Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Cultivation of Acaryochloris
2.2. Pigment Extraction
2.3. Extraction of Genomic DNA
2.4. Whole Genome Sequencing of A. marina MBIC10699
2.5. Quality Control of Sequencing Reads
2.6. Genome Assembly and Gene Annotation
2.7. Comparative Genomics
2.8. Core genome Phylogenic Analysis
3. Results and Discussion
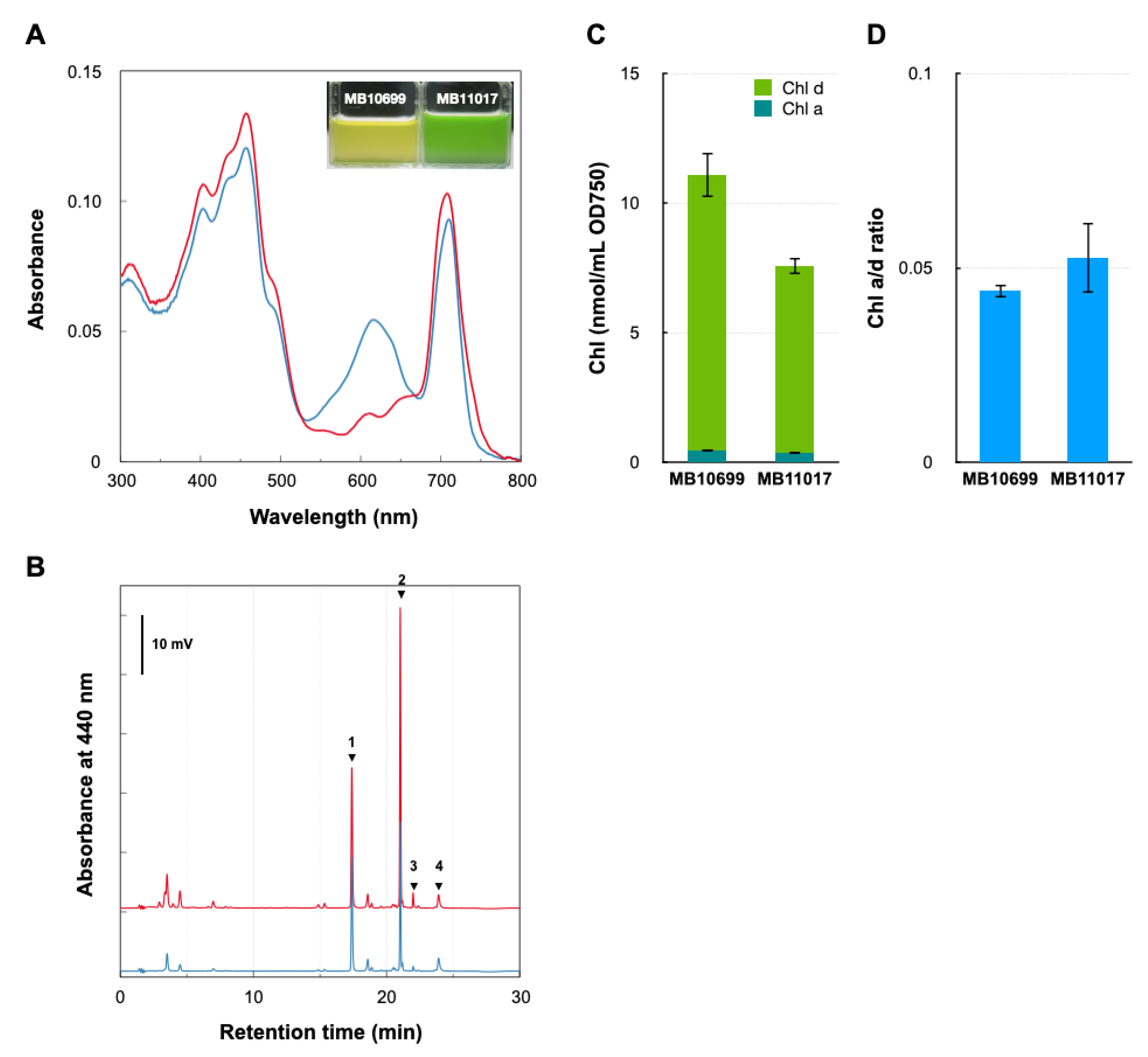
3.1. Comparative Analysis of Pigment Contents of Two Acaryochloris Strains
3.2. Overall View of the Complete Genome of A. marina MBIC10699
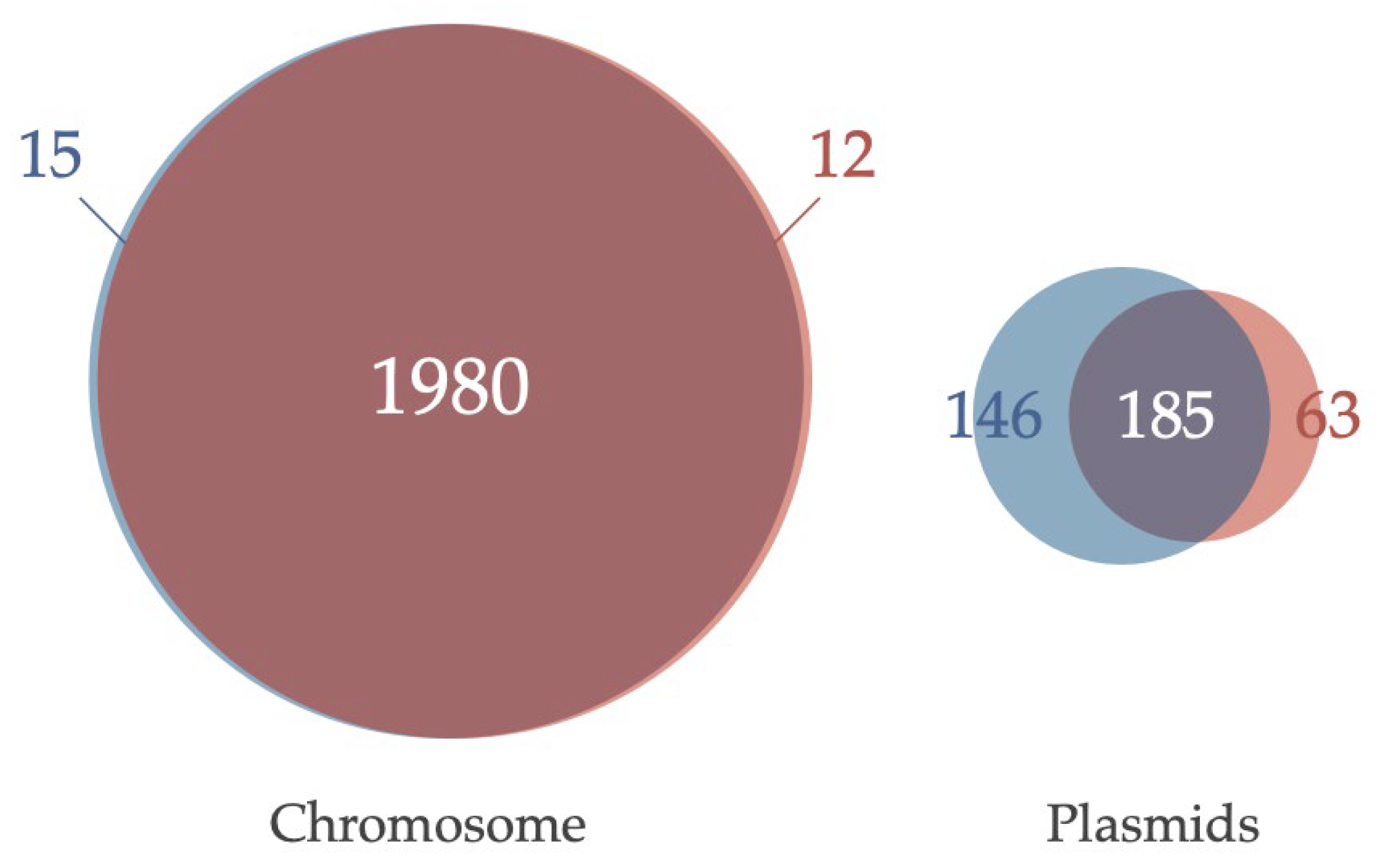
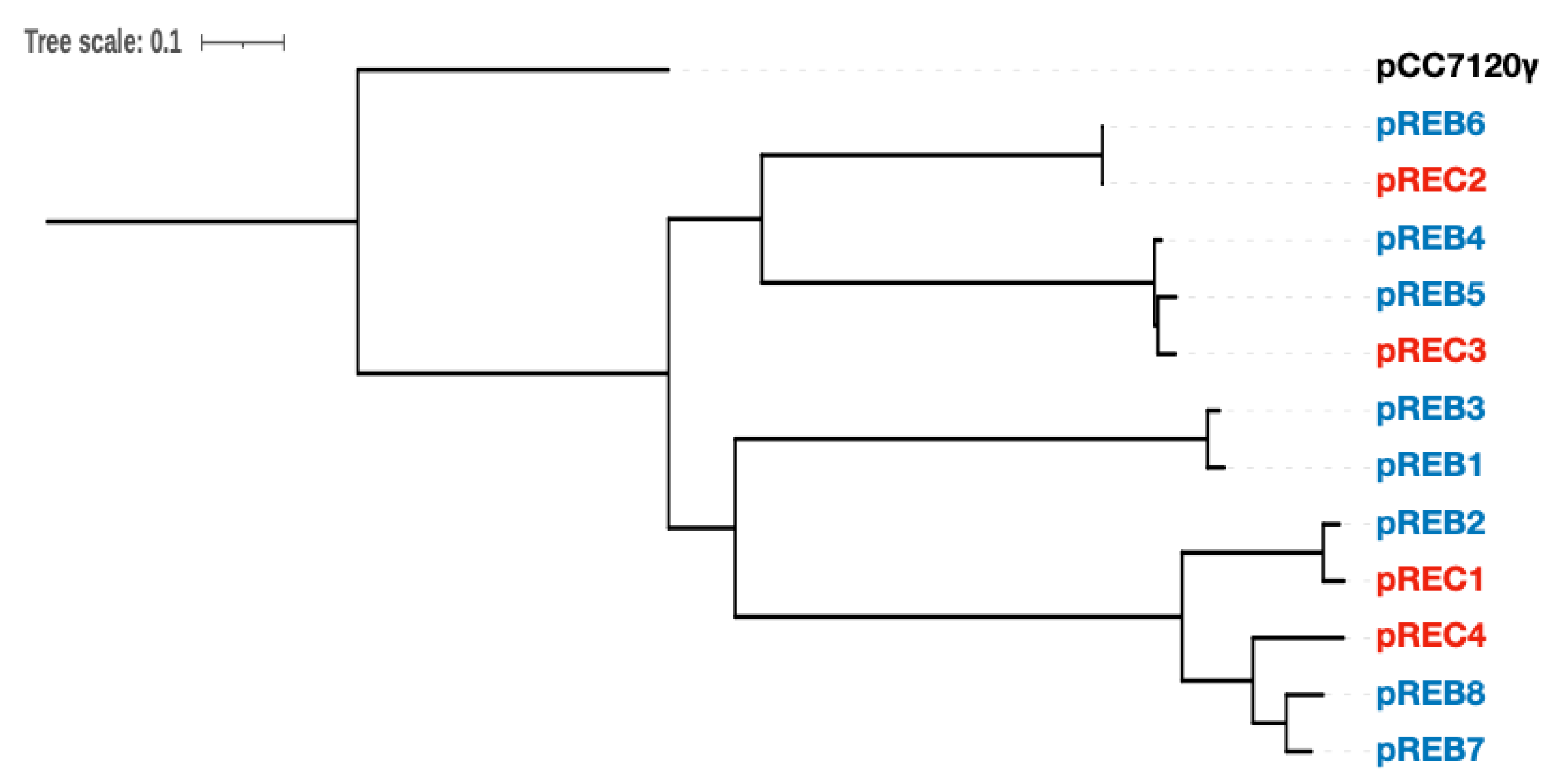
3.3. Comparative Analysis of Genes Conserved in Plasmids between Two Acaryochloris Species
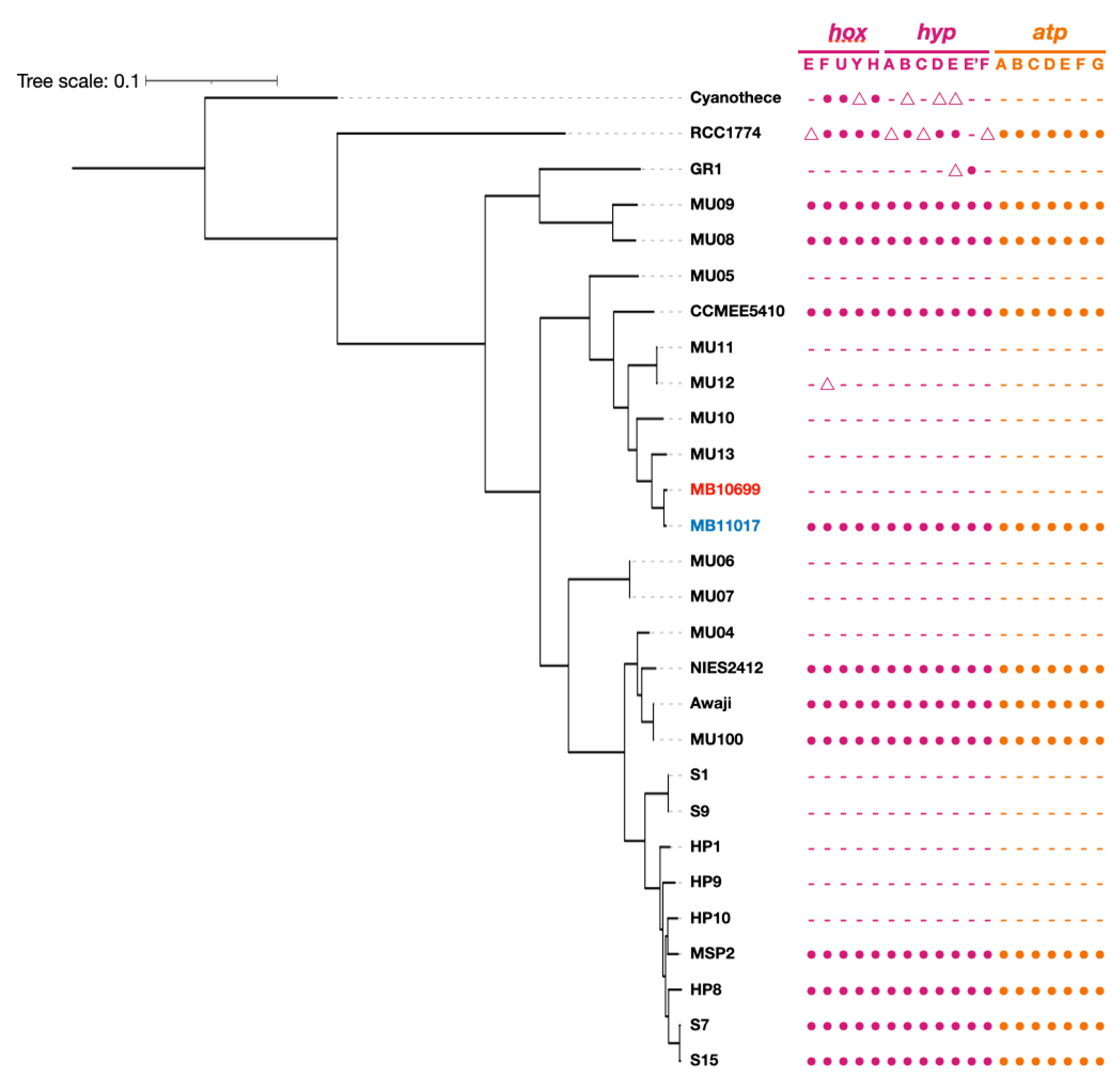
3.4. PBP-Related Genes
3.5. Genes Related to Alternative ATP Synthase and Hydrogenase
3.6. Genes Related to Photosynthetic Reaction Centers, Electron Transfer, and Chl-Binding Antenna Complex
3.7. Availability of Carbon Sources in Two Acaryochloris Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, M. Chlorophyll modifications and their spectral extension in oxygenic photosynthesis. Annu. Rev. Biochem. 2014, 83, 317–340. [Google Scholar] [CrossRef]
