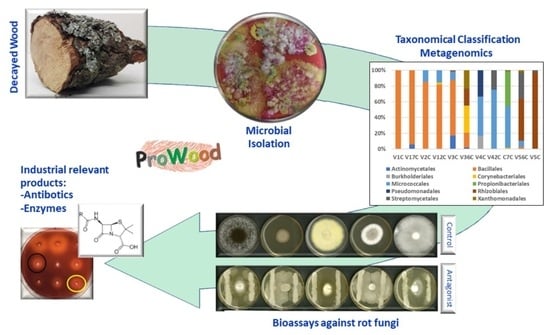
Characterization of Microbial Diversity in Decayed Wood from a Spanish Forest: An Environmental Source of Industrially Relevant Microorganisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling of Decayed Wood
2.2. DNA Extraction for Metagenome Analyses
2.3. Metagenome Sequencing
2.4. Isolation and Identification of Cultivable Microbes from Decaying Wood
2.5. Assessment of Microbial Diversity
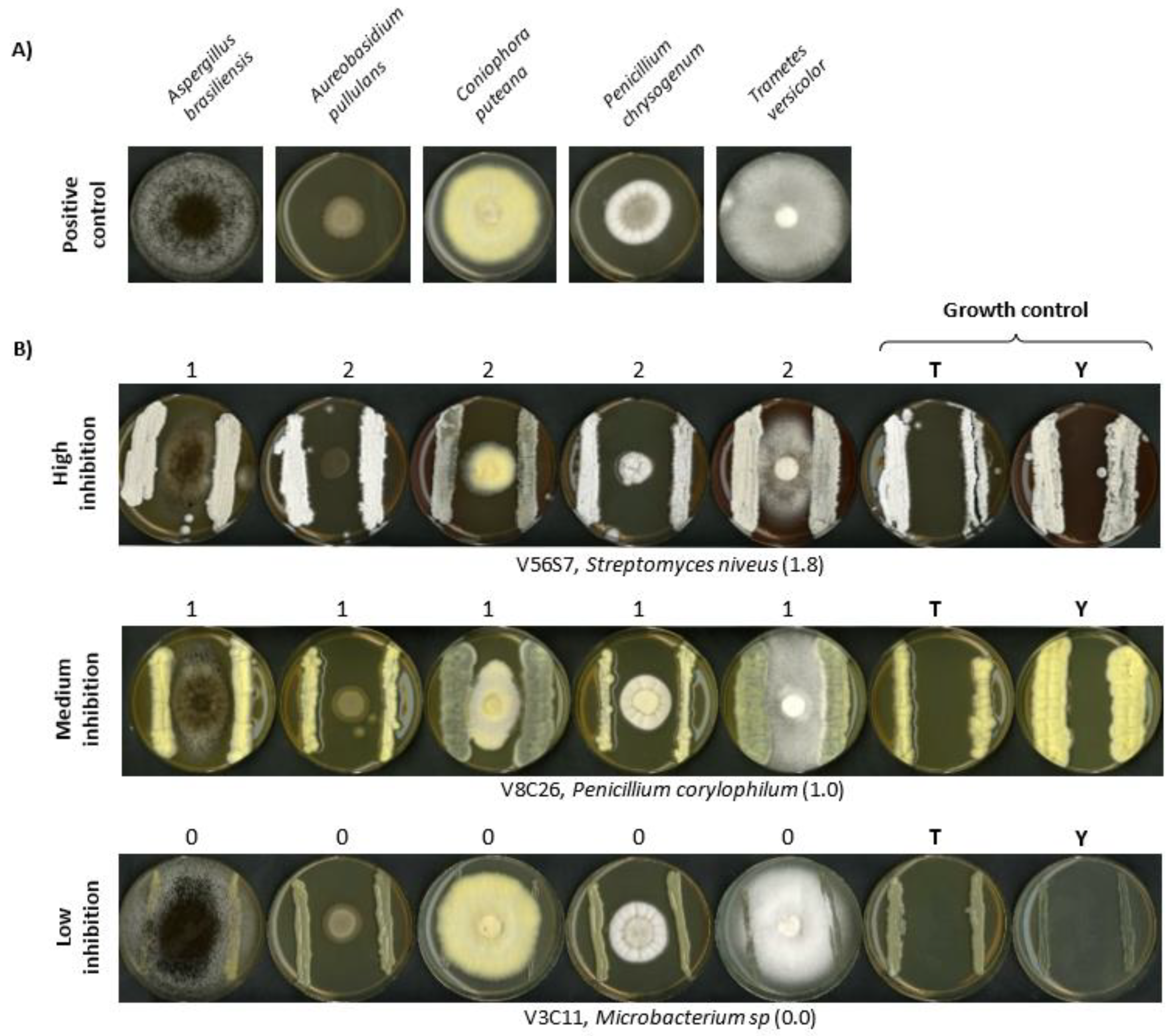
2.6. Evaluation of Microbial Antagonism against Wood Decay Fungi (Competitive Bioassays)
2.7. Polyketide Synthases and Non-Ribosomal Peptide Synthetases Gene Screening
2.8. Enzymatic Assays: Cellulolytic and Feruloyl Esterase Activity Assays
3. Results
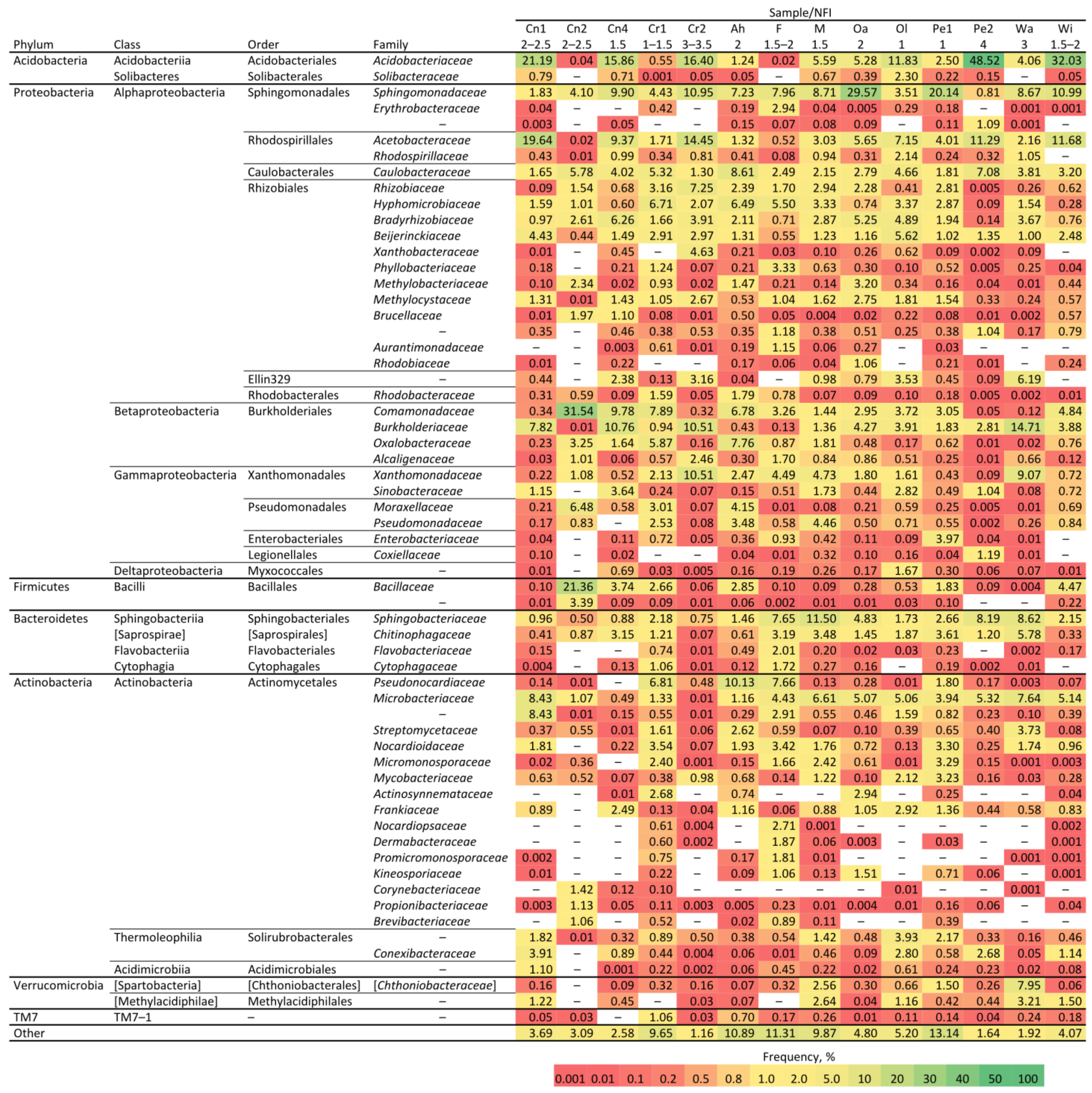
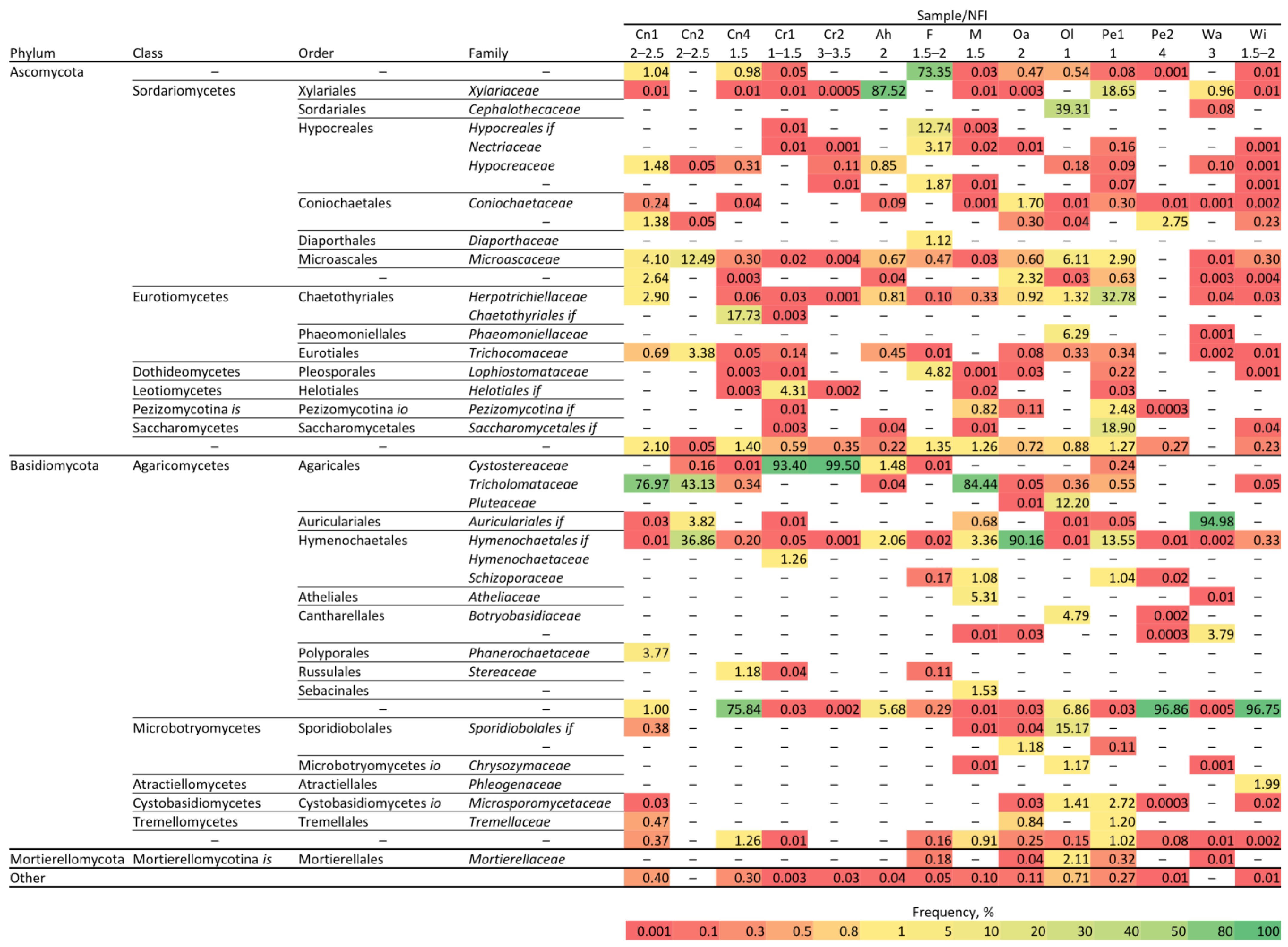
3.1. Metagenome Analyses of Different Species and Rotting Stages of Wood
3.2. Isolation of Cultivable Microorganisms from Decayed Wood
3.3. Characterization of the Cultivable Isolates: Antimicrobial Potential (NRPS and PKS)
3.4. Validation of In Vivo Antibiotic Production
3.5. Enzyme Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, Z.; Shen, Y.; Shen, F.; Luo, Y.; Su, X. Herbivory-induced volatiles elicit defence genes in lima bean leaves. Trees 2009, 23, 512–515. [Google Scholar] [CrossRef]
- Baldrian, P. Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 2017, 41, 109–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoppe, B.; Krüger, D.; Kahl, T.; Arnstadt, T.; Buscot, F.; Bauhus, J.; Wubet, T. A pyrosequencing insight into sprawling bacterial diversity and community dynamics in decaying deadwood logs of Fagus sylvatica and Picea abies. Sci. Rep. 2015, 5, 9456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarze, F.W.M.R. Wood decay under the microscope. Fungal Biol. Rev. 2007, 21, 133–170. [Google Scholar] [CrossRef]
- Schmidt, O. Wood and Tree Fungi: Biology, Damage, Protection, and Use; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 9783540321392. [Google Scholar]
- Johnston, S.R.; Boddy, L.; Weightman, A.J. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol. Ecol. 2016, 92, fiw179. [Google Scholar] [CrossRef] [Green Version]
- Daniel, G. Fungal and bacterial biodegradation: White rots, brown rots, soft rots, and bacteria. In ACS Symposium Series; ACS: Washington, DC, USA, 2014; Volume 1158, pp. 23–58. ISBN 9780841230040. [Google Scholar]
- Ringman, R. Biochemical Mechanisms of Brown Rot Decay: A Study on the Mode of Action of Modified Wood. Ph.D. Thesis, Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, München, Germany, 2017. [Google Scholar]
- Martínez, A.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martínez, M.J.; Gutiérrez, A.; del Río, J.C. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar]
- Rayner, A.D.M.; Boddy, L. Fungal Decomposition of Wood; John Wiley Sons: Hoboken, NJ, USA, 1988; p. 587. [Google Scholar]
- Aboagye, S.Y.; Amarh, V.; Lartey, P.A.; Arthur, P.K. Wood-decaying fungi found in Southern Ghana: A potential source of new anti-infective compounds. AAS Open Res. 2019, 2, 20. [Google Scholar] [CrossRef] [Green Version]
