2007, 50, 376–388. [Google Scholar] [CrossRef] [PubMed]
- McLean, P.G.; Borman, R.A.; Lee, K. 5-HT in the enteric nervous system: Gut function and neuropharmacology. Trends Neurosci. 2007, 30, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Desbonnet, L.; Garrett, L.; Clarke, G.; Bienenstock, J.; Dinan, T.G. The probiotic Bifidobacteria infantis: An assessment of potential antidepressant properties in the rat. J. Psychiatr. Res. 2008, 43, 164–174. [Google Scholar] [CrossRef]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Aizawa, E.; Tsuji, H.; Asahara, T.; Takahashi, T.; Teraishi, T.; Yoshida, S.; Ota, M.; Koga, N.; Hattori, K.; Kunugi, H. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord. 2016, 202, 254–257. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Golubeva, A.V.; Joyce, S.A.; Moloney, G.; Burokas, A.; Sherwin, E.; Arboleya, S.; Flynn, I.; Khochanskiy, D.; Moya-Pérez, A.; Peterson, V.; et al. Microbiota-related Changes in Bile Acid & Tryptophan Metabolism are Associated with Gastrointestinal Dysfunction in a Mouse Model of Autism. EBioMedicine 2017, 24, 166–178. [Google Scholar] [CrossRef] [Green Version]
- Flowers, S.A.; Evans, S.J.; Ward, K.M.; McInnis, M.G.; Ellingrod, V.L. Interaction between Atypical Antipsychotics and the Gut Microbiome in a Bipolar Disease Cohort. Pharmacotherapy 2017, 37, 261–267. [Google Scholar] [CrossRef]
- Sun, J.; Wang, F.; Hu, X.; Yang, C.; Xu, H.; Yao, Y.; Liu, J. Clostridium butyricum Attenuates Chronic Unpredictable Mild Stress-Induced Depressive-Like Behavior in Mice via the Gut-Brain Axis. J. Agric. Food Chem. 2018, 66, 8415–8421. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Fang, X.; Zhan, G.; Huang, N.; Li, S.; Bi, J.; Jiang, R.; Yang, L.; Miao, L.; Zhu, B.; et al. Key role of gut microbiota in anhedonia-like phenotype in rodents with neuropathic pain. Transl. Psychiatry 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rook, G.A.W.; Raison, C.L.; Lowry, C.A. Childhood Microbial Experience, Immunoregulation, Inflammation, and Adult Susceptibility to Psychosocial Stressors and Depression. In Inflammation and Immunity in Depression Basic Science and Clinical Applications; Baune, B.T., Ed.; Academic Press: Washington, DC, USA, 2018; pp. 17–44. [Google Scholar]
- Dahiya, D.K.; Dangi, A.K.; Shandilya, U.K.; Puniya, A.K.; Shukla, P. New-Generation Probiotics: Perspectives and Applications. In Microbiome and Metabolome in Diagnosis, Therapy, and Other Strategic Applications; Faintuch, E.D.S.J., Faintuch, S., Eds.; Academic Press: Washington, DC, USA, 2019; pp. 417–424. [Google Scholar]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-H.; Bai, J.; Wu, D.; Yu, S.-F.; Qiang, X.-L.; Bai, H.; Wang, H.-N.; Peng, Z.-W. Association between fecal microbiota and generalized anxiety disorder: Severity and early treatment response. J. Affect. Disord. 2019, 259, 56–66. [Google Scholar] [CrossRef]
- Averina, O.V.; Zorkina, Y.A.; Yunes, R.A.; Kovtun, A.S.; Ushakova, V.M.; Morozova, A.Y.; Kostyuk, G.P.; Danilenko, V.N.; Chekhonin, V.P. Bacterial Metabolites of Human Gut Microbiota Correlating with Depression. Int. J. Mol. Sci. 2020, 21, 9234. [Google Scholar] [CrossRef]
- Fukumoto, S.; Tatewaki, M.; Yamada, T.; Fujimiya, M.; Mantyh, C.; Voss, M.; Eubanks, S.; Harris, M.; Pappas, T.; Takahashi, T. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, R1269–R1276. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.-C.; Shen, M.-H.; Liu, C.-Y.; Pu, C.-M.; Hu, J.-M.; Huang, C.-J. A gut butyrate-producing bacterium Butyricicoccus pullicaecorum regulates short-chain fatty acid transporter and receptor to reduce the progression of 1,2-dimethylhydrazine-associated colorectal cancer. Oncol. Lett. 2020, 20, 327. [Google Scholar] [CrossRef]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.-C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Gerritsen, J.; Hornung, B.; Ritari, J.; Paulin, L.; Rijkers, G.T.; Schaap, P.J.; de Vos, W.M.; Smidt, H. A Comparative and functional ge-nomics analysis of the genus romboutsia provides insight into adaptation to an intestinal lifestyle. BioRxiv 2019, 845511. [Google Scholar] [CrossRef] [Green Version]
