Efficiency of Combining Strains Ag87 (Bacillus megaterium) and Ag94 (Lysinibacillus sp.) as Phosphate Solubilizers and Growth Promoters in Maize
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening of Bacterial Isolates—Phosphate Solubilization and Indole Acetic Acid Production
2.2. Preparation of Bacterial Isolates
2.3. Laboratory and Greenhouse Tests
2.4. Genomic Sequencing of the Strains Ag87 and Ag94
2.5. Field Trials—Harvest (2020/2021)
2.6. Field Trials—Second Harvest (2021)
2.7. Data Analysis
3. Results
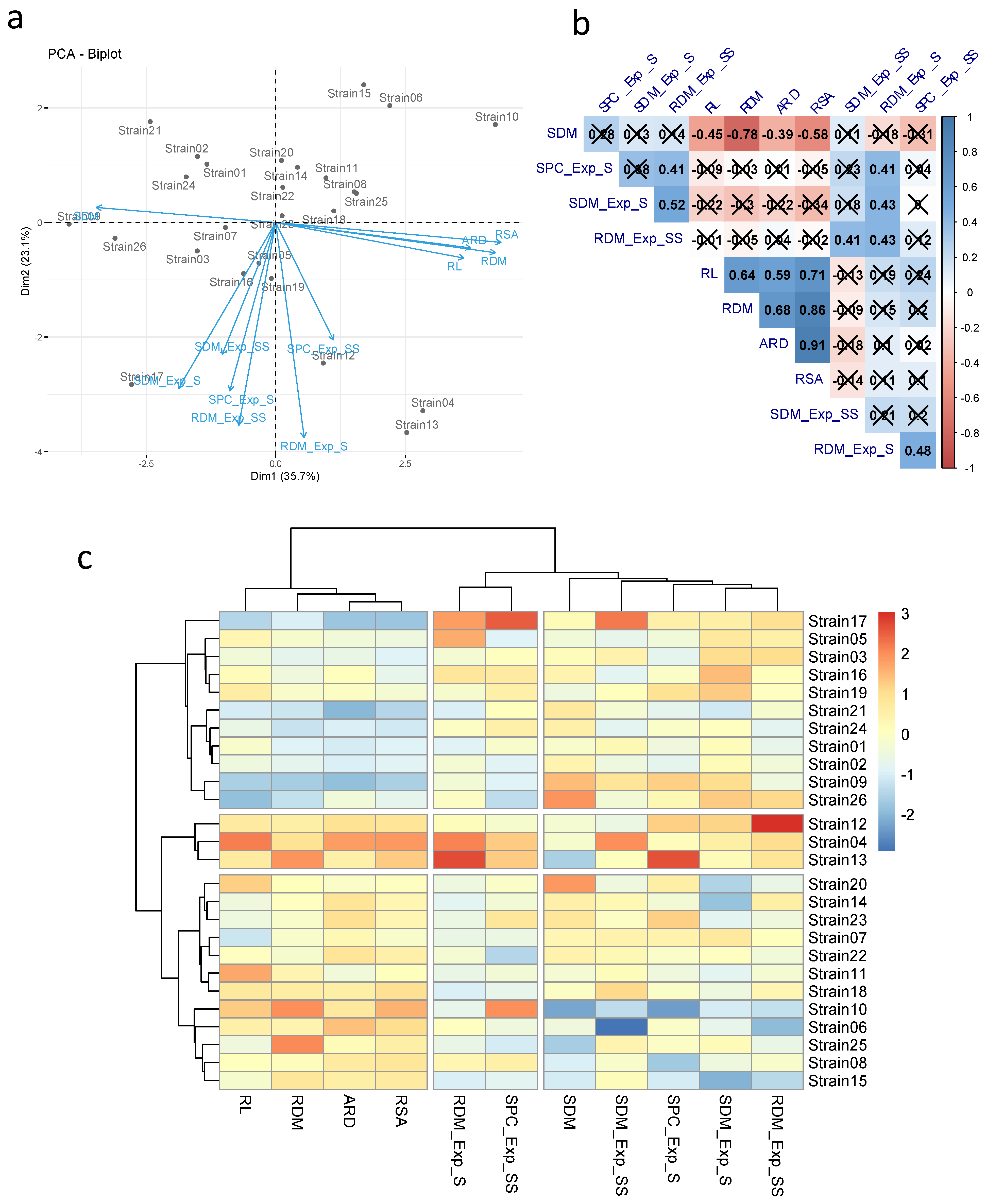
3.1. Screening of Bacterial Strains
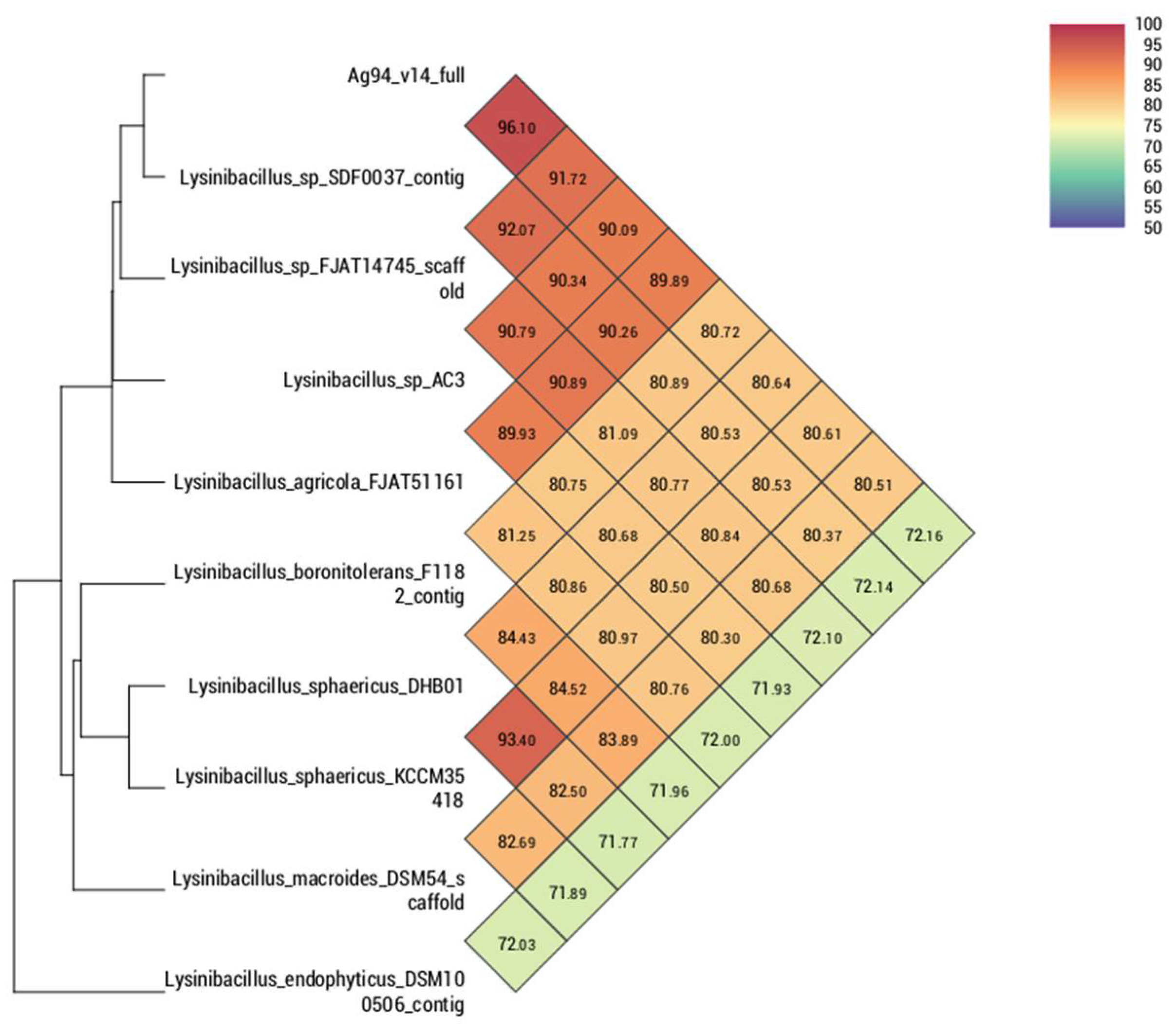
3.2. Genomic Analysis of Strains Ag87 and Ag94
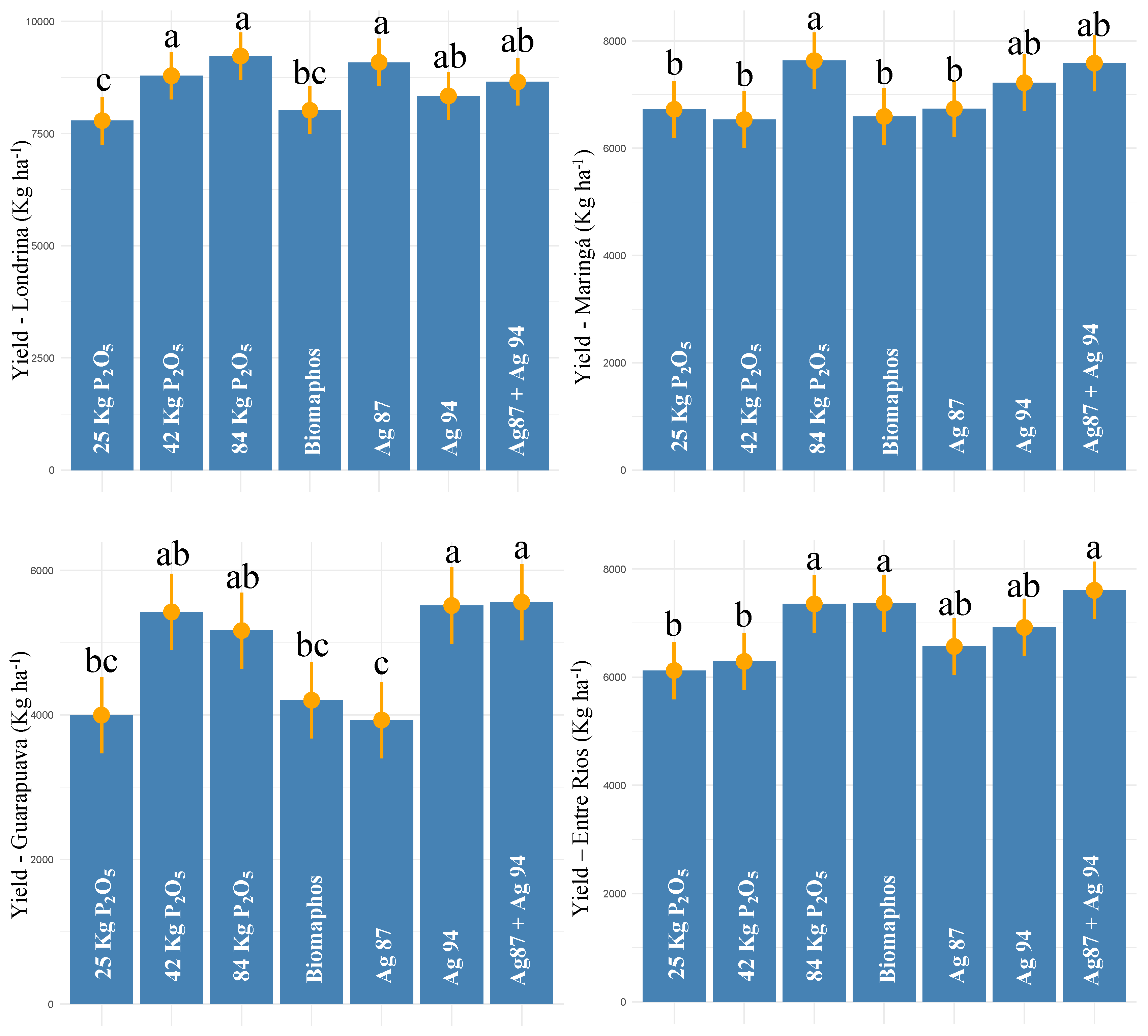
3.3. Field Data—Harvest 2020/2021
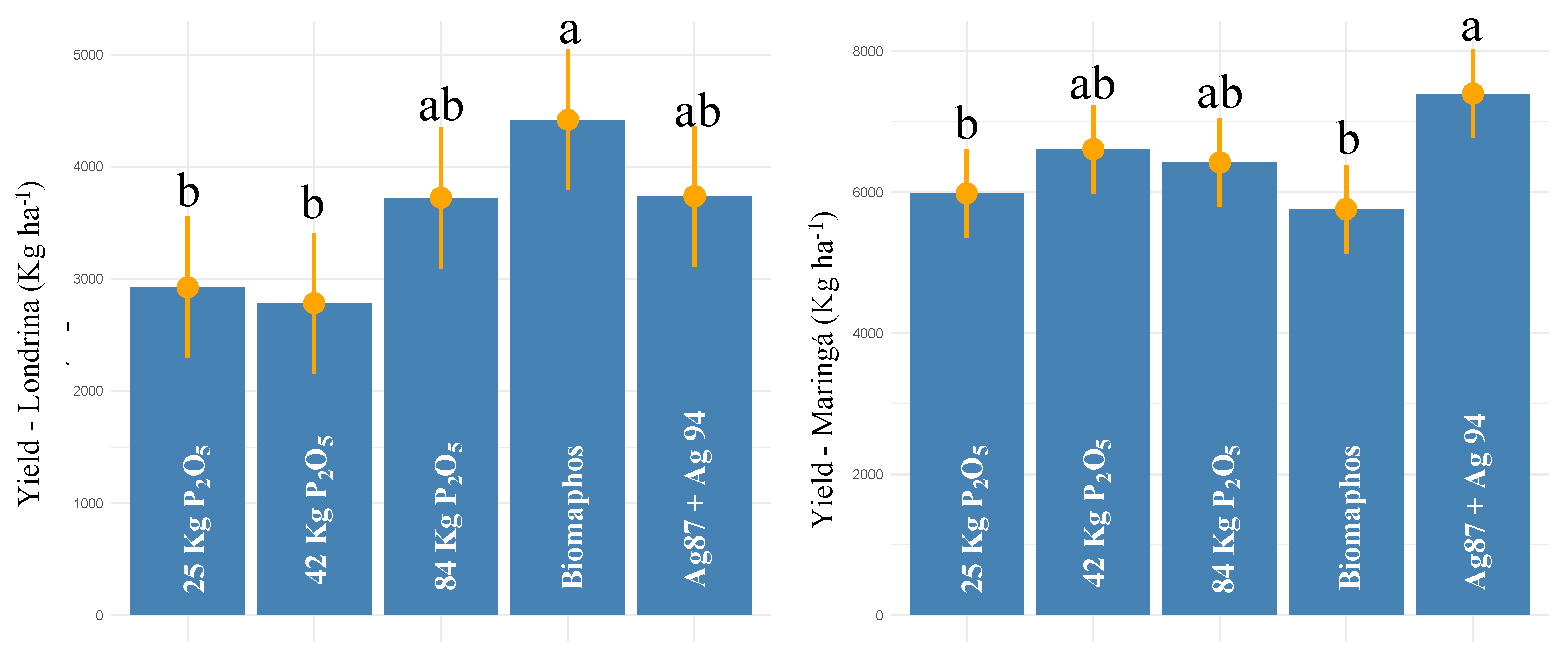
3.4. Field Data—Second Harvest 2021
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Taiz, L.; Zeiger Ian Max Møller, E.; Murphy, A. Fisiologia e Desenvolvimento Vegetal—6a Edição; Sinauer Associates, Inc.: Sunderland, MA, USA, 2016; ISBN 9788582713679. [Google Scholar]
- Billah, M.; Khan, M.; Bano, A.; Hassan, T.U.; Munir, A.; Gurmani, A.R. Phosphorus and Phosphate Solubilizing Bacteria: Keys for Sustainable Agriculture. Geomicrobiol. J. 2019, 36, 904–916. [Google Scholar] [CrossRef]
- Brownlie, W.J.; Sutton, M.A.; Reay, D.S.; Heal, K.V.; Hermann, L.; Kabbe, C.; Spears, B.M. Global Actions for a Sustainable Phosphorus Future. Nat. Food 2021, 2, 71–74. [Google Scholar] [CrossRef]
- Baveye, P.C. Looming Scarcity of Phosphate Rock and Intensification of Soil Phosphorus Research. Rev. Bras. Ciência Solo 2015, 39, 637–642. [Google Scholar] [CrossRef] [Green Version]