- Sawadsitang, S.; Suwannasai, N.; Mongkolthanaruk, W.; Ahmadi, P.; McCloskey, S. A new amino amidine derivative from the wood-decaying fungus Xylaria cf. cubensis SWUF08-86. Nat. Prod. Res. 2018, 32, 2260–2267. [Google Scholar] [CrossRef]
- Perlatti, B.; Nichols, C.B.; Alspaugh, J.A.; Gloer, J.B.; Bills, G.F. Sphaerostilbellins, new antimicrobial aminolipopeptide peptaibiotics from Sphaerostilbella toxica. Biomolecules 2020, 10, 1371. [Google Scholar] [CrossRef]
- Webster, G.; Mullins, A.J.; Bettridge, A.S.; Jones, C.; Cunningham-Oakes, E.; Connor, T.R.; Parkhill, J.; Mahenthiralingam, E. The genome sequences of three Paraburkholderia sp. strains isolated from wood-decay fungi reveal them as novel species with antimicrobial biosynthetic potential. Microbiol. Resour. Announc. 2019, 8, e00778-19. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, C.; Swenson, D.C.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. Novel antiinsectan oxalicine alkaloids from two undescribed fungicolous Penicillium spp. Org. Lett. 2003, 5, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, C.; Barredo, J.L. (Eds.) Antimicrobial Therapies: Methods and Protocols; Springer Science+Business Media, LLC: New York, NY, USA, 2021; ISBN 978-1-0716-1357-3. [Google Scholar]
- Oliverio, A.M.; Power, J.F.; Washburne, A.; Cary, S.C.; Stott, M.B.; Fierer, N. The ecology and diversity of microbial eukaryotes in geothermal springs. ISME J. 2018, 12, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.N.; Cantrell, C.L.; Wedge, D.E.; Ferreira, M.C.; Soares, M.A.; Jacob, M.R.; Oliveira, F.S.; Galante, D.; Rodrigues, F.; Alves, T.M.A.; et al. Fungi associated with rocks of the Atacama Desert: Taxonomy, distribution, diversity, ecology and bioprospection for bioactive compounds. Environ. Microbiol. 2016, 18, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Li, J.; Chen, H.H.; Zhao, G.Z.; Zhu, W.Y.; Jiang, C.L.; Xu, L.H.; Li, W.J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna China. Appl. Environ. Microbiol. 2009, 75, 6176–6186. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Jiang, Y.; Wang, X.; Chen, D.; Chen, X.; Wang, L.; Han, L.; Huang, X.; Jiang, C. Diversity, Antimicrobial Activity, and Biosynthetic Potential of Cultivable Actinomycetes Associated with Lichen Symbiosis. Microb. Ecol. 2017, 74, 570–584. [Google Scholar] [CrossRef]
- Pereira, L.B.; Palermo, B.R.Z.; Carlos, C.; Ottoboni, L.M.M. Diversity and antimicrobial activity of bacteria isolated from different Brazilian coral species. FEMS Microbiol. Lett. 2017, 364, fnx164. [Google Scholar] [CrossRef] [Green Version]
- Harald, B.; Galatenko, O.A.; Engelhardt, K.; Fjaervik, E.; Terekhova, L.P.; Zotchev, S.B. Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: Isolation, diversity and biological activity. Environ. Microbiol. 2007, 9, 2756–2764. [Google Scholar] [CrossRef]
- Xi, L.; Ruan, J.; Huang, Y. Diversity and biosynthetic potential of culturable actinomycetes associated with marine sponges in the China seas. Int. J. Mol. Sci. 2012, 13, 5917–5932. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef] [Green Version]
