Sequence-Specific Electrochemical Genosensor for Rapid Detection of blaOXA-51-like Gene in Acinetobacter baumannii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Bacterial Culture and Growth
2.3. Development and Pretreatment of Screen-Printed Carbon Electrode (SPCE)
2.4. Preparation of DNA Samples for PCR Amplification
2.4.1. Lysate DNA Preparation
2.4.2. Genomic DNA Preparation
2.4.3. Preparation of Internal Control (IC) Plasmid DNA
2.5. Preparation of aPCR Reaction Mixture
2.6. Electrochemical Detection of Synthetic DNA and Amplicons
2.7. Analytical Evaluation of the Electrochemical Genosensor
2.7.1. Analytical Specificity
2.7.2. Analytical Sensitivity at DNA and Cell Levels
2.8. Clinical Application of the Electrochemical Genosensor
2.8.1. Calculation of Sample Size for Spiked Blood Samples
2.8.2. Preparation of Spiked Blood Sample
2.9. Stability Evaluation of the Modified SPCEs
3. Results
3.1. Determination of Amperometric Cut-Off Value
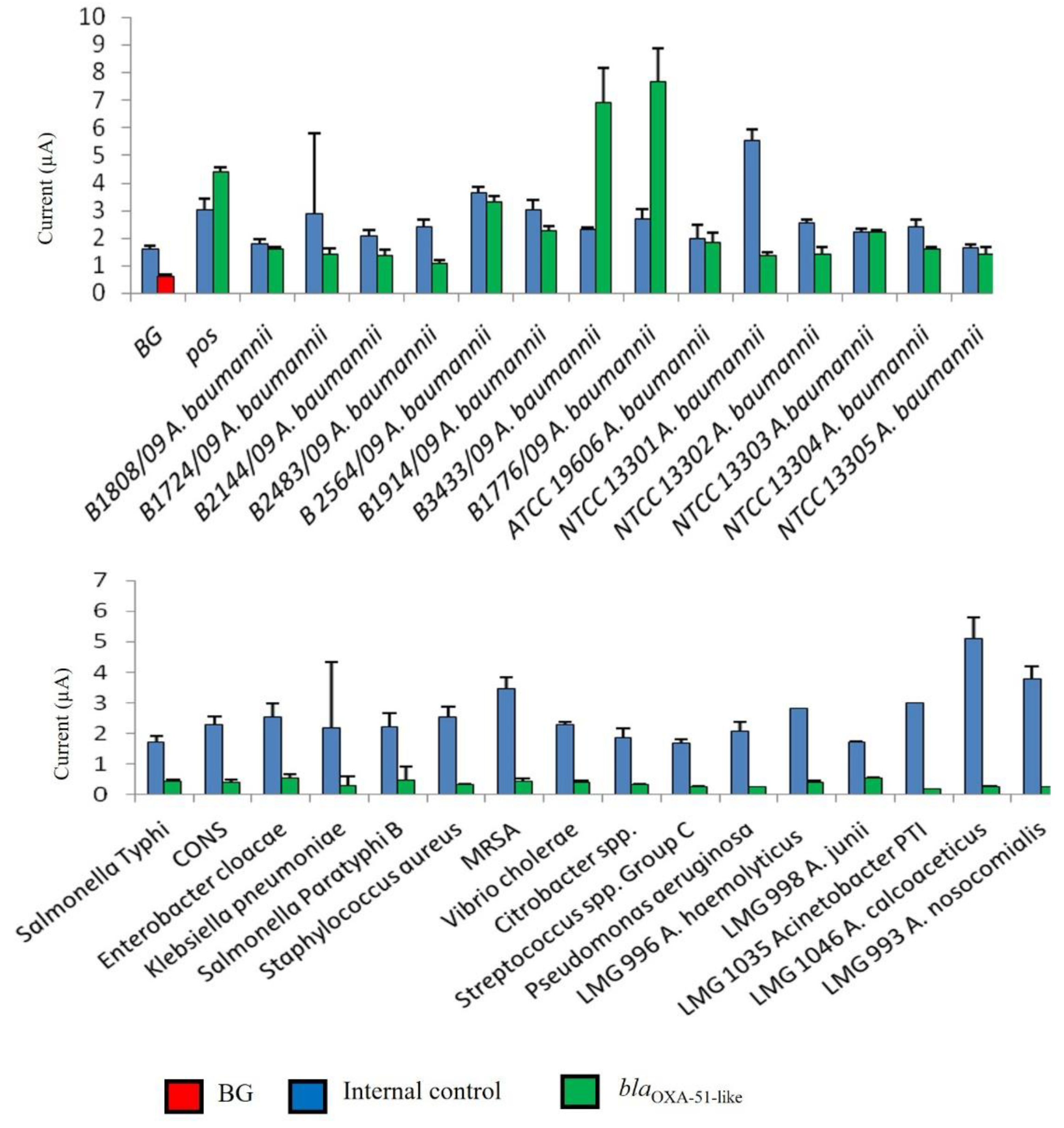
3.2. Evaluation of Analytical Specificity of Enzyme-Based Electrochemical DNA Biosensor
3.3. Evaluation of Analytical Sensitivity of Enzyme-Based Electrochemical DNA Biosensor
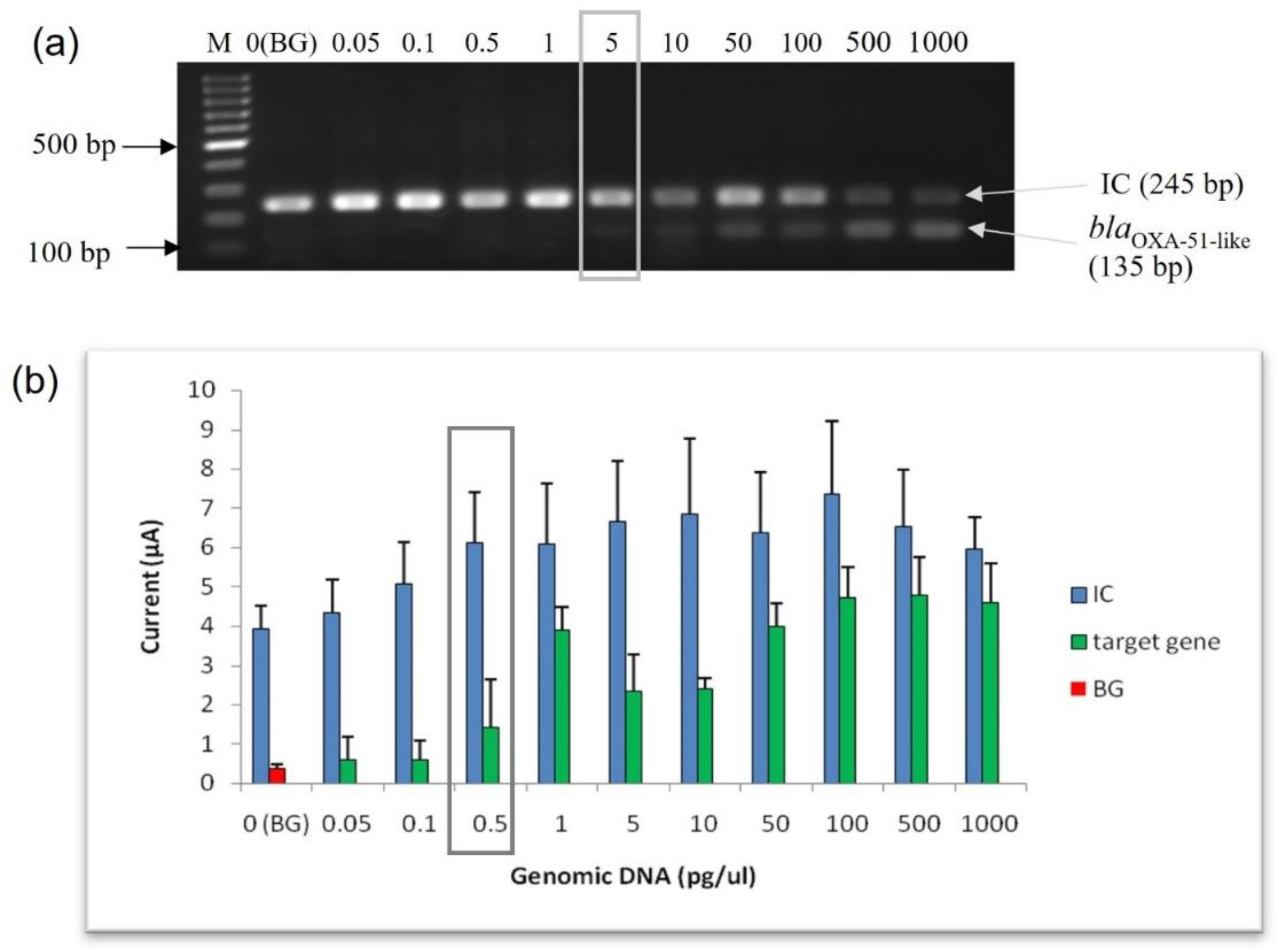
3.3.1. Limit of Detection (LoD) at Genomic DNA Level
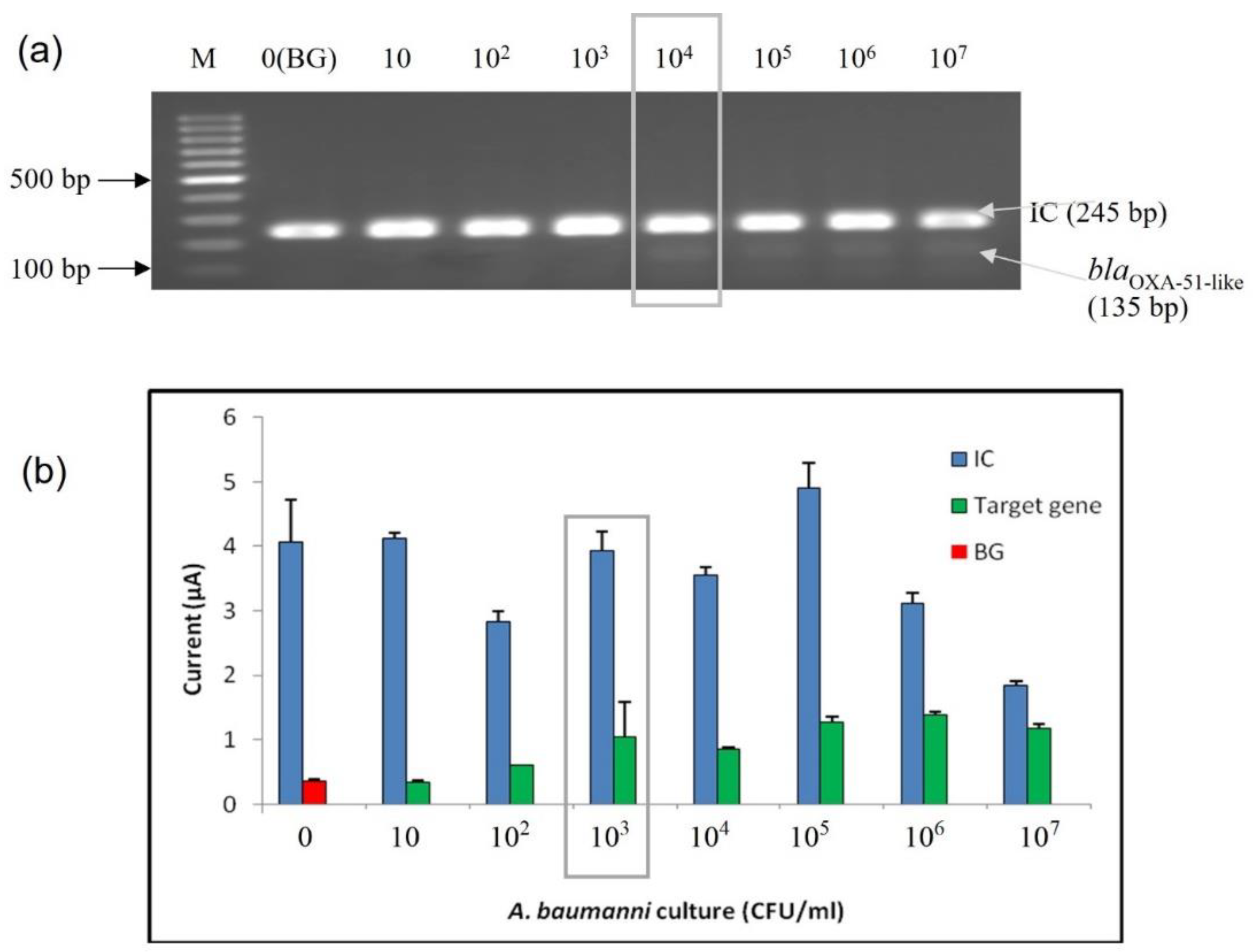
3.3.2. Limit of Detection (LoD) at Bacterial Cell Level
3.4. Diagnostic Evaluation of Enzyme-Based Electrochemical DNA Biosensor Assay Using Spiked Blood Samples
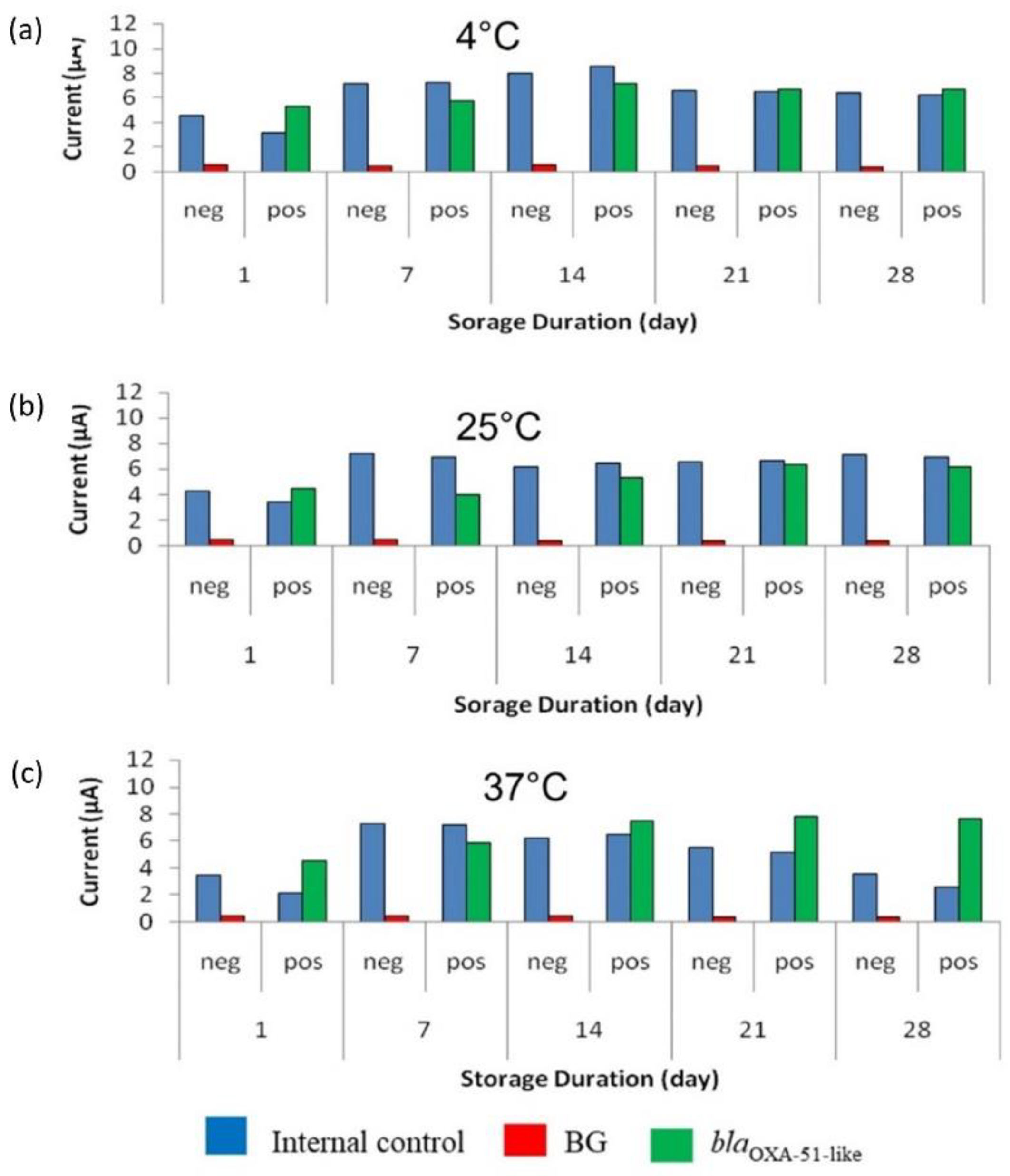
3.5. Accelerated Stability Evaluation of Enzyme-Based Electrochemical DNA Biosensor
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bergogne-Bérézin, E.; Towner, K.J. Acinetobacter Spp. as Nosocomial Pathogens: Microbiological, Clinical, and Epidemiological Features. Clin. Microbiol. Rev. 1996, 9, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Hujer, A.M.; Hujer, K.M.; Decker, B.K.; Rather, P.N.; Bonomo, R.A. Global Challenge of Multidrug-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 3471–3484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andriamanantena, T.S.; Ratsima, E.; Rakotonirina, H.C.; Randrianirina, F.; Ramparany, L.; Carod, J.F.; Richard, V.; Talarmin, A. Dissemination of Multidrug Resistant Acinetobacter baumannii in Various Hospitals of Antananarivo Madagascar. