The Role of Early Life Microbiota Composition in the Development of Allergic Diseases
Abstract
:1. Introduction
2. Maternal Influencing Factors for the Development of Allergic Diseases in Infants
3. Non-Maternal Influencing Factors for the Development of Allergic Diseases in Infants
4. The Role of Lung and Gut Microbiota in Asthma
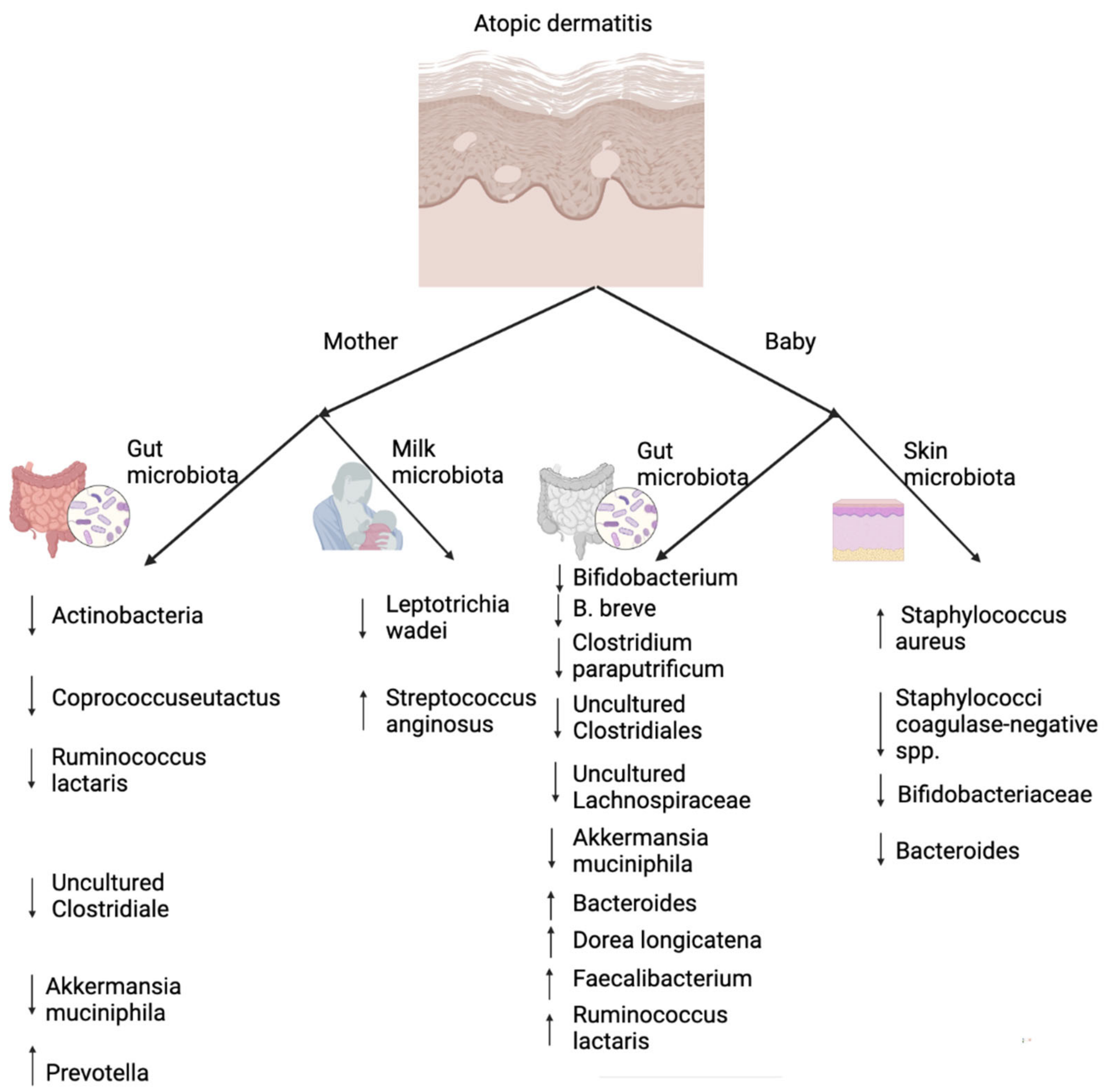
5. The Role of Skin and Gut Microbiota in Atopic Dermatitis
6. The Role of (Oral and) Gut Microbiota in Food Allergy
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bach, J.-F. The Effect of Infections on Susceptibility to Autoimmune and Allergic Diseases. N. Engl. J. Med. 2002, 347, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Xu, J.; Zhang, P.; Bao, Y. Prevalence and Risk Factors of Asthma in Preschool Children in Shanghai, China: A Cross-Sectional Study. Front. Pediatr. 2022, 9, 793452. [Google Scholar] [CrossRef] [PubMed]
- Portelli, M.A.; Hodge, E.; Sayers, I. Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin. Exp. Allergy 2015, 45, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Gilles, S.; Akdis, C.; Lauener, R.; Schmid-Grendelmeier, P.; Bieber, T.; Schäppi, G.; Traidl-Hoffmann, C. The role of environmental factors in allergy: A critical reappraisal. Exp. Dermatol. 2018, 27, 1193–1200. [Google Scholar] [CrossRef] [Green Version]
- Özdemir, A.; Can, D.; Günay, I.; Nacaroglu, T.; Karkiner, C.; Üstyol, A.; Kamalı, H.; Ayanoğlu, M.; Günay, T.; Dogan, D. Effect of industrialization on allergic diseases in school children. J. Turgut Ozal Med. Cent. 2018, 25, 232–235. [Google Scholar] [CrossRef]
- Fogarty, A.W. What have studies of non-industrialized countries told us about the cause of allergic disease? Clin. Exp. Allergy 2015, 45, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Sbihi, H.; Boutin, R.C.; Cutler, C.; Suen, M.; Finlay, B.B.; Turvey, S.E. Thinking bigger: How early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy 2019, 74, 2103–2115. [Google Scholar] [CrossRef] [Green Version]
- Coyte, K.Z.; Rao, C.; Rakoff-Nahoum, S.; Foster, K.R. Ecological rules for the assembly of microbiome communities. PLoS Biol. 2021, 19, e3001116. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020, 202, 2147–2167. [Google Scholar] [CrossRef] [PubMed]
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome Composition and Its Impact on the Development of Allergic Diseases. Front. Immunol. 2020, 11, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugahara, H.; Odamaki, T.; Hashikura, N.; Abe, F.; Xiao, J.-Z. Differences in folate production by bifidobacteria of different origins. Biosci. Microbiota Food Health 2015, 34, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Early Development of the Gut Microbiota and Immune Health. Pathogens 2014, 3, 769–790. [Google Scholar] [CrossRef] [Green Version]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef] [Green Version]
- Gensollen, T.; Blumberg, R.S. Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 2017, 139, 1084–1091. [Google Scholar] [CrossRef] [Green Version]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Shu, S.-A.; Yuen, A.W.T.; Woo, E.; Chu, K.-H.; Kwan, H.-S.; Yang, G.-X.; Yang, Y.; Leung, P.S.C. Microbiota and Food Allergy. Clin. Rev. Allergy Immunol. 2019, 57, 83–97. [Google Scholar] [CrossRef]
- Kim, J.E.; Kim, H.S. Microbiome of the Skin and Gut in Atopic Dermatitis (AD): Understanding the Pathophysiology and Finding Novel Management Strategies. J. Clin. Med. Res. 2019, 8, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barcik, W.; Boutin, R.C.T.; Sokolowska, M.; Finlay, B.B. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020, 52, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokholm, J.; Thorsen, J.; Chawes, B.L.; Schjørring, S.; Krogfelt, K.A.; Bønnelykke, K.; Bisgaard, H. Cesarean section changes neonatal gut colonization. J. Allergy Clin. Immunol. 2016, 138, 881–889.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaterian, N.; Abdi, F.; Ghavidel, N.; Alidost, F. Role of cesarean section in the development of neonatal gut microbiota: A systematic review. Open Med. 2021, 16, 624–639. [Google Scholar] [CrossRef]
- Kim, G.; Bae, J.; Kim, M.J.; Kwon, H.; Park, G.; Kim, S.-J.; Choe, Y.H.; Kim, J.; Park, S.-H.; Choe, B.-H.; et al. Delayed Establishment of Gut Microbiota in Infants Delivered by Cesarean Section. Front. Microbiol. 2020, 11, 2099. [Google Scholar] [CrossRef]
- Ma, J.; Li, Z.; Zhang, W.; Zhang, C.; Zhang, Y.; Mei, H.; Zhuo, N.; Wang, H.; Wang, L.; Wu, D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: A study of 91 term infants. Sci. Rep. 2020, 10, 15792. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, X.-G.; Wang, J.; Chen, Y.-J.; Qin, H.-L.; Yang, R. Impact of probiotics supplement on the gut microbiota in neonates with antibiotic exposure: An open-label single-center randomized parallel controlled study. World J. Pediatr. 2021, 17, 385–393. [Google Scholar] [CrossRef]