- Cordell, D.; Drangert, J.O.; White, S. The Story of Phosphorus: Global Food Security and Food for Thought. Glob. Environ. Change 2009, 19, 292–305. [Google Scholar] [CrossRef]
- CONAB. Operational Logistics Superintendence—Logistic Bulletin; CONAB: Brasília, Brazil, 2022. [Google Scholar]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [Green Version]
- Bargaz, A.; Elhaissoufi, W.; Khourchi, S.; Benmrid, B.; Borden, K.A.; Rchiad, Z. Benefits of Phosphate Solubilizing Bacteria on Belowground Crop Performance for Improved Crop Acquisition of Phosphorus. Microbiol. Res. 2021, 252, 126842. [Google Scholar] [CrossRef]
- Fatima, F.; Ahmad, M.M.; Verma, S.R.; Pathak, N. Relevance of Phosphate Solubilizing Microbes in Sustainable Crop Production: A Review. Int. J. Environ. Sci. Technol. 2021. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate Solubilizing Microbes: Sustainable Approach for Managing Phosphorus Deficiency in Agricultural Soils. SpringerPlus 2013, 2, 587. [Google Scholar] [CrossRef] [Green Version]
- Kalayu, G. Phosphate Solubilizing Microorganisms: Promising Approach as Biofertilizers. Int. J. Agron. 2019, 2019, 4917256. [Google Scholar] [CrossRef]
- Alori, E.T.; Babalola, O.O. Microbial Inoculants for Improving Crop Quality and Human Health in Africa. Front. Microbiol. 2018, 9, 2213. [Google Scholar] [CrossRef] [Green Version]
- Conab Companhia Nacional de Abastecimento. Acompanhamento Da Safra Brasileira de Grãos, Brasília, DF, v. 8, Safra 2020/21, n. 12 Décimo Segundo Levantamento, Setembro. 2021. Available online: https://www.conab.gov.br/info-agro/safras/graos/boletim-da-safra-de-graos (accessed on 7 April 2022).
- Nautiyal, C.S. An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Sarwar, M.; Kremer, R.J. Determination of Bacterially Derived Auxins Using a Microplate Method. Lett. Appl. Microbiol. 1995, 20, 282–285. [Google Scholar] [CrossRef]