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, R.A. Nosocomial Infection Update. Emerg. Infect. Dis. 1998, 4, 416–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a Global Pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, Y.C.; Wang, S.H.; Chen, Y.Y.; Lin, T.L.; Shie, S.S.; Huang, C.T.; Lee, C.H.; Chen, Y.C.; Quyen, T.L.T.; Pan, Y.J. Association of Capsular Types with Carbapenem Resistance, Disease Severity, and Mortality in Acinetobacter baumannii. Emerg. Microbes Infect. 2020, 9, 2094–2104. [Google Scholar] [CrossRef]
- Khorsi, K.; Messai, Y.; Hamidi, M.; Ammari, H.; Bakour, R. High Prevalence of Multidrug-Resistance in Acinetobacter baumannii and Dissemination of Carbapenemase-Encoding Genes BlaOXA-23-like, BlaOXA-24-like and BlaNDM-1 in Algiers Hospitals. Asian Pac. J. Trop. Med. 2015, 8, 438–446. [Google Scholar] [CrossRef] [Green Version]
- Butler, D.A.; Biagi, M.; Tan, X.; Qasmieh, S.; Bulman, Z.P.; Wenzler, E. Multidrug Resistant Acinetobacter baumannii: Resistance by Any Other Name Would Still Be Hard to Treat. Curr. Infect. Dis. Rep. 2019, 21, 1–17. [Google Scholar] [CrossRef]
- Madaha, E.L.; Gonsu, H.K.; Bughe, R.N.; Fonkoua, M.C.; Ateba, C.N.; Mbacham, W.F. Occurrence of BlaTEM and BlaCTXM Genes and Biofilm-Forming Ability among Clinical Isolates of Pseudomonas Aeruginosa and Acinetobacter baumannii in Yaoundé, Cameroon. Microorganisms 2020, 8, 708. [Google Scholar] [CrossRef]
- Cai, Y.; Chai, D.; Wang, R.; Liang, B.; Bai, N. Colistin Resistance of Acinetobacter baumannii: Clinical Reports, Mechanisms and Antimicrobial Strategies. J. Antimicrob. Chemother. 2012, 67, 1607–1615. [Google Scholar] [CrossRef]
- We, I.; Adeniyi, B.A.; Soge, O.O. Prevalence of Multidrug Resistant Acinetobacter baumannii in Eight Tertiary Hospitals in Southwestern Nigeria. N. Y. Sci. J. 2014, 77, 86–93. [Google Scholar]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An Increasing Threat in Hospitals: Multidrug-Resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-Resistant Acinetobacter baumannii: In Pursuit of an Effective Treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef]