- Sajith, S.; Priji, P.; Sreedevi, S.; Benjamin, S. An overview on fungal cellulases with an industrial perspective. J. Nutr. Food Sci. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol. Adv. 2009, 27, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, N.; Singh, A.; Adsul, M.; Dixit, P.; Sandhu, S.K.; Mathur, A.; Puri, S.K.; Singhania, R.R. Penicillium: The next emerging champion for cellulase production. Bioresour. Technol. Rep. 2018, 2, 131–140. [Google Scholar] [CrossRef]
- Méndez-Líter, J.A.; de Eugenio, L.I.; Nieto-Domínguez, M.; Prieto, A.; Martínez, M.J. Hemicellulases from Penicillium and Talaromyces for lignocellulosic biomass valorization: A review. Bioresour. Technol. 2021, 324, 124623. [Google Scholar] [CrossRef]
- Palani Swamy, S.K.; Govindaswamy, V. Therapeutical properties of ferulic acid and bioavailability enhancement through feruloyl esterase. J. Funct. Foods 2015, 17, 657–666. [Google Scholar] [CrossRef]
- Zduńska, K.; Dana, A.; Kolodziejczak, A.; Rotsztejn, H. Antioxidant properties of ferulic acid and its possible application. Skin Pharmacol. Physiol. 2018, 31, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Kwok, K.-C. Ferulic acid: Pharmaceutical functions, preparation and applications in foods. J. Sci. Food Agric. 2004, 84, 1261–1269. [Google Scholar] [CrossRef]
- Kumar, N.; Pruthi, V. Potential applications of ferulic acid from natural sources. Biotechnol. Rep. 2014, 4, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Rivas-Martínez, S.; Sáenz, S.; Penas, A. Worldwide bioclimatic classification system. Glob. Geobot. 2011, 1, 1–634 + 4 Maps. [Google Scholar] [CrossRef]
- Sandström, F.; Petersson, H.; Kruys, N.; Ståhl, G. Biomass conversion factors (density and carbon concentration) by decay classes for dead wood of Pinus sylvestris, Picea abies and Betula spp. in boreal forests of Sweden. For. Ecol. Manag. 2007, 243, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic Press Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Rodríguez, O.; Fil, M.; García-Calvo, L.; Kosalková, K.; Barreiro, C. Microbial isolation and characterization of new antibiotic-producing strains from decayed wood. In Antimicrobial Therapies: Methods and Protocols; Barreiro, C., Barredo, J.L., Eds.; Springer US: New York, NY, USA, 2021; ISBN 978-1-0716-1357-3. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating deep phylogenetic relationships among Cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.H. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Hurlbert, S.H. The nonconcept of species diversity: A critique and alternative parameters. Ecology 1971, 52, 577–586. [Google Scholar] [CrossRef]
- Chen, B.; He, X.; Pan, B.; Zou, X.; You, N. Comparison of beta diversity measures in clustering the high-dimensional microbial data. PLoS ONE 2021, 16, e0246893. [Google Scholar] [CrossRef]
- Hamed, S.A.M. In-vitro studies on wood degradation in soil by soft-rot fungi: Aspergillus niger and Penicillium chrysogenum. Int. Biodeterior. Biodegrad. 2013, 78, 98–102. [Google Scholar] [CrossRef]