- Galkovskyi, T.; Mileyko, Y.; Bucksch, A.; Moore, B.; Symonova, O.; Price, C.A.; Topp, C.N.; Iyer-Pascuzzi, A.S.; Zurek, P.R.; Fang, S.; et al. GiA Roots: Software for the High Throughput Analysis of Plant Root System Architecture. BMC Plant Biol. 2012, 12, 116. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Armon, D.I. The Water-Culture Method for Growing Plants without Soil. Circular 1950, 347, 32. [Google Scholar] [CrossRef]
- Magnavaca, R.; Gardner, C.O.; Clark, R.B. Evaluation of Inbred Maize Lines for Aluminum Tolerance in Nutrient Solution. In Genetic Aspects of Plant Mineral Nutrition; Springer: Dordrecht, The Netherlands, 1987; pp. 255–265. [Google Scholar]
- Malavolta, E.; Vitti, G.C.; Oliveira, S.A. Evaluation of the Nutritional State of Plants: Principles and Applications—Avaliação Do Estado Nutricional de Plantas: Princípios e Aplicações, 2nd ed.; Potafos: Piracicaba, Brazil, 1997. [Google Scholar]
- Pradhan, S.; Pokhrel, M.R. Spectrophotometric Determination of Phosphate in Sugarcane Juice, Fertilizer, Detergent and Water Samples by Molybdenum Blue Method. Sci. World 2013, 11, 58–62. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 7 April 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Genome Analysis Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.-J.; Lilly, S.J.; Kovač, K.; Bolzonella, M.; Pozzetti, L.; Renzini, A.; Zamorani, G.; Ilbert, O.; Knobel, C.; Iovino, A.; et al. Mass and environment as drivers of galaxy evolution in SDSS and zCOSMOS and the origin of the schechter function. Astrophys. J. 2010, 721, 193–221. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Genome Analysis QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Alikhan, N.