- Qureshi, Z.A.; Hittle, L.E.; O’Hara, J.A.; Rivera, J.I.; Syed, A.; Shields, R.K.; Pasculle, A.W.; Ernst, R.K.; Doi, Y. Colistin-Resistant Acinetobacter baumannii: Beyond Carbapenem Resistance. Clin. Infect. Dis. 2015, 60, 1295–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, J.; Zander, E.; Stefanik, D.; Higgins, P.G.; Roca, I.; Vila, J.; McConnell, M.J.; Cisneros, J.M.; Seifert, H.; Garnacho-Montero, J.; et al. High Incidence of Pandrug-Resistant Acinetobacter baumannii Isolates Collected from Patients with Ventilator-Associated Pneumonia in Greece, Italy and Spain as Part of the MagicBullet Clinical Trial. J. Antimicrob. Chemother. 2017, 72, 3277–3282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amudhan, S.M.; Sekar, U.; Arunagiri, K.; Sekar, B. OXA Beta-Lactamase-Mediated Carbapenem Resistance in Acinetobacter baumannii. Indian J. Med. Microbiol. 2011, 29, 269–274. [Google Scholar] [CrossRef]
- Cai, X.; Sun, J.; Bao, L.; Li, W. Distribution and Antibiotic Resistance of Pathogens Isolated from Ventilator-Associated Pneumonia Patients in Pediatric Intensive Care Unit. World J. Emerg. Med. 2011, 2, 117. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Choi, C.H.; Kang, H.Y.; Lee, J.Y.; Kim, J.; Lee, Y.C.; Seol, S.Y.; Cho, D.T.; Kim, K.W.; Song, D.Y.; et al. Differences in Phenotypic and Genotypic Traits against Antimicrobial Agents between Acinetobacter baumannii and Acinetobacter Genomic Species 13TU. J. Antimicrob. Chemother. 2007, 59, 633–639. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Hanneken, J.; Houchins, D.; King, R.S.; Lee, P.; Richard, J.L. Validation of an ELISA Test Kit for the Detection of Ochratoxin A in Several Food Commodities by Comparison with HPLC. Mycopathologia 2005, 159, 265–272. [Google Scholar] [CrossRef]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The Role of ISAba1 in Expression of OXA Carbapenemase Genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, M.H.Y.; Chan, B.K.W.; Chan, E.W.C.; Chen, S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-β-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019, 