Expression of Macrophage Polarization Markers against the Most Prevalent Serotypes of Aggregatibacter actinomycetemomitans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Stains
2.2. THP-1-Derived Macrophages Culture
2.3. Infection Assay
2.4. RNA Extraction and RT-PCR
2.5. qPCR
2.6. Statistical Analysis
3. Results
3.1. Expression of M0 Markers in Response to Aggregatibacter actinomycetemcomitans Serotypes
3.2. Expression of M1 Markers in Response to Aggregatibacter actinomycetemcomitans Serotypes
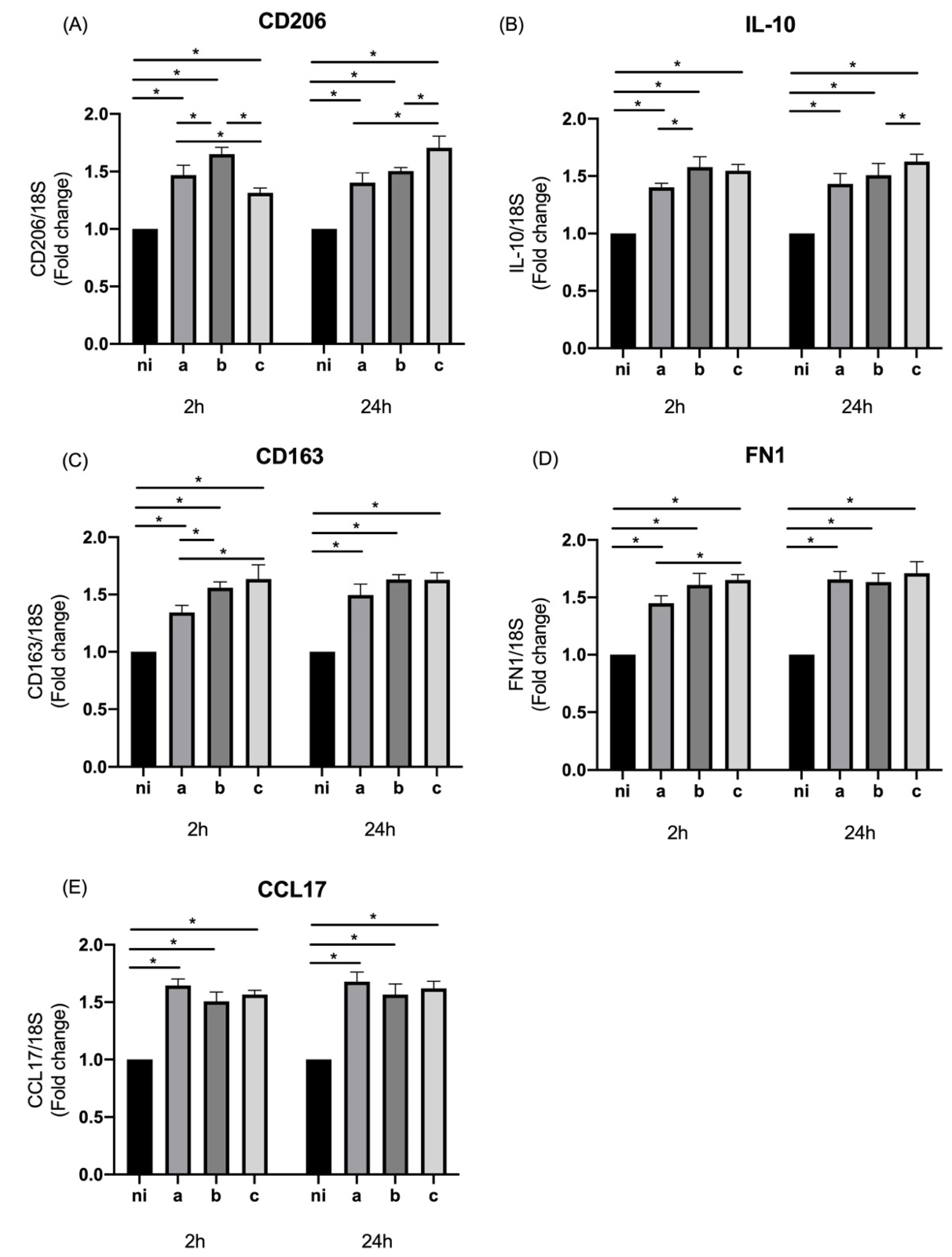
3.3. Expression of M2 Markers in Response to Aggregatibacter actinomycetemcomitans Serotypes
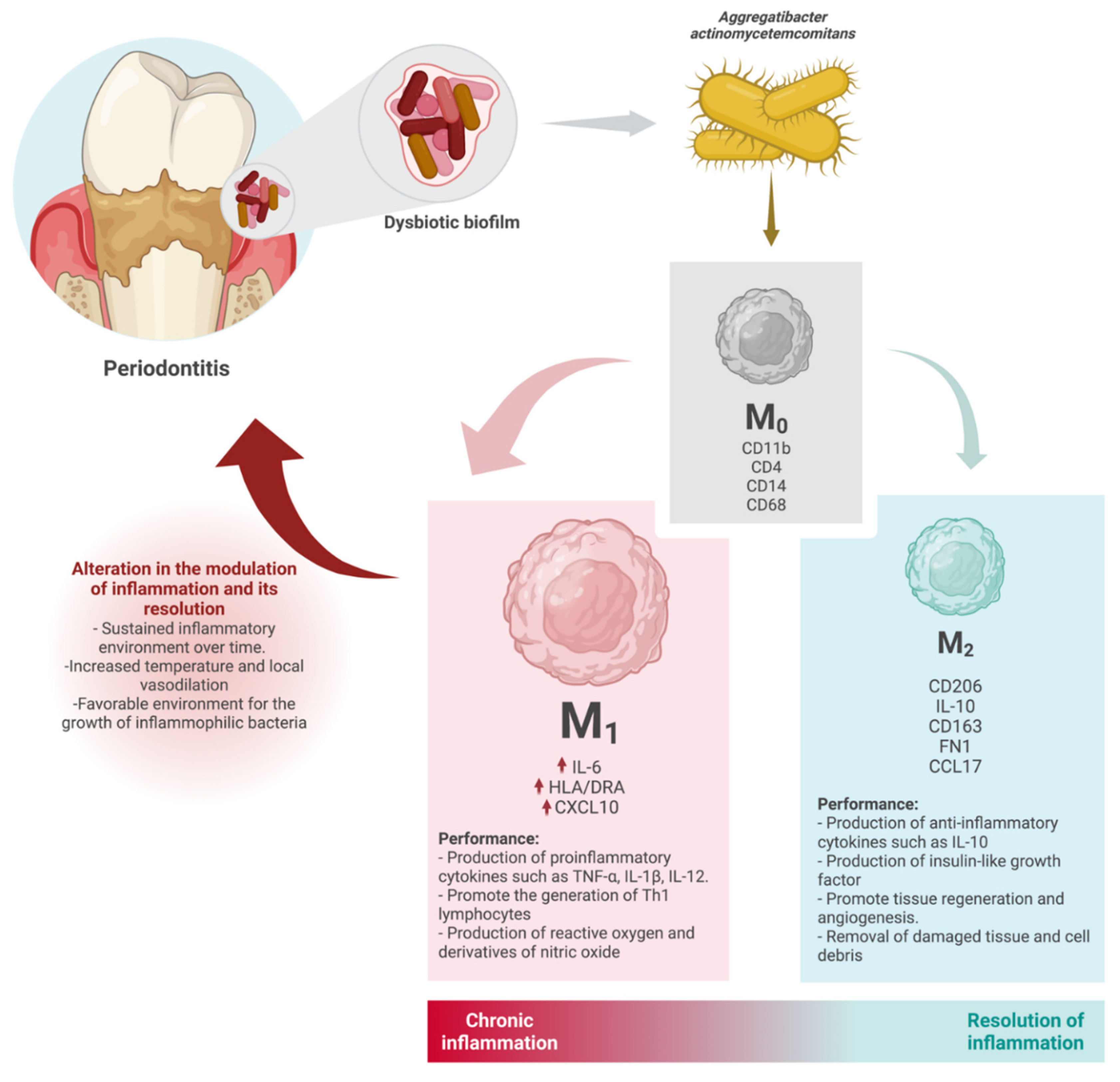
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Grassi, R.; Nardi, G.M.; Mazur, M.; Di Giorgio, R.; Ottolenghi, L.; Guerra, F. The Dental-BIOfilm Detection TECHnique (D-BioTECH): A Proof of Concept of a Patient-Based Oral Hygiene. Medicina 2022, 58, 537. [Google Scholar] [CrossRef]
- Bandara, H.M.H.N.; Panduwawala, C.P.; Samaranayake, L.P. Biodiversity of the human oral mycobiome in health and disease. Oral Dis. 2019, 25, 363–371. [Google Scholar] [CrossRef]
- Morse, D.J.; Wilson, M.J.; Wei, X.; Lewis, M.A.; Bradshaw, D.J.; Murdoch, C.; Williams, D.W. Denture-associated biofilm infection in three-dimensional oral mucosal tissue models. J. Med. Microbiol. 2018, 67, 364–375. [Google Scholar] [CrossRef] [Green Version]
- Simón-Soro, A.; Mira, A. Solving the etiology of dental caries. Trends Microbiol. 2015, 23, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the Oral Microbiota in Health: Mechanisms That Prevent Dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Radaic, A.; Kapila, Y.L. The oralome and its dysbiosis: New insights into oral microbiome-host interactions. Comput. Struct. Biotechnol. J. 2021, 19, 1335–1360. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Idris, A.; Hasnain, S.Z.; Huat, L.Z.; Koh, D. Human diseases, immunity and the oral microbiota—Insights gained from metagenomic studies. Oral Sci. Int. 2017, 14, 27–32. [Google Scholar]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef] [Green Version]
- Avila, M.; Ojcius, D.M.; Yilmaz, O. The oral microbiota: Living with a permanent guest. DNA Cell Biol. 2009, 28, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Mosaddad, S.A.; Tahmasebi, E.; Yazdanian, A.; Rezvani, M.B.; Seifalian, A.; Yazdanian, M.; Tebyanian, H. Oral microbial bio- films: An update. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2005–2019. [Google Scholar] [CrossRef] [PubMed]