- Yousuf, E.I.; Carvalho, M.; Dizzell, S.E.; Kim, S.; Gunn, E.; Twiss, J.; Giglia, L.; Stuart, C.; Hutton, E.K. Persistence of Suspected Probiotic Organisms in Preterm Infant Gut Microbiota Weeks After Probiotic Supplementation in the NICU. Front. Microbiol. 2020, 11, 574137. [Google Scholar] [CrossRef]
- Berardi, A.; Rossi, K.; Pizzi, C.; Baronciani, D.; Venturelli, C.; Ferrari, F.; Facchinetti, F. Absence of neonatal streptococcal colonization after planned cesarean section. Acta Obstet. Gynecol. Scand. 2006, 85, 1012–1013. [Google Scholar] [CrossRef]
- Wilson, B.C.; Butler, M.; Grigg, C.P.; Derraik, J.G.B.; Chiavaroli, V.; Walker, N.; Thampi, S.; Creagh, C.; Reynolds, A.J.; Vatanen, T.; et al. Oral administration of maternal vaginal microbes at birth to restore gut microbiome development in infants born by caesarean section: A pilot randomised placebo-controlled trial. eBioMedicine 2021, 69, 103443. [Google Scholar] [CrossRef]
- Korpela, K.; Helve, O.; Kolho, K.-L.; Saisto, T.; Skogberg, K.; Dikareva, E.; Stefanovic, V.; Salonen, A.; Andersson, S.; de Vos, W.M. Maternal Fecal Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell 2020, 183, 324–334.e5. [Google Scholar] [CrossRef] [PubMed]
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86 (Suppl. 1), 13–15. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Feldman, A.S.; Rosas-Salazar, C.; James, K.; Escobar, G.; Gebretsadik, T.; Li, S.X.; Carroll, K.N.; Walsh, E.; Mitchel, E.; et al. Relative Importance and Additive Effects of Maternal and Infant Risk Factors on Childhood Asthma. PLoS ONE 2016, 11, e0151705. [Google Scholar] [CrossRef]
- Xu, B.; Pekkanen, J.; Jarvelin, M.R.; Olsen, P.; Hartikainen, A.L. Maternal infections in pregnancy and the development of asthma among offspring. Int. J. Epidemiol. 1999, 28, 723–727. [Google Scholar] [CrossRef] [Green Version]
- Sly, P.D. Maternal Asthma, Pregnancy Complications, and Offspring Wheeze. Untangling the Web. Am. J. Respir. Crit. Care Med. 2019, 199, 1–2. [Google Scholar] [CrossRef]
- Blaser, M.J.; Bello, M.G.D. Maternal antibiotic use and risk of asthma in offspring. Lancet Respir. Med. 2014, 2, e16. [Google Scholar] [CrossRef]
- Loewen, K.; Monchka, B.; Mahmud, S.M.; Azad, M.B.; Jong, G. Prenatal antibiotic exposure and childhood asthma: A population-based study. Eur. Respir. J. 2018, 52, 1702070. [Google Scholar] [CrossRef]
- Gray, L.E.K.; O’Hely, M.; Ranganathan, S.; Sly, P.D.; Vuillermin, P. The Maternal Diet, Gut Bacteria, and Bacterial Metabolites during Pregnancy Influence Offspring Asthma. Front. Immunol. 2017, 8, 365. [Google Scholar] [CrossRef] [Green Version]
- Viljoen, K.; Segurado, R.; O’Brien, J.; Murrin, C.; Mehegan, J.; Kelleher, C.C. Pregnancy diet and offspring asthma risk over a 10-year period: The Lifeways Cross Generation Cohort Study, Ireland. BMJ Open 2018, 8, e017013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polloni, L.; Ferruzza, E.; Ronconi, L.; Lazzarotto, F.; Toniolo, A.; Bonaguro, R.; Muraro, A. Perinatal stress and food allergy: A preliminary study on maternal reports. Psychol. Health Med. 2015, 20, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Mägi, C.-A.O.; Bäcklund, A.B.; Carlsen, K.L.; Almqvist, C.; Carlsen, K.-H.; Granum, B.; Haugen, G.; Hilde, K.; Carlsen, O.C.L.; Jonassen, C.M.; et al. Allergic disease and risk of stress in pregnant women: A PreventADALL study. ERJ Open Res. 2020, 6, 00175-2020. [Google Scholar] [CrossRef]
- Dioun, A.F.; Harris, S.K.; Hibberd, P.L. Is maternal age at delivery related to childhood food allergy? Pediatr. Allergy Immunol. 2003, 14, 307–311. [Google Scholar] [CrossRef]
- Saito, K.; Yokoyama, T.; Miyake, Y.; Sasaki, S.; Tanaka, K.; Ohya, Y.; Hirota, Y. Maternal meat and fat consumption during pregnancy and suspected atopic eczema in Japanese infants aged 3-4 months: The Osaka Maternal and Child Health Study. Pediatr. Allergy Immunol. 2010, 21, 38–46. [Google Scholar] [CrossRef]
- Sestito, S.; D’Auria, E.; Baldassarre, M.E.; Salvatore, S.; Tallarico, V.; Stefanelli, E.; Tarsitano, F.; Concolino, D.; Pensabene, L. The Role of Prebiotics and Probiotics in Prevention of Allergic Diseases in Infants. Front. Pediatr. 2020, 8, 583946. [Google Scholar] [CrossRef]
- Schäfer, S.; Liu, A.; Campbell, D.; Nanan, R. Analysis of maternal and perinatal determinants of allergic sensitization in childhood. Allergy Asthma Clin. Immunol. 2020, 16, 71. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [Green Version]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: Mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Simpson, M.R.; Avershina, E.; Storrø, O.; Johnsen, R.; Rudi, K.; Øien, T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J. Dairy Sci. 2018, 101, 889–899. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe 2018, 24, 133–145.e5. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Xu, L.; Qian, Y.; Sun, Z.; Yu, D.; Huang, J.; Zhou, X.; Wang, Y.; Zhang, T.; Ren, R.; et al. Evolution of the Gut Microbiome in Early Childhood: A Cross-Sectional Study of Chinese Children. Front. Microbiol. 2020, 11, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenplas, Y.; Carnielli, V.P.; Ksiazyk, J.; Luna, M.S.; Migacheva, N.; Mosselmans, J.M.; Picaud, J.C.; Possner, M.; Singhal, A.; Wabitsch, M. Factors affecting early-life intestinal microbiota development. Nutrition 2020, 78, 110812. [Google Scholar] [CrossRef] [PubMed]
