Investigation of the Phageome and Prophages in French Cider, a Fermented Beverage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cider Sample Collection for Phageome and Prophage Investigation
2.2. Methodological Developments for Collecting Phage Particles from Cider and for Phageome DNA Extraction
2.2.1. Validation of Methods on Cider Samples Spiked with Phages
2.2.2. Application to Non-Spiked Cider Samples
2.2.3. Optimisation of the Method to Collect Phage Particles from Cider
2.3. Microbial DNA Extraction for Prophage Investigation
2.4. DNA Sequencing and Bioinformatics Analysis
2.5. Prophage Prediction
3. Results
3.1. Development of Methods for Phageome DNA Extraction from Cider
3.1.1. Validation of the Extraction Method after Collection of Phage Particles from Spiked Cider Samples
3.1.2. Extraction of Phage Particles from Non-Spiked Cider Samples
3.1.3. Viral Concentration Methods Were Tested to Improve the Phageome DNA Extraction
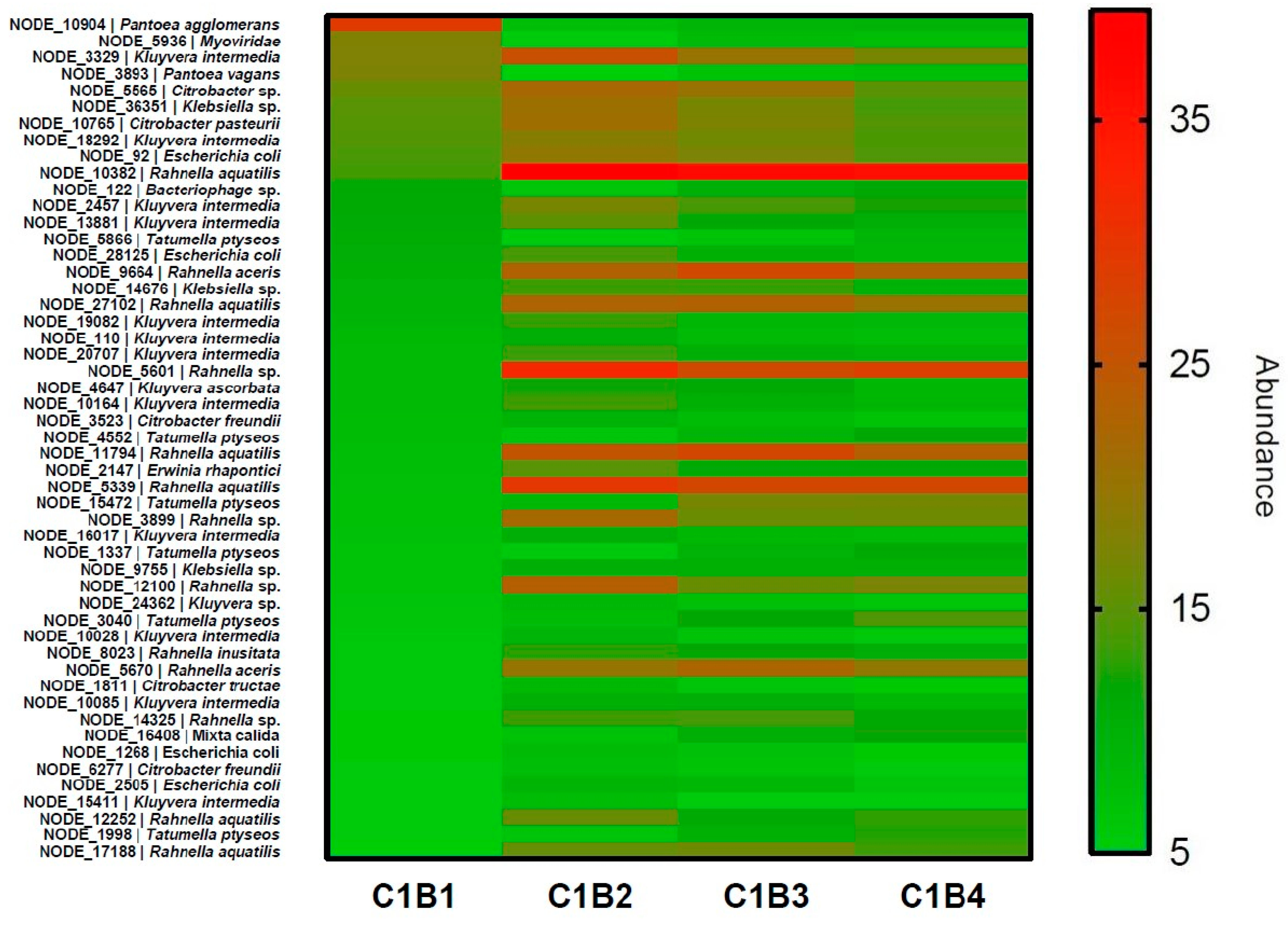
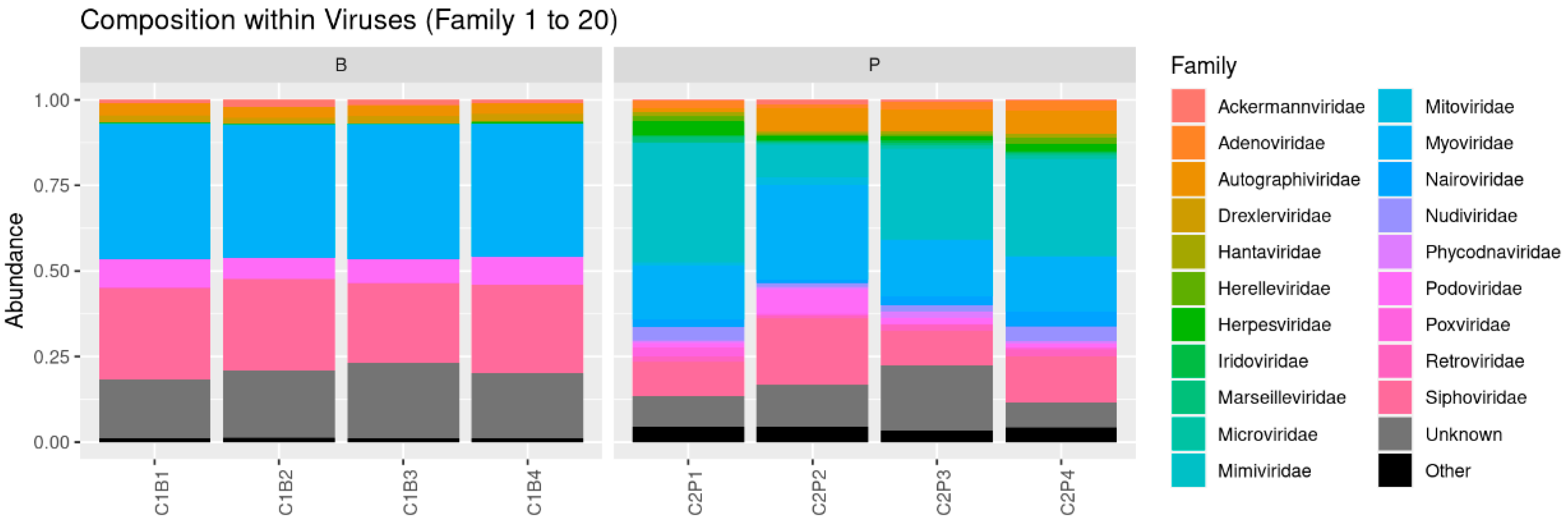
3.2. Prophage Investigation in Two Cider Tanks during Cider Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Manrique, P.; Bolduc, B.; Walk, S.T.; van der Oost, J.; de Vos, W.M.; Young, M.J. Healthy Human Gut Phageome. Proc. Natl. Acad. Sci. USA 2016, 113, 10400–10405. [Google Scholar] [CrossRef] [Green Version]
- Shkoporov, A.N.; Ryan, F.J.; Draper, L.A.; Forde, A.; Stockdale, S.R.; Daly, K.M.; McDonnell, S.A.; Nolan, J.A.; Sutton, T.D.S.; Dalmasso, M.; et al. Reproducible Protocols for Metagenomic Analysis of Human Faecal Phageomes. Mi-Crobiome 2018, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, M.; Hill, C.; Ross, R.P. Exploiting Gut Bacteriophages for Human Health. Trends Microbiol. 2014, 22, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Carr, V.R.; Shkoporov, A.; Gomez-Cabrero, D.; Mullany, P.; Hill, C.; Moyes, D.L. The Human Oral Phageome Is Highly Diverse and Rich in Jumbo Phages. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ma, Y.; Marais, A.; Lefebvre, M.; Theil, S.; Svanella-Dumas, L.; Faure, C.; Candresse, T. Phytovirome Analysis of Wild Plant Populations: Comparison of Double-Stranded RNA and Virion-Associated Nucleic Acid Metagenomic Approaches. J. Virol. 2019, 94, e01462-19. [Google Scholar] [CrossRef]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and Potential Biogeochemical Impacts of Globally Abundant Ocean Viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.-P.; Tian, F.; Roux, S.; Gazitúa, M.C.; Solonenko, N.E.; Li, Y.-F.; Davis, M.E.; Van Etten, J.L.; Mosley-Thompson, E.; Rich, V.I.; et al. Glacier Ice Archives Nearly 15,000-Year-Old Microbes and Phages. Microbiome 2021, 9, 160. [Google Scholar] [CrossRef]