- Fil, M.; Velasco-Rodríguez, O.; García-Calvo, L.; Sola-Landa, A.; Barreiro, C. Screening of antibiotic gene clusters in microorganisms isolated from wood. In Antimicrobial Therapies-Methods and Protocols; Barreiro, C., Barredo, J.-L., Eds.; Springer: New York, NY, USA, 2021; pp. 151–165. ISBN 978-1-0716-1357-3. [Google Scholar]
- Ayuso-Sacido, A.; Genilloud, O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005, 49, 10–24. [Google Scholar] [CrossRef]
- Metsä-Ketelä, M.; Salo, V.; Halo, L.; Hautala, A.; Hakala, J.; Mäntsälä, P.; Ylihonko, K. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 1999, 180, 1–6. [Google Scholar] [CrossRef]
- Amnuaykanjanasin, A.; Punya, J.; Paungmoung, P.; Rungrod, A.; Tachaleat, A.; Pongpattanakitshote, S.; Cheevadhanarak, S.; Tanticharoen, M. Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiol. Lett. 2005, 251, 125–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, T.P.; Rudd, B.A.M.; Dawson, M.; Lazarus, C.M.; Simpson, T.J.; Cox, R.J. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 2001, 8, 157–178. [Google Scholar] [CrossRef] [Green Version]
- Bingle, L.E.H.; Simpson, T.J.; Lazarus, C.M. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 1999, 26, 209–223. [Google Scholar] [CrossRef]
- Slightom, J.L.; Metzger, B.P.; Luu, H.T.; Elhammer, A.P. Cloning and molecular characterization of the gene encoding the Aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 2009, 431, 67–79. [Google Scholar] [CrossRef]
- Barreiro, C.; Prieto, C.; Sola-Landa, A.; Solera, E.; Martínez-Castro, M.; Pérez-Redondo, R.; García-Estrada, C.; Aparicio, J.F.; Fernández-Martínez, L.T.; Santos-Aberturas, J.; et al. Draft genome of Streptomyces tsukubaensis NRRL 18488, the producer of the clinically important immunosuppressant tacrolimus (FK506). J. Bacteriol. 2012, 194, 3756–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreiro, C.; Martínez-Castro, M. Trends in the biosynthesis and production of the immunosuppressant tacrolimus (FK506). Appl. Microbiol. Biotechnol. 2014, 98, 497–507. [Google Scholar] [CrossRef]
- Fierro, F.; Barredo, J.L.; Díez, B.; Gutierrez, S.; Fernández, F.J.; Martín, J.F. The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc. Natl. Acad. Sci. USA 1995, 92, 6200–6204. [Google Scholar] [CrossRef] [Green Version]
- Barreiro, C.; García-Estrada, C. Proteomics and Penicillium chrysogenum: Unveiling the secrets behind penicillin production. J. Proteom. 2019, 198, 119–131. [Google Scholar] [CrossRef]
- Casqueiro, J.; Bañuelos, O.; Gutiérrez, S.; Hijarrubia, M.J.; Martín, J.F. Intrachromosomal recombination between direct repeats in Penicillium chrysogenum: Gene conversion and deletion events. Mol. Gen. Genet. 1999, 261, 994–1000. [Google Scholar] [CrossRef]
- Jami, M.-S.; Barreiro, C.; García-Estrada, C.; Martín, J.-F. Proteome analysis of the penicillin producer Penicillium chrysogenum: Characterization of protein changes during the industrial strain improvement. Mol. Cell. Proteom. 2010, 9, 1182–1198. [Google Scholar] [CrossRef] [Green Version]
- Padmavathi, T.; Nandy, V.; Agarwal, P. Optimization of the medium for the production of cellulases by Aspergillus terreus and Mucor plumbeus. Eur. J. Exp. Biol. 2012, 2, 1161–1170. [Google Scholar]