- Richards, P.; Fothergill, J.; Bernardeau, M.; Wigley, P. Development of the Caecal Microbiota in Three Broiler Breeds. Front. Vet. Sci. 2019, 6, 201. [Google Scholar] [CrossRef]
- Moreno-Arrones, O.; Serrano-Villar, S.; Perez-Brocal, V.; Saceda-Corralo, D.; Morales-Raya, C.; Rodrigues-Barata, R.; Moya, A.; Jaen-Olasolo, P.; Vano-Galvan, S. Analysis of the gut microbiota in alopecia areata: Identification of bacterial biomarkers. J. Eur. Acad. Dermatol. Venereol. 2019, 34, 400–405. [Google Scholar] [CrossRef]
- Leonard, B.E. Stress, Norepinephrine and depression. J. Psychiatry Neurosci. 2001, 26, S11–S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, S.K.; Valentino, R.J. The brain norepinephrine system, stress and cardiovascular vulnerability. Neurosci. Biobehav. Rev. 2017, 74, 393–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, S.K.; Bhatnagar, S. Resilience to the effects of social stress: Evidence from clinical and preclinical studies on the role of coping strategies. Neurobiol. Stress 2015, 1, 164–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, S.K.; McFadden, K.V.; Grigoriadis, D.; Bhatnagar, S.; Valentino, R.J. Depressive and cardiovascular disease comorbidity in a rat model of social stress: A putative role for corticotropin-releasing factor. Psychopharmacology 2012, 222, 325–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, M.; Kubera, M.; Leunis, J.-C.; Berk, M.; Geffard, M.; Bosmans, E. In depression, bacterial translocation may drive inflammatory responses, oxidative and nitrosative stress (O&NS), and autoimmune responses directed against O&NS-damaged neoepitopes. Acta Psychiatr. Scand. 2013, 127, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Cao, Q.; Cheng, Y.; Zhao, D.; Wang, Z.; Yang, H.; Wu, Q.; You, L.; Wang, Y.; Lin, Y.; et al. Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc. Natl. Acad. Sci. USA 2018, 115, E2960–E2969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengmark, S. Gut microbiota, immune development and function. Pharmacol. Res. 2013, 69, 87–113. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Cohen, L.J.; Esterhazy, D.; Kim, S.-H.; Lemetre, C.; Aguilar, R.; Gordon, E.A.; Pickard, A.; Cross, J.R.; Emiliano, A.B.; Han, S.M.; et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017, 549, 48–53. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; ter Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016, 167, 1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandsma, E.; Kloosterhuis, N.J.; Koster, M.; Dekker, D.C.; Gijbels, M.J.; van der Velden, S.; Ríos-Morales, M.; Van Faassen, M.J.; Loreti, M.G.; De Bruin, A.; et al. A Proinflammatory Gut Microbiota Increases Systemic Inflammation and Accelerates Atherosclerosis. Circ. Res. 2019, 124, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, A.; Fiorentino, M.; Buommino, E.; Donnarumma, G.; Losacco, A.; Bevilacqua, N. Lactobacillus crispatus mediates anti-inflammatory cytokine interleukin-10 induction in response to Chlamydia trachomatis infection in vitro. Int. J. Med. Microbiol. 2015, 305, 815–827. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, W.; Guo, R.; Liu, H. Faecalibacterium prausnitzii (ATCC 27766) has preventive and therapeutic effects on chronic unpredictable mild stress-induced depression-like and anxiety-like behavior in rats. Psychoneuroendocrinology 2019, 104, 132–142. [Google Scholar] [CrossRef]
- Yang, C.; Chalasani, G.; Ng, Y.-H.; Robbins, P.D. Exosomes released from mycoplasma infected tumor cells activate inhibitory B cells. PLoS ONE 2012, 7, e36138. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; He, Z.; Zhuo, Y.; Li, S.; Yang, W.; Hu, L.; Zhong, H. Rubidium chloride modulated the fecal microbiota community in mice. BMC Microbiol. 2021, 21, 46. [Google Scholar] [CrossRef]