- Ledormand, P.; Desmasures, N.; Dalmasso, M. Phage Community Involvement in Fermented Beverages: An Open Door to Technological Advances? Crit. Rev. Food Sci. Nutr. 2020, 61, 2911–2920. [Google Scholar] [CrossRef]
- Jung, M.-J.; Kim, M.-S.; Yun, J.-H.; Lee, J.-Y.; Kim, P.S.; Lee, H.-W.; Ha, J.-H.; Roh, S.W.; Bae, J.-W. Viral Community Predicts the Geographical Origin of Fermented Vegetable Foods More Precisely than Bacterial Community. Food Microbiol. 2018, 76, 319–327. [Google Scholar] [CrossRef]
- Park, E.-J.; Kim, K.-H.; Abell, G.C.; Kim, M.-S.; Roh, S.W.; Bae, J.-W. Metagenomic Analysis of the Viral Communities in Fermented Foods. Appl. Environ. Microbiol. 2011, 77, 1284–1291. [Google Scholar] [CrossRef] [Green Version]
- Dugat-Bony, E.; Lossouarn, J.; De Paepe, M.; Sarthou, A.-S.; Fedala, Y.; Petit, M.-A.; Chaillou, S. Viral Metagenomic Analysis of the Cheese Surface: A Comparative Study of Rapid Procedures for Extracting Viral Particles. Food Microbiol. 2020, 85, 103278. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Arioli, S.; Gargari, G.; Neri, E.; Della Scala, G.; Mora, D. Characterization of Airborne Viromes in Cheese Production Plants. J. Appl. Microbiol. 2018, 125, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A New Perspective on Lysogeny: Prophages as Active Regulatory Switches of Bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Howard-Varona, C.; Lindback, M.M.; Bastien, G.E.; Solonenko, N.; Zayed, A.A.; Jang, H.; Andreopoulos, B.; Brewer, H.M.; del Rio, T.G.; Adkins, J.N.; et al. Phage-Specific Metabolic Reprogramming of Virocells. ISME J. 2020, 14, 881–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, S.; Shanahan, F.; Stanton, C.; Hill, C.; Coffey, A.; Ross, R.P. Movers and Shakers: Influence of Bacteriophages in Shaping the Mammalian Gut Microbiota. Gut Microbes 2013, 4, 4–16. [Google Scholar] [CrossRef] [PubMed]
- LeLièvre, V.; Besnard, A.; Schlusselhuber, M.; Desmasures, N.; Dalmasso, M. Phages for Biocontrol in Foods: What Opportunities for Salmonella sp. Control along the Dairy Food Chain? Food Microbiol. 2019, 78, 89–98. [Google Scholar] [CrossRef]
- Pietrysiak, E.; Smith, S.; Ganjyal, G.M. Food Safety Interventions to Control Listeria monocytogenes in the Fresh Apple Packing Industry: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1705–1726. [Google Scholar] [CrossRef] [Green Version]