- Abreham, B.; Tariku, A.; Adane, H.; Addisalem; Fitala; Tigist, G.; Musin, K.; Admas, B. Isolation and characterization of efficient cellulolytic fungi from degraded wood and industrial samples. Afr. J. Biotechnol. 2016, 14, 3228–3234. [Google Scholar] [CrossRef] [Green Version]
- Donaghy, J.A.; McKay, A.M. Novel screening assay for the detection of phenolic acid esterases. World J. Microbiol. Biotechnol. 1994, 10, 41–44. [Google Scholar] [CrossRef] [PubMed]
- García-Calvo, L.; Ullán, R.V.; Fernández-Aguado, M.; García-Lino, A.M.; Balaña-Fouce, R.; Barreiro, C. Secreted protein extract analyses present the plant pathogen Alternaria alternata as a suitable industrial enzyme toolbox. J. Proteom. 2018, 177, 48–64. [Google Scholar] [CrossRef]
- Dieste, A.; Rodríguez, K.; Baño, V. Wood–water relations of chestnut wood used for structural purposes. Eur. J. Wood Wood Prod. 2013, 71, 133–134. [Google Scholar] [CrossRef]
- Aicher, S.; Christian, Z.; Dill-Langer, G. Hardwood glulams—Emerging timber products of superior mechanical properties. In Proceedings of the World Conference on Timber Engineering (WCTE 2014), Quebec City, QC, Canada, 10–14 August 2014. [Google Scholar]
- Martínez-Alonso, C.; Berdasco, L. Carbon footprint of sawn timber products of Castanea sativa Mill. in the north of Spain. J. Clean. Prod. 2015, 102, 127–135. [Google Scholar] [CrossRef]
- Buehlmann, U.; Bumgardner, M.; Alderman, D. Recent developments in US hardwood lumber markets and linkages to housing construction. Curr. For. Rep. 2017, 3, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Barrasa, J.M.; Esteve-Raventós, F.; Dähncke, R.M. Clitocybula canariensis (Tricholomataceae), a new brown-rot fungus from the Canary Islands (Spain). Fungal Divers. 2006, 22, 1–11. [Google Scholar]
- Gorjón, S. Genera of corticioid fungi: Keys, nomenclature and taxonomy. Stud. Fungi 2020, 5, 125–309. [Google Scholar] [CrossRef]
- García-Estrada, C.; Martín, J.F.; Cueto, L.; Barreiro, C. Omics approaches applied to Penicillium chrysogenum and penicillin production: Revealing the secrets of improved productivity. Genes 2020, 11, 712. [Google Scholar] [CrossRef]
- Błaszczyk, L.; Siwulski, M.; Sobieralski, K.; Lisiecka, J.; Jędryczka, M. Trichoderma spp.—Application and prospects for use in organic farming and industry. J. Plant Prot. Res. 2014, 54, 309–317. [Google Scholar] [CrossRef]
- Cappelletti, M.; Presentato, A.; Piacenza, E.; Firrincieli, A.; Turner, R.J.; Zannoni, D. Biotechnology of Rhodococcus for the production of valuable compounds. Appl. Microbiol. Biotechnol. 2020, 104, 8567–8594. [Google Scholar] [CrossRef] [PubMed]
- Prieto, C.; García-Estrada, C.; Lorenzana, D.; Martín, J.F. NRPSsp: Non-ribosomal peptide synthase substrate predictor. Bioinformatics 2012, 28, 426–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Huang, H.; Chen, Y.; Ding, J.; Zhang, Y.; Sun, A.; Zhang, W.; Ju, J. Cytotoxic and antibacterial marfuraquinocins from the deep south China sea-derived Streptomyces niveus SCSIO 3406. J. Nat. Prod. 2013, 76, 2263–2268. [Google Scholar] [CrossRef]