- Delcenserie, V.; Martel, D.; Lamoureux, M.; Amiot, J.; Boutin, Y.; Roy, D. Immunomodulatory effects of probiotics in the intestinal tract. Curr. Issues Mol. Biol. 2008, 10, 37–54. [Google Scholar]
- Li, N.; Russell, W.M.; Douglas-Escobar, M.; Hauser, N.; Lopez, M.; Neu, J. Live and Heat-Killed Lactobacillus rhamnosus GG: Effects on Proinflammatory and Anti-Inflammatory Cytokines/Chemokines in Gastrostomy-Fed Infant Rats. Pediatr. Res. 2009, 66, 203–207. [Google Scholar] [CrossRef] [Green Version]
- Oh, N.S.; Joung, J.Y.; Lee, J.Y.; Kim, Y. Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PLoS ONE 2018, 13, e0192021. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yan, H.; Lu, Y.; Li, X.; Wang, X.; Shan, Y.; Yi, Y.; Liu, B.; Zhou, Y.; Lü, X. Anti-obesity effect of Lactobacillus rhamnosus LS-8 and Lactobacillus crustorum MN047 on high-fat and high-fructose diet mice base on inflammatory response alleviation and gut microbiota regulation. Eur. J. Nutr. 2019, 59, 2709–2728. [Google Scholar] [CrossRef] [PubMed]
- Knott, P.J.; Curzon, G. Free Tryptophan in Plasma and Brain Tryptophan Metabolism. Nature 1972, 239, 452–453. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Takao, K.; Funakoshi, H.; Miyakawa, T. Comprehensive behavioral analysis of tryptophan 2,3-dioxygenase (Tdo2) knockout mice. Neuropsychopharmacol. Rep. 2018, 38, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.-Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, S.; Schwieler, L.; Imbeault, S.; Engberg, G. The kynurenine pathway in schizophrenia and bipolar disorder. Neuropharmacology 2017, 112 Pt B, 297–306. [Google Scholar] [CrossRef]
- Imbeault, S.; Goiny, M.; Liu, X.; Erhardt, S. Effects of IDO1 and TDO2 inhibition on cognitive deficits and anxiety following LPS-induced neuroinflammation. Acta Neuropsychiatr. 2020, 32, 43–53. [Google Scholar] [CrossRef]
- Oxenkrug, G.F. Tryptophan kynurenine metabolism as a common mediator of genetic and environmental impacts in major depressive disorder: The serotonin hypothesis revisited 40 years later. Isr. J. Psychiatry Relat. Sci. 2010, 47, 56–63. [Google Scholar] [CrossRef] [Green Version]
- Campbell, D.L.M.; de Haas, E.N.; Lee, C. A review of environmental enrichment for laying hens during rearing in relation to their behavioral and physiological development. Poult. Sci. 2019, 98, 9–28. [Google Scholar] [CrossRef]





| f | 5-HT (ng/mL) 1 | TRP (µmol/mL) 1 | CORT (ng/mL) 1 | H/L Ratio 1 |
|---|---|---|---|---|
| 63-hen | 61.38 | 171.52 a | 8.44 b | 0.16 b |
| 72-hen | 59.46 | 121.42 b | 9.75 a | 0.50 a |
| SEM | 3.79 | 15.37 | 1.51 | 0.04 |
| n 2 | 10 | 10 | 10 | 10 |
| p-Value | 0.73 | 0.03 | 0.05 | <0.0001 |
| IgG (mg/mL) 1 | IL-6 (pg/mL) 1 | IL-2 (pg/mL) 1 | IL-10 (pg/mL) 1 | TNF-α (ng/mL) 1 | |
|---|---|---|---|---|---|
| 63-hen | 12.0 | 28.14 | 60.09 | 9.37 | 36.65 |
| 72-hen | 12.9 | 27.56 | 71.65 | 13.13 | 30.73 |
| SEM | 0.73 | 1.63 | 12.8 | 1.64 | 2.37 |
| n 2 | 10 | 10 | 10 | 10 | 10 |
| p-value | 0.54 | 0.81 | 0.54 | 0.12 | 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Johnson, T.A.; Zhang, H.; Cheng, H.-W. The Microbiota–Gut–Brain Axis: Gut Microbiota Modulates Conspecific Aggression in Diversely Selected Laying Hens. Microorganisms 2022, 10, 1081. https://doi.org/10.3390/microorganisms10061081
Hu J, Johnson TA, Zhang H, Cheng H-W. The Microbiota–Gut–Brain Axis: Gut Microbiota Modulates Conspecific Aggression in Diversely Selected Laying Hens. Microorganisms. 2022; 10(6):1081. https://doi.org/10.3390/microorganisms10061081
Chicago/Turabian StyleHu, Jiaying, Timothy A. Johnson, Huanmin Zhang, and Heng-Wei Cheng. 2022. "The Microbiota–Gut–Brain Axis: Gut Microbiota Modulates Conspecific Aggression in Diversely Selected Laying Hens" Microorganisms 10, no. 6: 1081. https://doi.org/10.3390/microorganisms10061081
APA StyleHu, J., Johnson, T. A., Zhang, H., & Cheng, H.-W. (2022). The Microbiota–Gut–Brain Axis: Gut Microbiota Modulates Conspecific Aggression in Diversely Selected Laying Hens. Microorganisms, 10(6), 1081. https://doi.org/10.3390/microorganisms10061081