- Slots, J.; Ting, M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: Occurrence and treatment. Periodontology 2000 1999, 20, 82–121. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef]
- Fine, D.H.; Patil, A.G.; Velusamy, S.K. Aggregatibacter actinomycetemcomitans (Aa) Under the Radar: Myths and Misunderstand- ings of Aa and Its Role in Aggressive Periodontitis. Front. Immunol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Pormohammad, A.; Eslami, H.; Shokouhi, B.; Fakhrzadeh, V.; Kafil, H.S. Oral pathogenesis of Aggregatibacter actinomycetemcomitans. Microb. Pathog. 2017, 113, 303–311. [Google Scholar] [CrossRef]
- Raja, M.; Ummer, F.; Dhivakar, C.P. Aggregatibacter actinomycetemcomitans—A tooth killer? J. Clin. Diagn. Res. 2014, 8, ZE13–ZE16. [Google Scholar] [CrossRef]
- Tuuli, A.; Laura, K.; Terhi, M.; Jan, O.; Riikka, I. Aggregatibacter actinomycetemcomitans LPS binds human interleukin-8. J. Oral Microbiol. 2018, 11, 1549931. [Google Scholar] [CrossRef] [Green Version]
- Takada, K.; Saito, M.; Tsuzukibashi, O.; Kawashima, Y.; Ishida, S.; Hirasawa, M. Characterization of a new serotype g isolate of Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 2010, 25, 200–206. [Google Scholar] [CrossRef]
- Melgar-Rodríguez, S.; Díaz-Zúñiga, J.; Alvarez, C.; Rojas, L.; Monasterio, G.; Carvajal, P.; Escobar, A.; Sanz, M.; Vernal, R. Sero- type b of Aggregatibacter actinomycetemcomitans increases osteoclast and memory T-lymphocyte activation. Mol. Oral Microbiol. 2016, 31, 162–174. [Google Scholar] [CrossRef]
- Díaz-Zúñiga, J.; Monasterio, G.; Alvarez, C.; Melgar-Rodríguez, S.; Benítez, A.; Ciuchi, P.; García, M.; Arias, J.; Sanz, M.; Vernal, R. Variability of the dendritic cell response triggered by different serotypes of Aggregatibacter actinomycetemcomitans or Porphy- romonas gingivalis is toll-like receptor 2 (TLR2) or TLR4 dependent. J. Periodontol. 2015, 86, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, C.; Benítez, A.; Rojas, L.; Pujol, M.; Carvajal, P.; Díaz-Zúñiga, J.; Vernal, R. Differential expression of CC chemokines (CCLs) and receptors (CCRs) by human T lymphocytes in response to different Aggregatibacter actinomycetemcomitans serotypes. J. Appl. Oral Sci. 2015, 23, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Vernal, R.; Leon, R.; Herrera, D.; Garcia-Sanz, J.A.; Silva Sanz, M. Variability in the response of human dendritic cells stimulated with Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans. J. Periodontal Res. 2008, 43, 689–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Zúñiga, J.; Yáñez, J.P.; Alvarez, C.; Melgar-Rodríguez, S.; Hernández, M.; Sanz, M.; Vernal, R. Serotype-dependent re- sponse of human dendritic cells stimulated with Aggregatibacter actinomycetemcomitans. J. Clin. Periodontol. 2014, 41, 242–251. [Google Scholar] [CrossRef]