- Dekker, J.P.; Boekema, E.J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 2005, 1706, 12–39. [Google Scholar] [CrossRef]
- Miyashita, H.; Ikemoto, H.; Kurano, N.; Adachi, K.; Chihara, M.; Miyachi, S. Chlorophyll d as a major pigment. Nature 1996, 383, 402. [Google Scholar] [CrossRef]
- Hu, Q.; Miyashita, H.; Iwasaki, I.; Kurano, N.; Miyachi, S.; Iwaki, M.; Itoh, S. A photosystem I reaction center driven by chlorophyll d in oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 13319–13323. [Google Scholar] [CrossRef]
- Tomo, T.; Okubo, T.; Akimoto, S.; Yokono, M.; Miyashita, H.; Tsuchiya, T.; Noguchi, T.; Mimuro, M. Identification of the special pair of photosystem II in a chlorophyll d-dominated cyanobacterium. Proc. Natl. Acad. Sci. USA 2007, 104, 7283–7288. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.R.; Abresch, H.E.; Baroch, J.J.; Fishman Miller, C.K.; Garber, A.I.; Oman, A.R.; Ulrich, N.J. Genomic and Functional Variation of the Chlorophyll d-Producing Cyanobacterium Acaryochloris marina. Microorganisms 2022, 10, 569. [Google Scholar] [CrossRef]