- Hoyen, C. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Yearb. Neonatal Périnat. Med. 2007, 2007, 202–203. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; Van den Brandt, P.A.; Stobberingh, E.E. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics 2006, 118, 511–521. [Google Scholar] [CrossRef] [Green Version]
- Dierikx, T.H.; Visser, D.H.; Benninga, M.A.; van Kaam, A.H.L.C.; de Boer, N.K.H.; de Vries, R.; Van Limbergen, J.; de Meij, T.G.J. The influence of prenatal and intrapartum antibiotics on intestinal microbiota colonisation in infants: A systematic review. J. Infect. 2020, 81, 190–204. [Google Scholar] [CrossRef]
- Milliken, S.; Allen, R.M.; Lamont, R.F. The role of antimicrobial treatment during pregnancy on the neonatal gut microbiome and the development of atopy, asthma, allergy and obesity in childhood. Expert Opin. Drug Saf. 2019, 18, 173–185. [Google Scholar] [CrossRef]
- Panduru, M.; Epure, A.M.; Cimpoca, B.; Cozma, C.; Giuca, B.A.; Pop, A.; Pop, G.; Simon, L.G.; Robu, M.; Panduru, N.M. Antibiotics administration during last trimester of pregnancy is associated with atopic dermatitis—A cross-sectional study. Rom. J. Intern. Med. 2020, 58, 99–107. [Google Scholar] [CrossRef]
- Stensballe, L.G.; Simonsen, J.; Jensen, S.M.; Bønnelykke, K.; Bisgaard, H. Use of Antibiotics during Pregnancy Increases the Risk of Asthma in Early Childhood. J. Pediatr. 2013, 162, 832–838.e3. [Google Scholar] [CrossRef]
- Geng, M.; Tang, Y.; Liu, K.; Huang, K.; Yan, S.; Ding, P.; Zhang, J.; Wang, B.; Wang, S.; Li, S.; et al. Prenatal low-dose antibiotic exposure and children allergic diseases at 4 years of age: A prospective birth cohort study. Ecotoxicol. Environ. Saf. 2021, 225, 112736. [Google Scholar] [CrossRef]
- Junca, H.; Pieper, D.H.; Medina, E. The emerging potential of microbiome transplantation on human health interventions. Comput. Struct. Biotechnol. J. 2022, 20, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Grier, A.; McDavid, A.; Wang, B.; Qiu, X.; Java, J.; Bandyopadhyay, S.; Yang, H.; Holden-Wiltse, J.; Kessler, H.A.; Gill, A.L.; et al. Neonatal gut and respiratory microbiota: Coordinated development through time and space. Microbiome 2018, 6, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The Human Microbiome and Child Growth—First 1000 Days and Beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, I.; Corwin, E.J.; Brennan, P.A.; Jordan, S.; Murphy, J.R.; Dunlop, A. The Infant Microbiome: Implications for Infant Health and Neurocognitive Development. Nurs. Res. 2016, 65, 76–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aversa, Z.; Atkinson, E.J.; Schafer, M.J.; Theiler, R.N.; Rocca, W.A.; Blaser, M.J.; LeBrasseur, N.K. Association of Infant Antibiotic Exposure with Childhood Health Outcomes. Mayo Clin. Proc. 2021, 96, 66–77. [Google Scholar] [CrossRef]
- Moreau, M.C.; Ducluzeau, R.; Guy-Grand, D.; Muller, M.C. Increase in the Population of Duodenal Immunoglobulin A Plasmocytes in Axenic Mice Associated with Different Living or Dead Bacterial Strains of Intestinal Origin. Infect. Immun. 1978, 21, 532–539. [Google Scholar] [CrossRef] [Green Version]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [Green Version]
- Baba, N.; Samson, S.; Bourdet-Sicard, R.; Rubio, M.; Sarfati, M. Commensal bacteria trigger a full dendritic cell maturation program that promotes the expansion of non-Tr1 suppressor T cells. J. Leukoc. Biol. 2008, 84, 468–476. [Google Scholar] [CrossRef]
- Fulde, M.; Hornef, M.W. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol. Rev. 2014, 260, 21–34. [Google Scholar] [CrossRef]
- Wesemann, D.R.; Portuguese, A.J.; Meyers, R.M.; Gallagher, M.P.; Cluff-Jones, K.; Magee, J.M.; Panchakshari, R.A.; Rodig, S.J.; Kepler, T.B.; Alt, F.W. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 2013, 501, 112–115. [Google Scholar] [CrossRef]
- Inman, C.F.; Laycock, G.M.; Mitchard, L.; Harley, R.; Warwick, J.; Burt, R.; Van Diemen, P.M.; Stevens, M.; Bailey, M. Neonatal Colonisation Expands a Specific Intestinal Antigen-Presenting Cell Subset Prior to CD4 T-Cell Expansion, without Altering T-Cell Repertoire. PLoS ONE 2012, 7, e33707. [Google Scholar] [CrossRef] [PubMed]