- McCormick, T.S.; Weinberg, A. Epithelial cell-derived antimicrobial peptides are multifunctional agents that bridge innate and adaptive immunity. Periodontology 2000 2010, 54, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Ramage, G.; Lappin, D.F.; Millhouse, E.; Malcolm, J.; Jose, A.; Yang, J.; Bradshaw, D.J.; Pratten, J.R.; Culshaw, S. The epithelial cell response to health and disease associated oral biofilm models. J. Periodontal Res. 2017, 52, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Kochi, S.; Yamashiro, K.; Hongo, S.; Yamamoto, T.; Ugawa, Y.; Shimoe, M.; Kawamura, M.; Hirata-Yoshihara, C.; Ideguchi, H.; Maeda, H.; et al. Aggregatibacter actinomycetemcomitans regulates the expression of integrins and reduces cell adhesion via integrin α5 in human gingival epithelial cells. Mol. Cell. Biochem. 2017, 436, 39–48. [Google Scholar] [CrossRef]
- Suga, T.; Mitani, A.; Mogi, M.; Kikuchi, T.; Fujimura, T.; Takeda, H.; Hishikawa, T.; Yamamoto, G.; Hayashi, J.; Ishihara, Y.; et al. Aggregatibacter actinomycetemcomitans lipopolysaccharide stimulated epithelial cells produce interleukin-15 that regulates T cell activation. Arch. Oral Biol. 2013, 58, 1541–1548. [Google Scholar] [CrossRef]
- Betancur, D.; Muñoz Grez, C.; Oñate, A. Comparative Analysis of Cytokine Expression in Oral Keratinocytes and THP-1 Macrophages in Response to the Most Prevalent Serotypes of Aggregatibacter actinomycetemcomitans. Microorganisms 2021, 9, 622. [Google Scholar] [CrossRef]
- Park, E.K.; Jung, H.S.; Yang, H.I.; Yoo, M.C.; Kim, C.; Kim, K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 2007, 56, 45–50, Erratum in Inflamm. Res. 2020, 69, 1157. [Google Scholar] [CrossRef]
- Daigneault, M.; Preston, J.A.; Marriott, H.M.; Whyte, M.K.; Dockrell, D.H. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, X.; Mosser, D.M. Macrophage activation by endogenous danger signals. J. Pathol. 2008, 214, 161–178. [Google Scholar] [CrossRef]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [Green Version]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Zhou, L.N.; Bi, C.S.; Gao, L.N.; An, Y.; Chen, F.; Chen, F.M. Macrophage polarization in human gingival tissue in response to periodontal disease. Oral Dis. 2019, 25, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Verreck, F.A.; de Boer, T.; Langenberg, D.M.; Hoeve, M.A.; Kramer, M.; Vaisberg, E.; Kastelein, R.; Kolk, A.; de Waal-Malefyt, R.; Ottenhoff, T.H. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc. Natl. Acad. Sci. USA 2004, 101, 4560–4565. [Google Scholar] [CrossRef] [Green Version]