- Swingley, W.D.; Chen, M.; Cheung, P.C.; Conrad, A.L.; Dejesa, L.C.; Hao, J.; Honchak, B.M.; Karbach, L.E.; Kurdoglu, A.; Lahiri, S.; et al. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl. Acad. Sci. USA 2008, 105, 2005–2010. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, N.J.; Uchida, H.; Kanesaki, Y.; Hirose, E.; Murakami, A.; Miller, S.R. Reacquisition of light-harvesting genes in a marine cyanobacterium confers a broader solar niche. Curr. Biol. 2021, 31, 1539–1546.e1534. [Google Scholar] [CrossRef] [PubMed]
- Pfreundt, U.; Stal, L.J.; Voß, B.; Hess, W.R. Dinitrogen fixation in a unicellular chlorophyll d-containing cyanobacterium. ISME J. 2012, 6, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, Y.; Tsukatani, Y.; Yamakawa, H.; Itoh, S.; Fujita, Y.; Yamamoto, H. Effects of Light and Oxygen on Chlorophyll d Biosynthesis in a Marine Cyanobacterium Acaryochloris marina. Plants 2022, 11, 915. [Google Scholar] [CrossRef]
- Zapata, M.; Rodríguez, F.; Garrido, J.L. Separation of chlorophylls and carotenoids from marine phytoplankton: A new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 2000, 195, 29–45. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Cabanettes, F.; Klopp, C. D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PEERJ 2018, 6, e4958. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Miyashita, H.; Adachi, K.; Kurano, N.; Ikemot, H.; Chihara, M.; Miyach, S. Pigment Composition of a Novel Oxygenic Photosynthetic Prokaryote Containing Chlorophyll d as the Major Chlorophyll. Plant Cell Physiol. 1997, 38, 274–281. [Google Scholar] [CrossRef]
- Chen, M.; Quinnell, R.G.; Larkum, A.W. The major light-harvesting pigment protein of Acaryochloris marina. FEBS Lett. 2002, 514, 149–152. [Google Scholar] [CrossRef]
- Li, Z.K.; Yin, Y.C.; Zhang, L.D.; Zhang, Z.C.; Dai, G.Z.; Chen, M.; Qiu, B.S. The identification of IsiA proteins binding chlorophyll d in the cyanobacterium Acaryochloris marina. Photosynth. Res. 2018, 135, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.S.; Gao, Y.; Wu, M. Re-evaluating the evidence for a universal genetic boundary among microbial species. Nat. Commun. 