- Xing, M.; Zheng, L.; Deng, Y.; Xu, D.; Xi, P.; Li, M.; Kong, G.; Jiang, Z. Antifungal activity of natural volatile organic compounds against litchi downy blight pathogen Peronophythora litchii. Molecules 2018, 23, 358. [Google Scholar] [CrossRef] [Green Version]
- Björdal, C.G.; Dayton, P.K. First evidence of microbial wood degradation in the coastal waters of the Antarctic. Sci. Rep. 2020, 10, 12774. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Iqbal, H.M.N.; Hu, H.; Zhang, X. Biotransformation of lignocellulosic materials into value-added products—A review. Int. J. Biol. Macromol. 2017, 98, 447–458. [Google Scholar] [CrossRef]
- Karthäuser, J.; Biziks, V.; Mai, C.; Militz, H. Lignin and lignin-derived compounds for wood applications—A review. Molecules 2021, 26, 2533. [Google Scholar] [CrossRef]
- Barreiro, C.; Barredo, J.L. Worldwide clinical demand for antibiotics. Is it a real countdown? In Antimicrobial Therapies. Methods and Protocols; Barreiro, C., Barredo, J.L., Eds.; Springer: New York, NY, USA, 2021; in press. [Google Scholar]
- Granato, E.T.; Meiller-Legrand, T.A.; Foster, K.R. The Evolution and Ecology of Bacterial Warfare. Curr. Biol. 2019, 29, R521–R537. [Google Scholar] [CrossRef]
- Díaz González, T.E.; Penas, Á. The high mountain area of northwestern Spain: The Cantabrian range, the Galician-Leonese mountains and the Bierzo trench. In The Vegetation of the Iberian Peninsula: Volume 1; Loidi, J., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 251–321. ISBN 978-3-319-54784-8. [Google Scholar]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [Green Version]
- Costa, O.Y.A.; de Hollander, M.; Pijl, A.; Liu, B.; Kuramae, E.E. Cultivation-independent and cultivation-dependent metagenomes reveal genetic and enzymatic potential of microbial community involved in the degradation of a complex microbial polymer. Microbiome 2020, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- León, M.J.; Fernández, A.B.; Ghai, R.; Sánchez-Porro, C.; Rodriguez-Valera, F.; Ventosa, A. From metagenomics to pure culture: Isolation and characterization of the noderately halophilic bacterium Spiribacter salinus gen. nov., sp. nov. Appl. Environ. Microbiol. 2014, 80, 3850–3857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvin, J.; Lanong, S.; Syiem, D.; De Mandal, S.; Kayang, H.; Kumar, N.S.; Kiran, G.S. Culture-dependent and metagenomic analysis of lesser horseshoe bats’ gut microbiome revealing unique bacterial diversity and signatures of potential human pathogens. Microb. Pathog. 2019, 137, 103675. [Google Scholar] [CrossRef] [PubMed]
- Embacher, J.; Neuhauser, S.; Zeilinger, S.; Kirchmair, M. Microbiota associated with different developmental stages of the dry rot fungus Serpula lacrymans. J. Fungi 2021, 7, 354. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.J. The family Acidobacteriaceae. In The Prokaryotes: Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 405–415. ISBN 978-3-642-38954-2. [Google Scholar]
- Humar, M.; Petrič, M.; Pohleven, F. Changes of the pH value of impregnated wood during exposure to wood-rotting fungi. Holz als Roh- und Werkst. 2001, 59, 288–293. [Google Scholar] [CrossRef]