- Okogbule-Wonodi, A.C.; Li, G.; Anand, B.; Luzina, I.G.; Atamas, S.P.; Blanchard, T. Human foetal intestinal fibroblasts are hyper-responsive to lipopolysaccharide stimulation. Dig. Liver Dis. 2012, 44, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Sansonetti, P.J. To be or not to be a pathogen: That is the mucosally relevant question. Mucosal Immunol. 2011, 4, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, S.; Reizis, B.; Ravindran, R.; Nakaya, H.; Salazar-Gonzalez, R.M.; Wang, Y.-C.; Pulendran, B. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science 2010, 329, 849–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchi, L.; Kamada, N.; Nakamura, Y.; Burberry, A.; Kuffa, P.; Suzuki, S.; Shaw, M.H.; Kim, Y.-G.; Núñez, G. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 2012, 13, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Ni, J.; Friedman, H.; Boyd, B.C.; McGurn, A.; Babinski, P.; Markossian, T.; Dugas, L.R. Early antibiotic exposure and development of asthma and allergic rhinitis in childhood. BMC Pediatr. 2019, 19, 225. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, C.; Yang, A.; Zhang, R.; Gong, J.; Mo, F. Is preterm birth associated with asthma among children from birth to 17 years old? A study based on 2011-2012 US National Survey of Children’s Health. Ital. J. Pediatr. 2018, 44, 151. [Google Scholar] [CrossRef] [Green Version]
- El-Heneidy, A.; Abdel-Rahman, M.E.; Mihala, G.; Ross, L.J.; Comans, T.A. Milk Other Than Breast Milk and the Development of Asthma in Children 3 Years of Age. A Birth Cohort Study (2006–2011). Nutrients 2018, 10, 1798. [Google Scholar] [CrossRef] [Green Version]
- Roduit, C.; Scholtens, S.; De Jongste, J.C.; Wijga, A.H.; Gerritsen, J.; Postma, D.S.; Brunekreef, B.; Hoekstra, M.O.; Aalberse, R.; Smit, H.A. Asthma at 8 years of age in children born by caesarean section. Thorax 2009, 64, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, S.; Israeli, E.; Schwartz, S.; Shochat, T.; Izbicki, G.; Toker-Maimon, O.; Klement, E.; Picard, E. Asthma prevalence, family size, and birth order. Chest 2007, 131, 1747–1752. [Google Scholar] [CrossRef] [PubMed]
- Andersén, H.; Ilmarinen, P.; Honkamäki, J.; Tuomisto, L.E.; Hisinger-Mölkänen, H.; Backman, H.; Lundbäck, B.; Rönmark, E.; Lehtimäki, L.; Sovijärvi, A.; et al. Influence of Childhood Exposure to a Farming Environment on Age at Asthma Diagnosis in a Population-Based Study. J. Asthma Allergy 2021, 14, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Wooldridge, A.L.; McMillan, M.; Kaur, M.; Giles, L.C.; Marshall, H.S.; Gatford, K.L. Relationship between birth weight or fetal growth rate and postnatal allergy: A systematic review. J. Allergy Clin. Immunol. 2019, 144, 1703–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathias, J.M.G.; Zhang, H.; Karmaus, W.; Soto-Ramirez, N.; Shen, Y. Mixed Infant Feeding—Direct Breastfeeding, Pumping and Feeding, and Formula Food Poses a Risk for Food Allergy in Early Childhood. J. Allergy Clin. Immunol. 2017, 139, AB385. [Google Scholar] [CrossRef] [Green Version]
- Christmann, B.S.; Abrahamsson, T.R.; Bernstein, C.N.; Duck, L.W.; Mannon, P.J.; Berg, G.; Björkstén, B.; Jenmalm, M.C.; Elson, C.O. Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol. 2015, 136, 1378–1386.e5. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- West, C.E.; Rydén, P.; Lundin, D.; Engstrand, L.; Tulic, M.K.; Prescott, S.L. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin. Exp. Allergy 2015, 45, 1419–1429. [Google Scholar] [CrossRef] [Green Version]
- Yiu, J.H.C.; Dorweiler, B.; Woo, C.W. Interaction between gut microbiota and toll-like receptor: From immunity to metabolism. J. Mol. Med. 2017, 95, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Liu, W.; Zhang, L.-J. The Role of Toll-Like Receptors in Skin Host Defense, Psoriasis, and Atopic Dermatitis. J. Immunol. Res. 2019, 2019, 1824624. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; LeVan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [Green Version]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.-M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.L.; et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prescott, S.L. Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. J. Allergy Clin. Immunol. 2013, 131, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; Lieber, A.D.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef] [Green Version]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Geervliet, M. A Window of Opportunity: Modulation of the Porcine Gut Microbiota and Immune System by Feed Additives in Early Life. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2021. [Google Scholar] [CrossRef]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [Green Version]
- Bisgaard, H.; Li, N.; Bonnelykke, K.; Chawes, B.L.K.; Skov, T.; Paludan-Müller, G.; Stokholm, J.; Smith, B.; Krogfelt, K.A. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011, 128, 646–652.e5. [Google Scholar] [CrossRef]
- Nowak-Wegrzyn, A. Allergy Development and the Intestinal Microflora During the First Year of Life. Pediatrics 2002, 110, 431. [Google Scholar] [CrossRef]
- Ege, M.J.; Mayer, M.; Normand, A.-C.; Genuneit, J.; Cookson, W.O.C.M.; Braun-Fahrländer, C.; Heederik, D.; Piarroux, R.; von Mutius, E. GABRIELA Transregio 22 Study Group. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 2011, 364, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Gerhold, K.; Thijs, C.; Zimmermann, K.; Wahn, U.; Lau, S.; Hamelmann, E. New insights into the hygiene hypothesis in allergic diseases: Mediation of sibling and birth mode effects by the gut microbiota. Gut Microbes 2014, 5, 239–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef]