2021, 12, 4059. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Ishizuka, T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 2008, 7, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Fushimi, K.; Ni Ni, W.; Ikeuchi, M. Red-shifted red/green-type cyanobacteriochrome AM1_1870g3 from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Biochem. Biophys. Res. Commun. 2015, 461, 390–395. [Google Scholar] [CrossRef]
- Miyake, K.; Fushimi, K.; Kashimoto, T.; Maeda, K.; Ni Ni, W.; Kimura, H.; Sugishima, M.; Ikeuchi, M.; Narikawa, R. Functional diversification of two bilin reductases for light perception and harvesting in unique cyanobacterium Acaryochloris marina MBIC 11017. FEBS J. 2020, 287, 4016–4031. [Google Scholar] [CrossRef]
- Carrieri, D.; Wawrousek, K.; Eckert, C.; Yu, J.; Maness, P.C. The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 2011, 102, 8368–8377. [Google Scholar] [CrossRef]
- Veerman, J.; Bentley, F.K.; Eaton-Rye, J.J.; Mullineaux, C.W.; Vasil’ev, S.; Bruce, D. The PsbU subunit of photosystem II stabilizes energy transfer and primary photochemistry in the phycobilisome-photosystem II assembly of Synechocystis sp. PCC 6803. Biochemistry 2005, 44, 16939–16948. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Liu, X.; Zhao, J. FesM, a membrane iron-sulfur protein, is required for cyclic electron flow around photosystem I and photoheterotrophic growth of the cyanobacterium Synechococcus sp. PCC 7002. Plant Physiol. 2005, 138, 1586–1595. [Google Scholar] [CrossRef] [PubMed][Green Version]
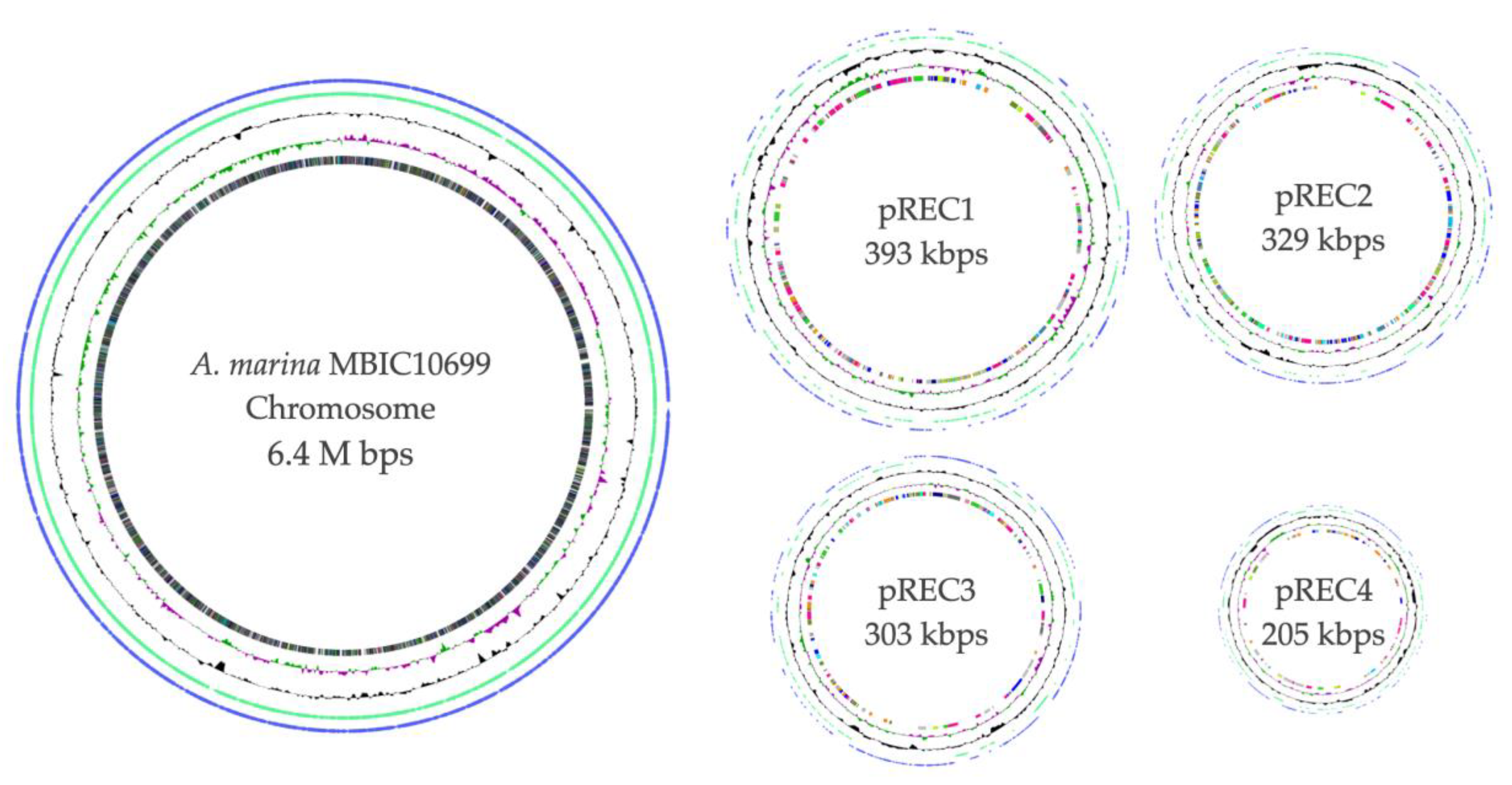


| Chr | pREC1 | pREC2 | pREC3 | pREC4 | Total | |
|---|---|---|---|---|---|---|
| Total Sequence Length (bp): | 6,415,507 | 393,608 | 329,949 | 303,490 | 205,174 | 7,647,728 |
| Number of Sequences: | 1 | 1 | 1 | 1 | 1 | 5 |
| Gap Ratio (%): | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| GC content (%): | 47.3 | 45.1 | 46.7 | 46.1 | 43.2 | 47.0 |
| Number of CDSs: | 5674 | 366 | 283 | 280 | 193 | 6813 |
| Coding Ratio (%): | 83.4 | 76.0 | 75.3 | 78.1 | 68.7 | 82.3 |