- Geffert, A.; Geffertova, J.; Dudiak, M. Direct method of measuring the pH value of wood. Forests 2019, 10, 852. [Google Scholar] [CrossRef] [Green Version]
- Tunca, S.; Barreiro, C.; Coque, J.-J.R.; Martín, J.F. Two overlapping antiparallel genes encoding the iron regulator DmdR1 and the Adm proteins control sidephore and antibiotic biosynthesis in Streptomyces coelicolor A3(2). FEBS J. 2009, 276, 4814–4827. [Google Scholar] [CrossRef]
- Flores, F.J.; Barreiro, C.; Coque, J.J.R.; Martín, J.F. Functional analysis of two divalent metal-dependent regulatory genes dmdR1 and dmdR2 in Streptomyces coelicolor and proteome changes in deletion mutants. FEBS J. 2005, 272, 725–735. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Dhakal, D.; Pham, V.T.T.; Nguyen, H.T.; Sohng, J.-K. Recent advances in strategies for activation and discovery/characterization of cryptic biosynthetic gene clusters in Streptomyces. Microorganisms 2020, 8, 616. [Google Scholar] [CrossRef] [Green Version]
- Becerril, A.; Álvarez, S.; Braña, A.F.; Rico, S.; Díaz, M.; Santamaría, R.I.; Salas, J.A.; Méndez, C. Uncovering production of specialized metabolites by Streptomyces argillaceus: Activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS ONE 2018, 13, e0198145. [Google Scholar] [CrossRef]
- Martínez-Burgo, Y.; Santos-Aberturas, J.; Rodríguez-García, A.; Barreales, E.G.; Tormo, J.R.; Truman, A.W.; Reyes, F.; Aparicio, J.F.; Liras, P. Activation of Secondary Metabolite Gene Clusters in Streptomyces clavuligerus by the PimM Regulator of Streptomyces natalensis. Front. Microbiol. 2019, 10, 580. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Schümann, J.; Scherlach, K.; Lange, C.; Brakhage, A.A.; Hertweck, C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007, 3, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Chávez, F.; Zwahlen, R.D.; Bovenberg, R.A.L.; Driessen, A.J.M. Engineering of the filamentous fungus Penicillium chrysogenum as cell factory for natural products. Front. Microbiol. 2018, 9, 2768. [Google Scholar] [CrossRef] [PubMed]
- Broda, M. Natural compounds for wood protection against fungi—A review. Molecules 2020, 25, 3538. [Google Scholar] [CrossRef]
- McKelvey, S.M.; Murphy, R.A. Biotechnological use of fungal enzymes. In Fungi: Biology and Applications; Kavanagh, K., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 201–225. [Google Scholar]
- Arnau, J.; Yaver, D.; Hjort, C.M. Strategies and challenges for the development of industrial enzymes using fungal cell factories. In Grand Challenges in Fungal Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 179–210. [Google Scholar]



| Primers | Metabarcode | Ref. | PCR Fragment |
|---|---|---|---|
| 16S-1 (Bakt_341F/S-D-Bact-0341-b-S-17) | 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3′ | [35,36] | ~402–427 bp |
| 16S-2 (Bakt_805R/S-D-Bact-0785-a-A-21) | 5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3′ | ||
| ITS2-1 (ITS4) | 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG TCC TCC GCT TAT TGA TAT GC-3′ | [37] | ~200–300 bp |