-F.; Petty, N.K.; ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Ågren, J.; Sundström, A.; Håfström, T.; Segerman, B. Gegenees: Fragmented Alignment of Multiple Genomes for Determining Phylogenomic Distances and Genetic Signatures Unique for Specified Target Groups. PLoS ONE 2012, 7, e39107. [Google Scholar] [CrossRef]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 1, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and Interpretation of Factors Which Contribute to Efficiency of Nitrogen Utilization. Agron. J. 1982, 74, 562–564. [Google Scholar] [CrossRef]
- Shimizu, G.D.; Marubayashi, R.Y.P.; Goncalves, L.S.A. Package AgroR version 1.2.0., Cran R. 2021. Available online: https://cran.r-project.org/web/packages/AgroR/index.html (accessed on 15 May 2022).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps., Cran R. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 15 May 2022).
- Wickham, H. Ggplot2; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-Solubilizing Microorganisms: Mechanism and Their Role in Phosphate Solubilization and Uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Kudoyarova, G.R.; Vysotskaya, L.B.; Arkhipova, T.N.; Kuzmina, L.Y.; Galimsyanova, N.F.; Sidorova, L.V.; Gabbasova, I.M.; Melentiev, A.I.; Veselov, S.Y. Effect of Auxin Producing and Phosphate Solubilizing Bacteria on Mobility of Soil Phosphorus, Growth Rate, and P Acquisition by Wheat Plants. Acta Physiol. Plant. 2017, 39, 253. [Google Scholar] [CrossRef]
- Zeffa, D.M.; Perini, L.J.; Silva, M.B.; de Sousa, N.V.; Scapim, C.A.; de Oliveira, A.L.M.; do Amaral Júnior, A.T.; Azeredo Gonçalves, L.S. Azospirillum brasilense Promotes Increases in Growth and Nitrogen Use Efficiency of Maize Genotypes. PLoS ONE 2019, 14, e0215332. [Google Scholar] [CrossRef] [Green Version]