10, 2809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jit, M.; Ng, D.H.L.; Luangasanatip, N.; Sandmann, F.; Atkins, K.E.; Robotham, J.V.; Pouwels, K.B. Quantifying the Economic Cost of Antibiotic Resistance and the Impact of Related Interventions: Rapid Methodological Review, Conceptual Framework and Recommendations for Future Studies. BMC Med. 2020, 18, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elshafie, S.; Taj-Aldeen, S.J. Emerging Resistant Serotypes of Invasive Streptococcus Pneumoniae. Infect. Drug Resist. 2016, 9, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Chaibun, T.; Thanasapburachot, P.; Chatchawal, P.; Yin, L.S.; Jiaranuchart, S.; Jearanaikoon, P.; Promptmas, C.; Buajeeb, W.; Lertanantawong, B. A Multianalyte Electrochemical Genosensor for the Detection of High-Risk HPV Genotypes in Oral and Cervical Cancers. Biosensors 2022, 12, 290. [Google Scholar] [CrossRef]
- Jain, S.; Santana, W.; Dolabella, S.S.; Santos, A.L.S.; Souto, E.B.; Severino, P. Are Nanobiosensors an Improved Solution for Diagnosis of Leishmania? Pharmaceutics 2021, 13, 491. [Google Scholar] [CrossRef]
- Sánchez-Salcedo, R.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Dual Electrochemical Genosensor for Early Diagnosis of Prostate Cancer through LncRNAs Detection. Biosens. Bioelectron. 2021, 192, 113520. [Google Scholar] [CrossRef]
- Wu, C.C.; Yen, H.Y.; Lai, L.T.; Perng, G.C.; Lee, C.R.; Wu, S.J. A Label-Free Impedimetric Genosensor for the Nucleic Acid Amplification-Free Detection of Extracted RNA of Dengue Virus. Sensors 2020, 20, 3728. [Google Scholar] [CrossRef]
- Mandong, G.; Yanqing, L.; Hongxia, G.; Xiaoqin, W.; Lifang, F. Electrochemical Detection of Short Sequences Related to the Hepatitis B Virus Using MB on Chitosan-Modified CPE. Bioelectrochemistry 2007, 70, 245–249. [Google Scholar] [CrossRef]
- Ilkhani, H.; Hughes, T.; Li, J.; Zhong, C.J.; Hepel, M. Nanostructured SERS-Electrochemical Biosensors for Testing of Anticancer Drug Interactions with DNA. Biosens. Bioelectron. 2016, 80, 257–264. [Google Scholar] [CrossRef]