- Philippe, C.; Krupovic, M.; Jaomanjaka, F.; Claisse, O.; Petrel, M.; Le Marrec, C. Bacteriophage GC1, a Novel Tectivirus Infecting Gluconobacter cerinus, an Acetic Acid Bacterium Associated with Wine-Making. Viruses 2018, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- Deasy, T.; Mahony, J.; Neve, H.; Heller, K.J.; Van Sinderen, D. Isolation of a Virulent Lactobacillus brevis Phage and Its Application in the Control of Beer Spoilage. J. Food Prot. 2011, 74, 2157–2161. [Google Scholar] [CrossRef]
- Mahony, J.; Van Sinderen, D. Current Taxonomy of Phages Infecting Lactic Acid Bacteria. Front. Microbiol. 2014, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Misery, B.; Legendre, P.; Rue, O.; Bouchart, V.; Guichard, H.; Laplace, J.M.; Cretenet, M. Diversity and Dynamics of Bacterial and Fungal Communities in Cider for Distillation. Int. J. Food Microbiol. 2021, 339, 108987. [Google Scholar] [CrossRef]
- Cousin, F.J.; Le Guellec, R.; Schlusselhuber, M.; Dalmasso, M.; Laplace, J.-M.; Cretenet, M. Microorganisms in Fermented Apple Beverages: Current Knowledge and Future Directions. Microorganisms 2017, 5, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledormand, P.; Desmasures, N.; Bernay, B.; Goux, D.; Monnet, C.; Dalmasso, M. Molecular Approaches to Uncover Phage-Lactic Acid Bacteria Interactions in a Model Community Simulating Fermented Beverages. Food Microbiol. 2022, in press. [CrossRef]
- Draper, L.A.; Ryan, F.J.; Dalmasso, M.; Casey, P.G.; McCann, A.; Velayudhan, V.; Ross, R.P.; Hill, C. Autochthonous Faecal Virome Transplantation (FVT) Reshapes the Murine Microbiome after Antibiotic Perturbation. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Kasajima, I.; Sasaki, K.; Tanaka, Y.; Terakawa, T.; Ohtsubo, N. Large-Scale Extraction of Pure DNA from Mature Leaves of Cyclamen Persicum Mill. and Other Recalcitrant Plants with Alkaline Polyvinylpolypyrrolidone (PVPP). Sci. Hortic. 2013, 164, 65–72. [Google Scholar] [CrossRef]
- John, S.G.; Mendez, C.B.; Deng, L.; Poulos, B.; Kauffman, A.K.M.; Kern, S.; Brum, J.; Polz, M.F.; Boyle, E.A.; Sullivan, M.B. A Simple and Efficient Method for Concentration of Ocean Viruses by Chemical Flocculation. Environ. Microbiol. Rep. 2011, 3, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. Fastqc: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 June 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and Sensitive Taxonomic Classification for Metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Catel-Ferreira, M.; Tnani, H.; Hellio, C.; Cosette, P.; Lebrun, L. Antiviral Effects of Polyphenols: Development