- Etesami, H.; Alikhani, H.A.; Hosseini, H.M. Indole-3-Acetic Acid (IAA) Production Trait, a Useful Screening to Select Endophytic and Rhizosphere Competent Bacteria for Rice Growth Promoting Agents. MethodsX 2015, 2, 72–78. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C. The Endophytic Continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [Green Version]
- Fesel, P.H.; Zuccaro, A. Dissecting Endophytic Lifestyle along the Parasitism/Mutualism Continuum in Arabidopsis. Curr. Opin. Microbiol. 2016, 32, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Kumar, A.; Patel, H. Role of Microbes in Phosphorus Availability and Acquisition by Plants. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1344–1347. [Google Scholar] [CrossRef]
- Huang, F.L.; Zhang, Y.; Zhang, L.-P.; Wang, S.; Feng, Y.; Rong, N.-H. Complete Genome Sequence of Bacillus megaterium JX285 Isolated from Camellia oleifera Rhizosphere. Comput. Biol. Chem. 2019, 79, 1–5. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Hernández, A.G.; Glick, B.R.; Rossi, M.J. Plant Growth-Promoting Activities and Genomic Analysis of the Stress-Resistant Bacillus megaterium STB1, a Bacterium of Agricultural and Biotechnological Interest. Biotechnol. Rep. 2020, 25, e00406. [Google Scholar] [CrossRef]
- de Freitas Duarte, N.; Oliveira Paiva, C.A.; Pagano, M.C.; Correa, E.J.A. Phosphate Solubilization by Microorganisms. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2022; pp. 257–282. [Google Scholar] [CrossRef]