- Smart, B.A. Childhood Asthma After Bacterial Colonization of the Airway in Neonates. Pediatrics 2008, 122, S206–S207. [Google Scholar] [CrossRef] [Green Version]
- Mansbach, J.M.; Luna, P.N.; Shaw, C.A.; Hasegawa, K.; Petrosino, J.F.; Piedra, P.A.; Sullivan, A.F.; Espinola, J.A.; Stewart, C.; Camargo, C.A., Jr. Increased Moraxella and Streptococcus species abundance after severe bronchiolitis is associated with recurrent wheezing. J. Allergy Clin. Immunol. 2020, 145, 518–527.e8. [Google Scholar] [CrossRef] [Green Version]
- Thorsen, J.; Rasmussen, M.A.; Waage, J.; Mortensen, M.; Brejnrod, A.; Bønnelykke, K.; Chawes, B.L.; Brix, S.; Sørensen, S.J.; Stokholm, J.; et al. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat. Commun. 2019, 10, 5001. [Google Scholar] [CrossRef] [Green Version]
- Ranucci, G.; Buccigrossi, V.; De Freitas, M.B.; Guarino, A.; Giannattasio, A. Early-Life Intestine Microbiota and Lung Health in Children. J. Immunol. Res. 2017, 2017, 8450496. [Google Scholar] [CrossRef]
- Hammad, H.; Chieppa, M.; Perros, F.; Willart, M.A.; Germain, R.N.; Lambrecht, B.N. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 2009, 15, 410–416. [Google Scholar] [CrossRef] [Green Version]
- Schuijs, M.J.; Willart, M.A.; Vergote, K.; Gras, D.; Deswarte, K.; Ege, M.J.; Madeira, F.B.; Beyaert, R.; van Loo, G.; Bracher, F.; et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 2015, 349, 1106–1110. [Google Scholar] [CrossRef]
- Le Noci, V.; Guglielmetti, S.; Tagliabue, E.; Sfondrini, L.; Arioli, S.; Camisaschi, C.; Bianchi, F.; Sommariva, M.; Storti, C.; Triulzi, T.; et al. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: A strategy to promote immunosurveillance against lung metastases. Cell Rep. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cicco, M.; Pistello, M.; Jacinto, T.; Ragazzo, V.; Piras, M.; Freer, G.; Pifferi, M.; Peroni, D. Does lung microbiome play a causal or casual role in asthma? Pediatr. Pulmonol. 2018, 53, 1340–1345. [Google Scholar] [CrossRef] [PubMed]
- Gollwitzer, E.S.; Saglani, S.; Trompette, A.; Yadava, K.; Sherburn, R.; McCoy, K.D.; Nicod, L.P.; Lloyd, C.M.; Marsland, B.J. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 2014, 20, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Genuneit, J. Exposure to farming environments in childhood and asthma and wheeze in rural populations: A systematic review with meta-analysis. Pediatr. Allergy Immunol. 2012, 23, 509–518. [Google Scholar] [CrossRef]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Mulder, B.; Pouwels, K.B.; Schuiling-Veninga, C.C.M.; Bos, H.J.; De Vries, T.W.; Jick, S.S.; Hak, E. Antibiotic use during pregnancy and asthma in preschool children: The influence of confounding. Clin. Exp. Allergy 2016, 46, 1214–1226. [Google Scholar] [CrossRef]
- Zhao, D.; Su, H.; Cheng, J.; Wang, X.; Xie, M.; Li, K.; Wen, L.; Yang, H. Prenatal antibiotic use and risk of childhood wheeze/asthma: A meta-analysis. Pediatr. Allergy Immunol. 2015, 26, 756–764. [Google Scholar] [CrossRef]
- Yoshida, S.; Ide, K.; Takeuchi, M.; Kawakami, K. Prenatal and early-life antibiotic use and risk of childhood asthma: A retrospective cohort study. Pediatr. Allergy Immunol. 2018, 29, 490–495. [Google Scholar] [CrossRef]
- Alhasan, M.M.; Cait, A.M.; Heimesaat, M.M.; Blaut, M.; Klopfleisch, R.; Wedel, A.; Conlon, T.M.; Yildirim, A.; Sodemann, E.B.; Mohn, W.W.; et al. Antibiotic use during pregnancy increases offspring asthma severity in a dose-dependent manner. Allergy 2020, 75, 1979–1990. [Google Scholar] [CrossRef] [Green Version]
- Durack, J.; Kimes, N.E.; Lin, D.L.; Rauch, M.; Mckean, M.; McCauley, K.; Panzer, A.R.; Mar, J.S.; Cabana, M.D.; Lynch, S.V. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 2018, 9, 707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durack, J.; Kimes, N.E.; Lin, D.; Mckean, M.; Rauch, M.; Cabana, M.D.; Lynch, S. Early-life Lactobacillus rhamnosus GG Supplementation of High-risk for Asthma Infants Reprograms Gut Microbiota Development and promotes regulatory T-cells. J. Allergy Clin. Immunol. 2017, 139, AB15. [Google Scholar] [CrossRef] [Green Version]
- Calder, P. Faculty of 1000 evaluation for Effects of early prebiotic and probiotic supplementation on development of gut microbiota and fussing and crying in preterm infants: A randomized, double-blind, placebo-controlled trial. F1000Prime Rep. 2013, 6, 793482506. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Cunnington, A.J.; Sim, K.; Deierl, A.; Kroll, J.S.; Brannigan, E.; Darby, J. “Vaginal seeding” of infants born by caesarean section. BMJ 2016, 352, i227. [Google Scholar] [CrossRef] [Green Version]