of Bio-Based Cleaning Wipes and Filters. J. Virol. Methods 2015, 212, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Philippe, C.; Chaïb, A.; Jaomanjaka, F.; Cluzet, S.; Lagarde, A.; Ballestra, P.; Decendit, A.; Petrel, M.; Claisse, O.; Goulet, A.; et al. Wine Phenolic Compounds Differently Affect the Host-Killing Activity of Two Lytic Bacteriophages Infecting the Lactic Acid Bacterium Oenococcus oeni. Viruses 2020, 12, 1316. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Yang, Y.; Jiao, N.; Zhang, R. Evaluation of Tangential Flow Filtration for the Concentration and Separation of Bacteria and Viruses in Contrasting Marine Environments. PLoS ONE 2015, 10, e0136741. [Google Scholar] [CrossRef]
- Jaomanjaka, F.; Claisse, O.; Blanche-Barbat, M.; Petrel, M.; Ballestra, P.; Le Marrec, C. Characterization of a New Virulent Phage Infecting the Lactic Acid Bacterium Oenococcus oeni. Food Microbiol. 2016, 54, 167–177. [Google Scholar] [CrossRef]
- Philippe, C.; Chaïb, A.; Jaomanjaka, F.; Claisse, O.; Lucas, P.M.; Samot, J.; Cambillau, C.; Le Marrec, C. Characterization of the First Virulent Phage Infecting Oenococcus oeni, the Queen of the Cellars. Front. Microbiol. 2021, 11, 596541. [Google Scholar] [CrossRef]
- Chaïb, A.; Claisse, O.; Delbarre, E.; Bosviel, J.; Le Marrec, C. Assessment of the Lysogenic Status in the Lactic Acid Bacterium O. oeni during the Spontaneous Malolactic Fermentation of Red Wines. Food Microbiol. 2022, 103, 103947. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Sadiq, F.A.; Han, X.; Zhao, J.; Zhang, H.; Ross, R.P.; Lu, W.; Chen, W. Comprehensive Scanning of Prophages in Lactobacillus: Distribution, Diversity, Antibiotic Resistance Genes, and Linkages with CRISPR-Cas Systems. mSystems 2021, 6, e0121120. [Google Scholar] [CrossRef]
- Claisse, O.; Chaïb, A.; Jaomanjaka, F.; Philippe, C.; Barchi, Y.; Lucas, P.M.; Le Marrec, C. Distribution of Prophages in the Oenococcus oeni Species. Microorganisms 2021, 9, 856. [Google Scholar] [CrossRef]
- Brouwer, S.; Barnett, T.C.; Ly, D.; Kasper, K.J.; De Oliveira, D.M.P.; Rivera-Hernandez, T.; Cork, A.J.; McIntyre, L.; Jespersen, M.G.; Richter, J.; et al. Prophage Exotoxins Enhance Colonization Fitness in Epidemic Scarlet Fever-Causing Streptococcus pyogenes. Nat. Commun. 2020, 11, 5018. [Google Scholar] [CrossRef] [PubMed]
- Wendling, C.C.; Refardt, D.; Hall, A.R. Fitness Benefits to Bacteria of Carrying Prophages and Prophage-Encoded Antibiotic-Resistance Genes Peak in Different Environments. Evolution 2021, 75, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; You, L.; Kwok, L.-Y.; Jin, H.; Peng, J.; Zhao, Z.; Sun, Z. Strain-Level Multiomics Analysis Reveals Significant Variation in Cheeses from Different Regions. LWT 2021, 151, 112043. [Google Scholar] [CrossRef]