- Vaníčková, M.; Lehotay, J.; Čižmárik, J.; Labuda, J. Kinetic Study of the Degradation of a Potential Local Anesthetic Drug in Serum Using the DNA-Based Electrochemical Biosensor. Bioelectrochemistry 2005, 66, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, S.A.; Kauffmann, J.-M.; Zuman, P. Electrochemical Biosensors for Drug Analysis; Springer: Berlin/Heidelberg, Germany, 2015; pp. 141–186. [Google Scholar] [CrossRef]
- Moura-Melo, S.; Miranda-Castro, R.; de-Los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Dos Santos Junior, J.R.; da Silva Fonseca, R.A.; Lobo-Castañón, M.J. A Quantitative PCR-Electrochemical Genosensor Test for the Screening of Biotech Crops. Sensors 2017, 17, 881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tichoniuk, M.; Ligaj, M.; Filipiak, M. Application of DNA Hybridization Biosensor as a Screening Method for the Detection of Genetically Modified Food Components. Sensors 2008, 8, 2118. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Sedeño, P.; Agüí, L.; Campuzano, S.; Pingarrón, J.M. What Electrochemical Biosensors Can Do for Forensic Science? Unique Features and Applications. Biosensors 2019, 9, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harshey, A.; Srivastava, A.; Das, T.; Nigam, K.; Shrivastava, R.; Yadav, V.K. Trends in Gunshot Residue Detection by Electrochemical Methods for Forensic Purpose. J. Anal. Test. 2021, 5, 258–269. [Google Scholar] [CrossRef]
- Zambry, N.S.; Obande, G.A.; Khalid, M.F.; Bustami, Y.; Hamzah, H.H.; Awang, M.S.; Aziah, I.; Manaf, A.A. Utilizing Electrochemical-Based Sensing Approaches for the Detection of SARS-CoV-2 in Clinical Samples: A Review. Biosensors 2022, 12, 473. [Google Scholar] [CrossRef]
- Ribeiro, B.V.; Cordeiro, T.A.R.; Oliveira e Freitas, G.R.; Ferreira, L.F.; Franco, D.L. Biosensors for the Detection of Respiratory Viruses: A Review. Talanta Open 2020, 2, 100007. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Y.; Yang, Q.; Yuan, N.; Zhang, W. Review of Electrochemical DNA Biosensors for Detecting Food Borne Pathogens. Sensors 2019, 19, 4916. [Google Scholar] [CrossRef] [Green Version]