- Galazzo, G.; van Best, N.; Bervoets, L.; Dapaah, I.O.; Savelkoul, P.H.; Hornef, M.W.; Lau, S.; Hamelmann, E.; Penders, J.; Hutton, E.K.; et al. Development of the Microbiota and Associations with Birth Mode, Diet, and Atopic Disorders in a Longitudinal Analysis of Stool Samples, Collected from Infancy Through Early Childhood. Gastroenterology 2020, 158, 1584–1596. [Google Scholar] [CrossRef]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 2014, 44, 842–850. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Nanan, R.; Macia, L.; Tan, J.; Sominsky, L.; Quinn, T.P.; O’Hely, M.; Ponsonby, A.-L.; Tang, M.L.K.; Collier, F.; et al. The maternal gut microbiome during pregnancy and offspring allergy and asthma. J. Allergy Clin. Immunol. 2021, 148, 669–678. [Google Scholar] [CrossRef]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519–1528. [Google Scholar] [CrossRef]
- Udkoff, J.; Waldman, A.; Ahluwalia, J.; Borok, J.; Eichenfield, L.F. Current and emerging topical therapies for atopic dermatitis. Clin. Dermatol. 2017, 35, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Brunner, P.M.; Leung, D.Y.M.; Guttman-Yassky, E. Immunologic, microbial, and epithelial interactions in atopic dermatitis. Ann. Allergy Asthma Immunol. 2018, 120, 34–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wollina, U. Microbiome in atopic dermatitis. Clin. Cosmet. Investig. Dermatol. 2017, 10, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, J.; Li, H.Y. The human skin microbiome. In The Human Skin Microbiome; CABI: Wallingford, UK, 2014; pp. 72–89. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Baurecht, H.; Rühlemann, M.C.; Rodríguez, E.; Thielking, F.; Harder, I.; Erkens, A.-S.; Stölzl, D.; Ellinghaus, E.; Hotze, M.; Lieb, W.; et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J. Allergy Clin. Immunol. 2018, 141, 1668–1676.e16. [Google Scholar] [CrossRef] [Green Version]
- Geoghegan, J.A.; Irvine, A.D.; Foster, T.J. Staphylococcus aureus and Atopic Dermatitis: A Complex and Evolving Relationship. Trends Microbiol. 2018, 26, 484–497. [Google Scholar] [CrossRef]
- Watanabe, S.; Narisawa, Y.; Arase, S.; Okamatsu, H.; Ikenaga, T.; Tajiri, Y.; Kumemura, M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 2003, 111, 587–591. [Google Scholar] [CrossRef]
- Gołębiewski, M.; Łoś-Rycharska, E.; Sikora, M.; Grzybowski, T.; Gorzkiewicz, M.; Krogulska, A. Mother’s Milk Microbiome Shaping Fecal and Skin Microbiota in Infants with Food Allergy and Atopic Dermatitis: A Pilot Analysis. Nutrients 2021, 13, 3600. [Google Scholar] [CrossRef]
- Park, Y.M.; Lee, S.Y.; Kang, M.J.; Kim, B.S.; Lee, M.J.; Jung, S.S.; Yoon, J.S.; Cho, H.J.; Lee, E.; Yang, S.I.; et al. Imbalance of Gut, and Determines the Natural Course of Atopic Dermatitis in Infant. Allergy Asthma Immunol. Res. 2020, 12, 322–337. [Google Scholar] [CrossRef]
- Cukrowska, B.; Bierła, J.B.; Zakrzewska, M.; Klukowski, M.; Maciorkowska, E. The Relationship between the Infant Gut Microbiota and Allergy. The Role of Bifidobacterium breve and Prebiotic Oligosaccharides in the Activation of Anti-Allergic Mechanisms in Early Life. Nutrients 2020, 12, 946. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.M.; Muraro, A.; Fleisher, T.A. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017, 139, 1099–1110. [Google Scholar] [PubMed] [Green Version]
- Malhotra, N.; Yoon, J.; Leyva-Castillo, J.M.; Galand, C.; Archer, N.; Miller, L.S.; Geha, R.S. IL-22 derived from γδ T cells restricts Staphylococcus aureus infection of mechanically injured skin. J. Allergy Clin. Immunol. 2016, 138, 1098–1107.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, E.A.; Connolly, J.; Hourihane, J.O.; Fallon, P.G.; McLean, W.H.I.; Murray, D.; Jo, J.-H.; Segre, J.A.; Kong, H.H.; Irvine, A.D. Skin microbiome before development of atopic dermatitis: Early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J. Allergy Clin. Immunol. 2017, 139, 166–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salava, A.; Lauerma, A. Role of the skin microbiome in atopic dermatitis. Clin. Transl. Allergy 2014, 4, 33. [Google Scholar] [CrossRef] [Green Version]
- Laubereau, B.; Filipiak-Pittroff, B.; von Berg, A.; Grübl, A.; Reinhardt, D.; Wichmann, H.E.; Koletzko, S.; GINI Study Group. Caesarean section and gastrointestinal symptoms, atopic dermatitis, and sensitisation during the first year of life. Arch. Dis. Child 2004, 89, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Richards, M.; Ferber, J.; Chen, H.; Swor, E.; Quesenberry, C.P.; Li, D.-K.; Darrow, L.A. Caesarean delivery and the risk of atopic dermatitis in children. Clin. Exp. Allergy 2020, 50, 805–814. [Google Scholar] [CrossRef]
- Lasley, M.V. Caesarean Delivery and the Risk of Atopic Dermatitis in Children. Pediatrics 2021, 148, S5–S6. [Google Scholar] [CrossRef]