- Tenorio-Salgado, S.; Castelán-Sánchez, H.G.; Dávila-Ramos, S.; Huerta-Saquero, A.; Rodríguez-Morales, S.; Merino-Pérez, E.; Roa de la Fuente, L.F.; Solis-Pereira, S.E.; Pérez-Rueda, E.; Lizama-Uc, G. Metagenomic Analysis and Antimicrobial Activity of Two Fermented Milk Kefir Samples. Microbiol. Open 2021, 10, e1183. [Google Scholar] [CrossRef] [PubMed]
- Abadias, M.; Cañamás, T.P.; Asensio, A.; Anguera, M.; Viñas, I. Microbial Quality of Commercial “Golden Delicious” Apples throughout Production and Shelf-Life in Lleida (Catalonia, Spain). Int. J. Food Microbiol. 2006, 108, 404–409. [Google Scholar] [CrossRef]
- Liu, J.; Abdelfattah, A.; Norelli, J.; Burchard, E.; Schena, L.; Droby, S.; Wisniewski, M. Apple Endophytic Microbiota of Different Rootstock/Scion Combinations Suggests a Genotype-Specific Influence. Microbiome 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Bae, J.-W. Lysogeny Is Prevalent and Widely Distributed in the Murine Gut Microbiota. ISME J. 2018, 12, 1127–1141. [Google Scholar] [CrossRef] [Green Version]
- Chevallereau, A.; Pons, B.J.; van Houte, S.; Westra, E.R. Interactions between Bacterial and Phage Communities in Natural Environments. Nat. Rev. Microbiol. 2021, 20, 49–62. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in Nature: Mechanisms, Impact and Ecology of Temperate Phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billaud, M.; Lamy-Besnier, Q.; Lossouarn, J.; Moncaut, E.; Dion, M.B.; Moineau, S.; Traoré, F.; Le Chatelier, E.; Denis, C.; Estelle, J.; et al. Analysis of Viromes and Microbiomes from Pig Fecal Samples Reveals That Phages and Prophages Rarely Carry Antibiotic Resistance Genes. ISME Commun. 2021, 1, 55. [Google Scholar] [CrossRef]
- Zünd, M.; Ruscheweyh, H.-J.; Field, C.M.; Meyer, N.; Cuenca, M.; Hoces, D.; Hardt, W.-D.; Sunagawa, S. High Throughput Sequencing Provides Exact Genomic Locations of Inducible Prophages and Accurate Phage-to-Host Ratios in Gut Microbial Strains. Microbiome 2021, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Trček, J.; Mahnič, A.; Rupnik, M. Diversity of the Microbiota Involved in Wine and Organic Apple Cider Submerged Vinegar Production as Revealed by DHPLC Analysis and Next-Generation Sequencing. Int. J. Food Microbiol. 2016, 223, 57–62. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | Producer ID (French Department) | Fermentation Time (Days) | Tank ID—Tank Volume (hL) | Sugar Concentration kg/m3 | pH |