- Pan, W.; Wang, Q.; Chen, Q. The cytokine network involved in the host immune response to periodontitis. Int. J. Oral Sci. 2019, 11, 30. [Google Scholar] [CrossRef] [Green Version]
- Tarique, A.A.; Logan, J.; Thomas, E.; Holt, P.G.; Sly, P.D.; Fantino, E. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am. J. Respir. Cell Mol. Biol. 2015, 53, 676–688. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S.; Locati, M.; Mantovani, A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: New molecules and patterns of gene expression. J. Immunol. 2006, 177, 7303–7311. [Google Scholar] [CrossRef] [Green Version]
- Garaicoa-Pazmino, C.; Fretwurst, T.; Squarize, C.H.; Berglundh, T.; Giannobile, W.V.; Larsson, L.; Castilho, R.M. Characterization of macrophage polarization in periodontal disease. J. Clin. Periodontol. 2019, 46, 830–839. [Google Scholar] [CrossRef]
- Liu, J.; Chen, B.; Bao, J.; Zhang, Y.; Lei, L.; Yan, F. Macrophage polarization in periodontal ligament stem cells enhanced periodontal regeneration. Stem Cell Res. Ther. 2019, 10, 320. [Google Scholar] [CrossRef]
- Vogel, D.Y.; Glim, J.E.; Stavenuiter, A.W.; Breur, M.; Heijnen, P.; Amor, S.; Dijkstra, C.D.; Beelen, R.H. Human macrophage polarization in vitro: Maturation and activation methods compared. Immunobiology 2014, 219, 695–703. [Google Scholar] [CrossRef]
- O’Brien, E.M.; Spiller, K.L. Pro-inflammatory polarization primes Macrophages to transition into a distinct M2-like phenotype in response to IL-4. J. Leukoc. Biol. 2022, 111, 989–1000. [Google Scholar] [CrossRef]
- Sima, C.; Glogauer, M. Macrophage subsets and osteoimmunology: Tuning of the immunological recognition and effector systems that maintain alveolar bone. Periodontol 2000 2013, 63, 80–101. [Google Scholar] [CrossRef]
- Pandruvada, S.; Farrar, N.; Al-Kakhan, W.; Emecen-Huja, P.; Yilmaz, O. Modeling macrophage response to periodontal infections in vitro. FASEB J. 2022, 36. [Google Scholar] [CrossRef]
- Chen, G.; Sun, Q.; Cai, Q.; Zhou, H. Outer Membrane Vesicles From Fusobacterium nucleatum Switch M0-Like Macrophages Toward the M1 Phenotype to Destroy Periodontal Tissues in Mice. Front. Microbiol. 2022, 13, 815638. [Google Scholar] [CrossRef]
- Dikilitas, A.; Karaaslan, F.; Aydin, E.Ö.; Yigit, U.; Ertugrul, A.S. Granulocyte-macrophage colony-stimulating factor (GM-CSF) in subjects with different stages of periodontitis according to the new classification. J. Appl. Oral Sci. 2022, 30, e20210423. [Google Scholar] [CrossRef]