- Shin, H.; Pei, Z.; Martinez, K.A., 2nd; Rivera-Vinas, J.I.; Mendez, K.; Cavallin, H.; Dominguez-Bello, M.G. The first microbial environment of infants born by C-section: The operating room microbes. Microbiome 2015, 3, 59. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, A.J.; Mills, B.W.; Shi, V.Y. Skin bacterial transplant in atopic dermatitis: Knowns, unknowns and emerging trends. J. Dermatol. Sci. 2019, 95, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Craig, J.M. Atopic dermatitis and the intestinal microbiota in humans and dogs. Vet. Med. Sci. 2016, 2, 95–105. [Google Scholar] [CrossRef]
- Zuo, W.; Sun, C. The Role of the Intestinal Microbiota in Atopic Dermatitis. Int. J. Dermatol. Venereol. 2021. ahead of print. [Google Scholar] [CrossRef]
- Penders, J.; Thijs, C.; van den Brandt, P.A.; Kummeling, I.; Snijders, B.; Stelma, F.; Adams, H.; van Ree, R.; Stobberingh, E.E. Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut 2007, 56, 661–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 2012, 129, 434–440.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forno, E.; Onderdonk, A.; McCracken, J.; Litonjua, A.; Laskey, D.; Delaney, M.; Dubois, A.; Gold, D.R.; Ryan, L.; Weiss, S.; et al. Diversity of the Gut Microbiota and Eczema in Infants. Am. J. Respir. Crit. Care Med. 2009, 179, A5981. [Google Scholar] [CrossRef]
- Rø, A.D.B.; Rø, T.B.; Storrø, O.; Johnsen, R.; Videm, V.; Øien, T.; Simpson, M.R. Reduced Th22 cell proportion and prevention of atopic dermatitis in infants following maternal probiotic supplementation. Clin. Exp. Allergy 2017, 47, 1014–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-H.; Kim, K.; Kim, W. Gut microbiota restoration through fecal microbiota transplantation: A new atopic dermatitis therapy. Exp. Mol. Med. 2021, 53, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Mashiah, J.; Karady, T.; Fliss-Isakov, N.; Sprecher, E.; Slodownik, D.; Artzi, O.; Samuelov, L.; Ellenbogen, E.; Godneva, A.; Segal, E.; et al. Clinical efficacy of fecal microbial transplantation treatment in adults with moderate-to-severe atopic dermatitis. Immun. Inflamm. Dis. 2022, 10, e570. [Google Scholar] [CrossRef] [PubMed]
- Allam, J.-P.; Duan, Y.; Winter, J.; Stojanovski, G.; Fronhoffs, F.; Wenghoefer, M.; Bieber, T.; Peng, W.-M.; Novak, N. Tolerogenic T cells, Th1/Th17 cytokines and TLR2/TLR4 expressing dendritic cells predominate the microenvironment within distinct oral mucosal sites. Allergy 2011, 66, 532–539. [Google Scholar] [CrossRef]
- Moingeon, P. Update on Immune Mechanisms Associated with Sublingual Immunotherapy: Practical Implications for the Clinician. J. Allergy Clin. Immunol. Pract. 2013, 1, 228–241. [Google Scholar] [CrossRef]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Fiscella, K.A.; Gill, S.R. Oral microbiome: Possible harbinger for children’s health. Int. J. Oral Sci. 2020, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Lif Holgerson, P.; Harnevik, L.; Hernell, O.; Tanner, A.C.R.; Johansson, I. Mode of birth delivery affects oral microbiota in infants. J. Dent. Res. 2011, 90, 1183–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, R.; Yama, K.; Aita, Y.; Tsutsumi, K.; Ishihara, C.; Maruyama, M.; Takeda, K.; Nishinaga, E.; Shibasaki, K.-I.; Morishima, S. Comparison of oral microbiome profiles in 18-month-old infants and their parents. Sci. Rep. 2021, 11, 861. [Google Scholar] [CrossRef] [PubMed]
- Von Mutius, E.; Vercelli, D. Farm living: Effects on childhood asthma and allergy. Nat. Rev. Immunol. 2010, 10, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.; Chinn, S.; Luczynska, C.; Burney, P. The association of family size with atopy and atopic disease. Clin. Exp. Allergy 1997, 27, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Mortha, A.; Chudnovskiy, A.; Hashimoto, D.; Bogunovic, M.; Spencer, S.P.; Belkaid, Y.; Merad, M. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science 2014, 343, 1249288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuillermin, P.J.; Macia, L.; Nanan, R.; Tang, M.L.; Collier, F.; Brix, S. The maternal microbiome during pregnancy and allergic disease in the offspring. Semin. Immunopathol. 2017, 39, 669–675. [Google Scholar] [CrossRef] [Green Version]
- Vuillermin, P.J.; O’Hely, M.; Collier, F.; Allen, K.J.; Tang, M.L.K.; Harrison, L.C.; Carlin, J.B.; Saffery, R.; Ranganathan, S.; Sly, P.D.; et al. Maternal carriage of Prevotella during pregnancy associates with protection against food allergy in the offspring. Nat. Commun. 2020, 11, 1452. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wei, Y.; Liu, L.; Li, Z. Association Between Breastmilk Microbiota and Food Allergy in Infants. Front. Cell. Infect. Microbiol. 2022, 11, 770913. [Google Scholar] [CrossRef]