- Zhao, Y.; Mao, X.; Zhang, M.; Yang, W.; Di, H.J.; Ma, L.; Liu, W.; Li, B. The Application of Bacillus megaterium Alters Soil Microbial Community Composition, Bioavailability of Soil Phosphorus and Potassium, and Cucumber Growth in the Plastic Shed System of North China. Agric. Ecosyst. Environ. 2021, 307, 107236. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans Gen. Nov. Sp. Nov., and Transfer of Bacillus Fusiformis to Lysinibacillus fusiformis Comb. Nov. and Bacillus sphaericus to Lysinibacillus sphaericus Comb. Nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Shabanamol, S.; Divya, K.; George, T.K.; Rishad, K.S.; Sreekumar, T.S.; Jisha, M.S. Characterization and in Planta Nitrogen Fixation of Plant Growth Promoting Endophytic Diazotrophic Lysinibacillus sphaericus Isolated from Rice (Oryza sativa). Physiol. Mol. Plant Pathol. 2018, 102, 46–54. [Google Scholar] [CrossRef]
- Aguirre-Monroy, A.M.; Santana-Martínez, J.C.; Dussán, J. Lysinibacillus sphaericus as a Nutrient Enhancer during Fire-Impacted Soil Replantation. Appl. Environ. Soil Sci. 2019, 2019, 3075153. [Google Scholar] [CrossRef] [Green Version]
- Lelapalli, S.; Baskar, S.; Jacob, S.M.; Paranthaman, S. Characterization of Phosphate Solubilizing Plant Growth Promoting Rhizobacterium Lysinibacillus pakistanensis Strain PCPSMR15 Isolated from Oryza sativa. Curr. Res. Microb. Sci. 2021, 2, 100080. [Google Scholar] [CrossRef]
- Withers, P.J.A.; Rodrigues, M.; Soltangheisi, A.; de Carvalho, T.S.; Guilherme, L.R.G.; Benites, V.D.M.; Gatiboni, L.C.; de Sousa, D.M.G.; Nunes, R.D.S.; Rosolem, C.A.; et al. Transitions to Sustainable Management of Phosphorus in Brazilian Agriculture. Sci. Rep. 2018, 8, 2537. [Google Scholar] [CrossRef]


| Source of Variation | First Harvest 2020/2021 Paraná | Second Harvest 2021 Paraná | Second Harvest 2021 Mato Grosso | |||
|---|---|---|---|---|---|---|