- Naginyte, M.; Do, T.; Meade, J.; Devine, D.A.; Marsh, P.D. Enrichment of periodontal pathogens from the biofilms of healthy adults. Sci. Rep. 2019, 9, 5491. [Google Scholar] [CrossRef]
- Chen, C.; Hemme, C.; Beleno, J.; Shi, Z.J.; Ning, D.; Qin, Y.; Tu, Q.; Jorgensen, M.; He, Z.; Wu, L.; et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018, 12, 1210–1224. [Google Scholar] [CrossRef]
- Van Dyke, T.E.; Bartold, P.M.; Reynolds, E.C. The Nexus between Periodontal Inflammation and Dysbiosis. Front. Immunol. 2020, 11, 511. [Google Scholar] [CrossRef] [PubMed]


| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CD11b | cagcctttgaccttatgtcatgg | cctgtgctgtagtcgcact |
| CD4 | cctcctgcttttcattgggctag | tgaggacactggcaggtcttct |
| CD14 | ctggaacaggtgcctaaaggac | gtccagtgtcaggttatccacc |
| CD68 | cgagcatcattctttcaccagct | atgagaggcagcaagatggacc |
| IL-6 | agacagccactcacctcttcag | ttctgccagtgcctctttgctg |
| HLA-DRA | aaaaggagggagttacactcagg | gctgtgagggacacatcagag |
| CXCL10 | ggtgagaagagatgtctgaatcc | gtccatccttggaagcactgca |
| CD206 | gcaaagtggattacgtgtcttg | ctgttatgtcgctggcaaatg |
| IL-10 | tctccgagatgccttcagcaga | tcagacaaggcttggcaaccca |
| CD163 | ccagtcccaaacactgtcct | atgccagtgagcttcccgttcagc |
| FN1 | acaacaccgaggtgactgagac | ggacacaacgatgcttcctgag |
| CCL17 | ccagggatgccatcgtttttgtaactgtgc | cctcactgtggctcttcttcgtccctggaa |
| 18S | ctcaacacgggaaacctcac | cgctccaccaactaagaacg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betancur, D.; Muñoz, C.; Oñate, A. Expression of Macrophage Polarization Markers against the Most Prevalent Serotypes of Aggregatibacter actinomycetemomitans. Microorganisms 2022, 10, 1384. https://doi.org/10.3390/microorganisms10071384
Betancur D, Muñoz C, Oñate A. Expression of Macrophage Polarization Markers against the Most Prevalent Serotypes of Aggregatibacter actinomycetemomitans. Microorganisms. 2022; 10(7):1384. https://doi.org/10.3390/microorganisms10071384
Chicago/Turabian StyleBetancur, Daniel, Camila Muñoz, and Angel Oñate. 2022. "Expression of Macrophage Polarization Markers against the Most Prevalent Serotypes of Aggregatibacter actinomycetemomitans" Microorganisms 10, no. 7: 1384. https://doi.org/10.3390/microorganisms10071384
APA StyleBetancur, D., Muñoz, C., & Oñate, A. (2022). Expression of Macrophage Polarization Markers against the Most Prevalent Serotypes of Aggregatibacter actinomycetemomitans. Microorganisms, 10(7), 1384. https://doi.org/10.3390/microorganisms10071384