- Thompson-Chagoyán, O.C.; Vieites, J.M.; Maldonado, J.; Edwards, C.; Gil, A. Changes in faecal microbiota of infants with cow’s milk protein allergy—A Spanish prospective case-control 6-month follow-up study. Pediatr. Allergy Immunol. 2010, 21, e394–e400. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.; Bunyavanich, S.; Zhou, Y.; O’Connor, G.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E.; et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018, 73, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Goedert, J.J.; Pu, A.; Yu, G.; Shi, J. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project. eBioMedicine 2016, 3, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Hong, S.M.C.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Lyons, A.; O’Mahony, D.; O’Brien, F.; MacSharry, J.; Sheil, B.; Ceddia, M.; Russell, W.M.; Forsythe, P.; Bienenstock, J.; Kiely, B.; et al. Bacterial strain-specific induction of Foxp3+T regulatory cells is protective in murine allergy models. Clin. Exp. Allergy 2010, 40, 811–819. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.-Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef] [Green Version]
- Berni Canani, R.; Sangwan, N.; Stefka, A.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef]
- Guest, J.F.; Fuller, G.W. Effectiveness of using an extensively hydrolyzed casein formula supplemented with Lactobacillus rhamnosus GG compared with an extensively hydrolysed whey formula in managing cow’s milk protein allergic infants. J. Comp. Eff. Res. 2019, 8, 1317–1326. [Google Scholar] [CrossRef] [Green Version]
- Luu, M.; Visekruna, A. Short-chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur. J. Immunol. 2019, 49, 842–848. [Google Scholar] [CrossRef] [Green Version]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450, Erratum in Nature, 2014, 506, 254. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.-M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cait, A.; Cardenas, E.; Dimitriu, P.A.; Amenyogbe, N.; Dai, D.; Cait, J.; Sbihi, H.; Stiemsma, L.; Subbarao, P.; Mandhane, P.J.; et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J. Allergy Clin. Immunol. 2019, 144, 1638–1647.e3. [Google Scholar] [CrossRef] [Green Version]
- Urschel, D.; Hernandez-Trujillo, V. Butyrate as a Bioactive Human Milk Protective Component Against Food Allergy. Pediatrics 2021, 148, S21–S22. [Google Scholar] [CrossRef]
- Luu, M.; Monning, H.; Visekruna, A. Exploring the Molecular Mechanisms Underlying the Protective Effects of Microbial SCFAs on Intestinal Tolerance and Food Allergy. Front. Immunol. 2020, 11, 1225. [Google Scholar] [CrossRef]
- Di Costanzo, M.; Carucci, L.; Canani, R.B.; Biasucci, G. Gut Microbiome Modulation for Preventing and Treating Pediatric Food Allergies. Int. J. Mol. Sci. 2020, 21, 5275. [Google Scholar] [CrossRef]
- Berni Canani, R.; Di Costanzo, M.; Bedogni, G.; Amoroso, A.; Cosenza, L.; Di Scala, C.; Granata, V.; Nocerino, R. Extensively hydrolyzed casein formula containing Lactobacillus rhamnosus GG reduces the occurrence of other allergic manifestations in children with cow’s milk allergy: 3-year randomized controlled trial. J. Allergy Clin. Immunol. 2017, 139, 1906–1913.e4. [Google Scholar] [CrossRef] [Green Version]
- Wopereis, H.; Van Ampting, M.T.J.; Yavuz, A.C.; Slump, R.; Candy, D.C.A.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; et al. A specific synbiotic-containing amino acid-based formula restores gut microbiota in non-IgE mediated cow’s milk allergic infants: A randomized controlled trial. Clin. Transl. Allergy 2019, 9, 27. [Google Scholar] [CrossRef]
- Fox, A.T.; Wopereis, H.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Candy, D.C.A.; Shah, N.; West, C.E.; et al. A specific synbiotic-containing amino acid-based formula in dietary management of cow’s milk allergy: A randomized controlled trial. Clin. Transl. Allergy 2019, 9, 5. [Google Scholar] [CrossRef]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr. Res. 2018, 83, 677–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, M.L.K.; Ponsonby, A.-L.; Orsini, F.; Tey, D.; Robinson, M.; Su, E.L.; Licciardi, P.; Burks, W.; Donath, S. Administration of a probiotic with peanut oral immunotherapy: A randomized trial. J. Allergy Clin. Immunol. 2015, 135, 737–744.e8. [Google Scholar] [CrossRef] [PubMed]
- Borody, T.J.; Khoruts, A. Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 2011, 9, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal microbiota transplantation: In perspective. Ther. Adv. Gastroenterol. 2015, 9, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuniyazi, M.; Li, S.; Hu, X.; Fu, Y.; Zhang, N. The Role of Early Life Microbiota Composition in the Development of Allergic Diseases. Microorganisms 2022, 10, 1190. https://doi.org/10.3390/microorganisms10061190
Tuniyazi M, Li S, Hu X, Fu Y, Zhang N. The Role of Early Life Microbiota Composition in the Development of Allergic Diseases. Microorganisms. 2022; 10(6):1190. https://doi.org/10.3390/microorganisms10061190
Chicago/Turabian StyleTuniyazi, Maimaiti, Shuang Li, Xiaoyu Hu, Yunhe Fu, and Naisheng Zhang. 2022. "The Role of Early Life Microbiota Composition in the Development of Allergic Diseases" Microorganisms 10, no. 6: 1190. https://doi.org/10.3390/microorganisms10061190
APA StyleTuniyazi, M., Li, S., Hu, X., Fu, Y., & Zhang, N. (2022). The Role of Early Life Microbiota Composition in the Development of Allergic Diseases. Microorganisms, 10(6), 1190. https://doi.org/10.3390/microorganisms10061190