| ITS2-2 (based on fITS7) | 5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGT GAA TCA TCG AAT CTT TG-3′ | [38] |
| Primers | Sanger Sequencing | Ref. | PCR Fragment |
|---|---|---|---|
| 27F | 5′-AGA GTT TGA TCC TGG CTC AG-3′ | [41] | 1000–1500 bp |
| 1492R | 5′-GGT TAC CTT GTT ACG ACT T-3′ | [42] | |
| ITS1 | 5′-TCC GTA GGT GAA CCT GCG G-3′ | [37] | 400–600 bp |
| ITS4 | 5′-TCC TCC GCT TAT TGA TAT GC-3′ |
| Primers | Amplicon | Ref. | PCR Fragment |
|---|---|---|---|
| Polyketide Synthases (PKSs) | |||
| K1F | 5′-TSA AGT CSA ACA TCG GBC A-3′ | [48] | PKSI (1200–1400 bp) |
| M6R | 5′-CGC AGG TTS CSG TAC CAG TA-3′ | ||
| KSF | 5′-TSG CST GCT TGG AYG CSA TC-3′ | [49] | PKSII (600–650 bp) |
| KSR | 5′TGG AAN CCG CCG AAB CCT CT-3′ | ||
| Non-Ribosomal Peptide Synthetase (NRPS) | |||
| A3F | 5′-GCS TAC SYS ATS TAC ACS TCS G-3′ | [48] | NRPS (700–800 bp) |
| A7R | 5′-SAS GTC VCC SGT SCG GTA S-3′ |
| Primers | Amplicon | Ref. | PCR Fragment |
|---|---|---|---|
| Polyketide Synthases (PKSs) | |||
| KA (ketosynthase-acyltransferase) | |||
| KAF1F | 5′-GAR KSI CAY GGI ACI GGI AC-3′ | [50] | PKS (700–800 bp) Highly reduced lovastatin-type PKSs (HR-PKS) |
| KAR2R | 5′-CCA YTG IGC ICC YTG ICC IGT RAA-3′ | ||
| KAF2F | 5′-GAR GCI CAY GCI ACI TCI AC-3′ | [50] | PKS (700 bp) Methylsalicylic acid synthase type (MSAS-type) |
| KAR1R | 5′-CCA YTG IGC ICC RTG ICC IGA RAA-3′ | ||
| PKS ketosynthase domains | |||
| LC3F | 5′-GCI GAR CAR ATG GAY CCI CA-3′ | [52] | PKS (680 bp) |
| LC5R | 5′-GTI GAI GTI GCR TGI GCY TC-3′ | ||
| LC1F | 5′-GAY CCI MGI TTY TTY AAY ATG-3′ | [52] | PKS (720 bp) |
| LC2R | 5′-GTI CCI GTI CCR TGC ATY TC-3′ | ||
| Non-Ribosomal Peptide Synthetase (NRPS) | |||
| AUG003F | 5′-CCG GCA CCA CCG GNA ARC CHA A-3′ | [53] | NRPS (1100 bp) |
| AUG007R | 5′-GCT GCA TGG CGG TGA TGS WRT SNC CBC C-3′ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velasco-Rodríguez, Ó.; Fil, M.; Heggeset, T.M.B.; Degnes, K.F.; Becerro-Recio, D.; Kolsaková, K.; Haugen, T.; Jønsson, M.; Toral-Martínez, M.; García-Estrada, C.; et al. Characterization of Microbial Diversity in Decayed Wood from a Spanish Forest: An Environmental Source of Industrially Relevant Microorganisms. Microorganisms 2022, 10, 1249. https://doi.org/10.3390/microorganisms10061249
Velasco-Rodríguez Ó, Fil M, Heggeset TMB, Degnes KF, Becerro-Recio D, Kolsaková K, Haugen T, Jønsson M, Toral-Martínez M, García-Estrada C, et al. Characterization of Microbial Diversity in Decayed Wood from a Spanish Forest: An Environmental Source of Industrially Relevant Microorganisms. Microorganisms. 2022; 10(6):1249. https://doi.org/10.3390/microorganisms10061249
Chicago/Turabian StyleVelasco-Rodríguez, Óscar, Mariana Fil, Tonje M. B. Heggeset, Kristin F. Degnes, David Becerro-Recio, Katarina Kolsaková, Tone Haugen, Malene Jønsson, Macarena Toral-Martínez, Carlos García-Estrada, and et al. 2022. "Characterization of Microbial Diversity in Decayed Wood from a Spanish Forest: An Environmental Source of Industrially Relevant Microorganisms" Microorganisms 10, no. 6: 1249. https://doi.org/10.3390/microorganisms10061249