- Nazari-Vanani, R.; Sattarahmady, N.; Yadegari, H.; Heli, H. A Novel and Ultrasensitive Electrochemical DNA Biosensor Based on an Ice Crystals-like Gold Nanostructure for the Detection of Enterococcus Faecalis Gene Sequence. Colloids Surf. B Biointerfaces 2018, 166, 245–253. [Google Scholar] [CrossRef]
- Yeh, C.H.; Chang, Y.H.; Chang, T.C.; Lin, H.P.; Lin, Y.C. Electro-Microchip DNA-Biosensor for Bacteria Detection. Analyst 2010, 135, 2717–2722. [Google Scholar] [CrossRef]
- Wang, Y.; He, H.; Liu, H.; Feng, C.; Yao, Z. An Electrochemical DNA-Hybridization Assay for Acinetobacter baumannii Detection. Int. J. Electrochem. Sci. 2018, 13, 1051–1061. [Google Scholar] [CrossRef]
- Roushani, M.; Sarabaegi, M.; Rostamzad, A. Novel Electrochemical Sensor Based on Polydopamine Molecularly Imprinted Polymer for Sensitive and Selective Detection of Acinetobacter baumannii. J. Iran. Chem. Soc. 2020, 17, 2407–2413. [Google Scholar] [CrossRef]
- Cinti, S.; Volpe, G.; Piermarini, S.; Delibato, E.; Palleschi, G. Electrochemical Biosensors for Rapid Detection of Foodborne Salmonella: A Critical Overview. Sensors 2017, 17, 1910. [Google Scholar] [CrossRef] [PubMed]
- Eksin, E. An Electrochemical Assay for Sensitive Detection of Acinetobacter baumannii Gene. Talanta 2022, 249, 123696. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.Y.; Kamarudin, B.; Ozkan, D.A.; Lee, S.Y.; Lalitha, P.; Ismail, A.; Ozsoz, M.; Ravichandran, M. Enzyme-Linked Amperometric Electrochemical Genosensor Assay for the Detection of PCR Amplicons on a Streptavidin-Treated Screen-Printed Carbon Electrode. Anal. Chem. 2008, 80, 2774–2779. [Google Scholar] [CrossRef]
- Low, K.F.; Karimah, A.; Yean, C.Y. A Thermostabilized Magnetogenosensing Assay for DNA Sequence-Specific Detection and Quantification of Vibrio Cholerae. Biosens. Bioelectron. 2013, 47, 38–44. [Google Scholar] [CrossRef]
- Stavropoulou, E.; Voidarou, C.; Rozos, G.; Vaou, N.; Bardanis, M.; Konstantinidis, T.; Vrioni, G.; Tsakris, A. Antimicrobial Evaluation of Various Honey Types against Carbapenemase-Producing Gram-Negative Clinical Isolates. Antibiotics 2022, 11, 422. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H. Clinical and Economic Evaluation of Multidrug-Resistant Acinetobacter baumannii Colonization in the Intensive Care Unit. Infect. Chemother. 2016, 48, 174–180. [Google Scholar] [CrossRef]