|---|---|---|---|---|---|
| C1B1 | B (Calvados) | 0 | C1—120 | 1060 | 4.02 |
| C1B2 | B (Calvados) | 7 | C1—120 | 1056 | 4.05 |
| C1B3 | B (Calvados) | 14 | C1—120 | 1048 | 4.03 |
| C1B4 | B (Calvados) | 27 | C1—120 | 1036 | 4.01 |
| C2P1 | P (Manche) | 0 | C2—10 | 1056 | 3.32 |
| C2P2 | P (Manche) | 12 | C2—10 | 1049 | 3.35 |
| C2P3 | P (Manche) | 25 | C2—10 | 1040 | 3.52 |
| C2P4 | P (Manche) | 36 | C2—10 | 1036 | 3.58 |
| Tested Conditions to Collect Viral Particles from Cider | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII Tangential Flow Filtration | IX Flocculation with FeCl3 | |
| Tested volumes of cider (mL) | 50 | 20, 50, 1000 | 400 | ||||||
| Sample spiked with phage UCMA 21115 (PFU/mL) | Yes (104–109) | Yes (105) | Yes (104–108) | ||||||
| pH adjusted to 5.5 | Yes | Yes | |||||||
| Precipitation with 10% PEG-8000 and 0.5 M NaCl | Yes | Yes | Yes | Yes | Yes | ||||
| Centrifugation step | Yes | Yes | Yes | Yes | Yes | Yes | Yes | ||
| DNA extraction from | Pellet | Supernatant | Pellet | Pellet | Supernatant | Pellet | Pellet | Retentate | Flocculate |
| Inoculated Quantity of Phage UCMA 21115 in 400 mL of Cider (i.e., Concentration) | Retrieved Quantity of Phage UCMA 21115 after Flocculation in Cider |
|---|---|
| 4 × 1010 (1 × 108 PFU/mL) | 8.5 × 108 |
| 4 × 109 (1 × 107 PFU/mL) | 9.9 × 107 |
| 4 × 108 (1 × 106 PFU/mL) | 6.6 × 106 |
| 4 × 107 (1 × 105 PFU/mL) | 7.6 × 105 |
| 4 × 106 (1 × 104 PFU/mL) | 5.0 × 103 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ledormand, P.; Desmasures, N.; Midoux, C.; Rué, O.; Dalmasso, M. Investigation of the Phageome and Prophages in French Cider, a Fermented Beverage. Microorganisms 2022, 10, 1203. https://doi.org/10.3390/microorganisms10061203
Ledormand P, Desmasures N, Midoux C, Rué O, Dalmasso M. Investigation of the Phageome and Prophages in French Cider, a Fermented Beverage. Microorganisms. 2022; 10(6):1203. https://doi.org/10.3390/microorganisms10061203
Chicago/Turabian StyleLedormand, Pierre, Nathalie Desmasures, Cédric Midoux, Olivier Rué, and Marion Dalmasso. 2022. "Investigation of the Phageome and Prophages in French Cider, a Fermented Beverage" Microorganisms 10, no. 6: 1203. https://doi.org/10.3390/microorganisms10061203
APA StyleLedormand, P., Desmasures, N., Midoux, C., Rué, O., & Dalmasso, M. (2022). Investigation of the Phageome and Prophages in French Cider, a Fermented Beverage. Microorganisms, 10(6), 1203. https://doi.org/10.3390/microorganisms10061203