| DF | MS (Yield) | DF | MS (Yield) | DF | MS (Yield) | |
| Repetitions/E | 12 | 1,077,861 | 6 | 471,722 | 9 | 304,887 |
| Environment (E) | 3 | 65,585,288 ** | 1 | 85,089,692 ** | 2 | 8,320,357 ** |
| Treatments (T) | 6 | 3,104,344 ** | 4 | 1,443,243 * | 4 | 2,022,543 ** |
| E × T | 18 | 837,210 * | 4 | 1,970,383 ** | 8 | 563,600 ** |
| Error | 81 | 455,739 | 24 | 394,322 | 36 | 156,392 |
| CV(%) | 9.90 | 12.62 | 7.78 | |||
| Mean Londrina | 8558.09 | 3516.60 | - | |||
| Mean Maringá | 7004.05 | 6434.40 | - | |||
| Mean Guarapuava | 4828.13 | - | - | |||
| Mean Entre Rios | 6888.12 | - | - | |||
| Mean Itiquira | - | - | 6321.41 | |||
| Mean Sorriso | - | - | 6653.19 | |||
| Mean Sapezal | - | - | 7553.43 | |||
| Shapiro–Wilk | 0.98 ns | 0.97 ns | 0.98 ns | |||
| Bartlett | 31.10 ns | 22.60 ns | 14.55 ns | |||
| Durbin–Watson | 2.24 ns | 2.63 ns | 2.51 ns | |||
| Treatments | Londrina 1/ | Treatments | Maringá | ||||
|---|---|---|---|---|---|---|---|
| PUpE_g | PUtE_g | PUE_g | PUpE_g | PUtE_g | PUE_g | ||
| 25 kg P2O5 | 1.31 b | 395 ab | 516 a | 25 kg P2O5 | 0.72 b | 757 a | 545 ab |
| 42 kg P2O5 | 0.78 c | 494 a | 385 b | 42 kg P2O5 | 0.47 c | 689 a | 323 b |
| 84 kg P2O5 | 0.45 d | 382 ab | 172 c | 84 kg P2O5 | 0.23 d | 698 a | 160 c |
| Biomaphos | 0.92 b | 489 a | 451 ab | Biomaphos | 0.71 b | 808 a | 573 ab |
| Ag 87 | 1.27 b | 475 a | 603 a | Ag 87 | 0.76 b | 552 a | 420 b |
| Ag 94 | 1.38 a | 412 ab | 568 a | Ag 94 | 0.90 a | 593 a | 533 ab |
| Ag 87 + 94 | 1.58 a | 347 b | 548 a | Ag 87 + 94 | 0.80 ab | 790 a | 632 a |
| Treatments | Guarapuava | Treatments | Entre Rios | ||||
| PUpE_g | PUtE_g | PUE_g | PUpE_g | PUtE_g | PUE_g | ||
| 25 kg P2O5 | 0.42 c | 1836 a | 771 b | 25 kg P2O5 | 0.50 ab | 1908 a | 954 a |