- Liu, Q.; Li, W.; Du, X.; Li, W.; Zhong, T.; Tang, Y.; Feng, Y.; Tao, C.; Xie, Y. Risk and Prognostic Factors for Multidrug-Resistant Acinetobacter baumannii Complex Bacteremia: A Retrospective Study in a Tertiary Hospital of West China. PLoS ONE 2015, 10, e0130701. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Gheorghe, I.; Czobor, I.B.; Chifiriuc, M.C. Antibiotic Resistance Profiles, Molecular Mechanisms and Innovative Treatment Strategies of Acinetobacter baumannii. Microorganisms 2020, 8, 1–40. [Google Scholar] [CrossRef]
- García-Garmendia, J.L.; Ortiz-Leyba, C.; Garnacho-Montero, J.; Jiménez-Jiménez, F.J.; Monterrubio-Villar, J.; Gili-Miner, M. Mortality and the Increase in Length of Stay Attributable to the Acquisition of Acinetobacter in Critically Ill Patients. Crit. Care Med. 1999, 27, 1794–1799. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Wisplinghoff, H.; Krut, O.; Seifert, H. A PCR-Based Method to Differentiate between Acinetobacter baumannii and Acinetobacter Genomic Species 13TU. Clin. Microbiol. Infect. 2007, 13, 1199–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, M.C.; Kuo, S.C.; Chen, Y.C.; Lee, Y.T.; Chen, T.L.; Fung, C.P. Polymerase Chain Reaction Assay for the Detection of Acinetobacter baumannii in Endotracheal Aspirates from Patients in the Intensive Care Unit. J. Microbiol. Immunol. Infect. 2011, 44, 106–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.B.; Livermore, D.M. Multiplex PCR for Genes Encoding Prevalent OXA Carbapenemases in Acinetobacter Spp. Int. J. Antimicrob. Agents 2006, 27, 351–353. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Chang, S.C.; Wang, W.K. High and Increasing Oxa-51 DNA Load Predict Mortality in Acinetobacter baumannii Bacteremia: Implication for Pathogenesis and Evaluation of Therapy. PLoS ONE 2010, 5, e14133. [Google Scholar] [CrossRef] [Green Version]
- Matthew, M.A.; Christie, J.; Yang, N.; Yao, C. A Loop-Mediated Isothermal Amplification (LAMP) Assay Specific to Trichomonas Tenax Is Suitable for Use at Point-of-Care. Microorganisms 2022, 10, 594. [Google Scholar] [CrossRef]