| Number of rRNAs: | 6 | 0 | 0 | 0 | 0 | 6 |
| Number of tRNAs: | 72 | 0 | 0 | 0 | 0 | 72 |
| Number of CRISPRs: | 1 | 2 | 0 | 0 | 0 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, H.; Uesaka, K.; Tsuzuki, Y.; Yamakawa, H.; Itoh, S.; Fujita, Y. Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids. Microorganisms 2022, 10, 1374. https://doi.org/10.3390/microorganisms10071374
Yamamoto H, Uesaka K, Tsuzuki Y, Yamakawa H, Itoh S, Fujita Y. Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids. Microorganisms. 2022; 10(7):1374. https://doi.org/10.3390/microorganisms10071374
Chicago/Turabian StyleYamamoto, Haruki, Kazuma Uesaka, Yuki Tsuzuki, Hisanori Yamakawa, Shigeru Itoh, and Yuichi Fujita. 2022. "Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids" Microorganisms 10, no. 7: 1374. https://doi.org/10.3390/microorganisms10071374
APA StyleYamamoto, H., Uesaka, K., Tsuzuki, Y., Yamakawa, H., Itoh, S., & Fujita, Y. (2022). Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids. Microorganisms, 10(7), 1374. https://doi.org/10.3390/microorganisms10071374