| 42 kg P2O5 | 0.44 c | 1726 a | 759 b | 42 kg P2O5 | 0.34 b | 1328 b | 451 c |
| 84 kg P2O5 | 0.15 d | 1242 a | 186 c | 84 kg P2O5 | 0.19 c | 1604 a | 304 c |
| Biomaphos | 0.79 a | 1334 a | 1053 a | Biomaphos | 0.73 a | 1372 b | 1001 a |
| Ag 87 | 0.78 a | 1638 a | 1278 a | Ag 87 | 0.59 ab | 1513 b | 892 b |
| Ag 94 | 0.35 c | 2012 a | 704 b | Ag 94 | 0.52 ab | 1964 a | 1021 a |
| Ag 87 + 94 | 0.61 b | 1763 a | 1075 a | Ag 87 + 94 | 0.63 ab | 1985 a | 1250 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massucato, L.R.; Almeida, S.R.d.A.; Silva, M.B.; Mosela, M.; Zeffa, D.M.; Nogueira, A.F.; de Lima Filho, R.B.; Mian, S.; Higashi, A.Y.; Teixeira, G.M.; et al. Efficiency of Combining Strains Ag87 (Bacillus megaterium) and Ag94 (Lysinibacillus sp.) as Phosphate Solubilizers and Growth Promoters in Maize. Microorganisms 2022, 10, 1401. https://doi.org/10.3390/microorganisms10071401
Massucato LR, Almeida SRdA, Silva MB, Mosela M, Zeffa DM, Nogueira AF, de Lima Filho RB, Mian S, Higashi AY, Teixeira GM, et al. Efficiency of Combining Strains Ag87 (Bacillus megaterium) and Ag94 (Lysinibacillus sp.) as Phosphate Solubilizers and Growth Promoters in Maize. Microorganisms. 2022; 10(7):1401. https://doi.org/10.3390/microorganisms10071401
Chicago/Turabian StyleMassucato, Luana Rainieri, Suelen Regina de Araújo Almeida, Mayara Barbosa Silva, Mirela Mosela, Douglas Mariani Zeffa, Alison Fernando Nogueira, Renato Barros de Lima Filho, Silas Mian, Allan Yukio Higashi, Gustavo Manoel Teixeira, and et al. 2022. "Efficiency of Combining Strains Ag87 (Bacillus megaterium) and Ag94 (Lysinibacillus sp.) as Phosphate Solubilizers and Growth Promoters in Maize" Microorganisms 10, no. 7: 1401. https://doi.org/10.3390/microorganisms10071401