- Ibarra-Meneses, A.V.; Chicharro, C.; Sánchez, C.; García, E.; Ortega, S.; Ndung’u, J.M.; Moreno, J.; Cruz, I.; Carrillo, E. Loop-Mediated Isothermal Amplification Allows Rapid, Simple and Accurate Molecular Diagnosis of Human Cutaneous and Visceral Leishmaniasis Caused by Leishmania Infantum When Compared to PCR. Microorganisms 2021, 9, 610. [Google Scholar] [CrossRef]
- Dent, L.L.; Marshall, D.R.; Pratap, S.; Hulette, R.B. Multidrug Resistant Acinetobacter baumannii: A Descriptive Study in a City Hospital. BMC Infect. Dis. 2010, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Zhou, L.; He, D.; Wang, K.; Qin, D. Biosensing Technologies for Mycobacterium Tuberculosis Detection: Status and New Developments. Clin. Dev. Immunol. 2011, 193963. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Dong, X.; Zhang, K.; Han, X.; Fang, X.; Zhang, Y. A Gold Nanorods-Based Fluorescent Biosensor for the Detection of Hepatitis B Virus DNA Based on Fluorescence Resonance Energy Transfer. Analyst 2013, 138, 642–650. [Google Scholar] [CrossRef]
- Rai, V.; Hapuarachchi, H.C.; Ng, L.C.; Soh, S.H.; Leo, Y.S.; Toh, C.S. Ultrasensitive CDNA Detection of Dengue Virus RNA Using Electrochemical Nanoporous Membrane-Based Biosensor. PLoS ONE 2012, 7, e42346. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.H.; Li, M.; Wang, Y.; Ouyang, H.X.; Wang, L.; Li, X.C.; Cao, Y.C.; Meng, Q.H.; Lu, J.X. Aptasensors for Rapid Detection of Escherichia Coli O157:H7 and Salmonella Typhimurium. Nanoscale Res. Lett. 2012, 7, 658–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerman, K.; Vestergaard, M.; Nagatani, N.; Takamura, Y.; Tamiya, E. Electrochemical Genosensor Based on Peptide Nucleic Acid-Mediated PCR and Asymmetric PCR Techniques: Electrostatic Interactions with a Metal Cation. Anal. Chem. 2006, 78, 2182–2189. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.Y.; Yin, L.S.; Ravichandran, M. Electrochemical Genosensor Assay for the Detection of Bacteria on Screen-Printed Chips. In Electrochemical DNA Biosensors; Jenny Stanford Publishing: Singapore, 2012; pp. 499–516. ISBN 9780429066412. [Google Scholar]
- Teles, F.R.R.; Fonseca, L.P. Trends in DNA Biosensors. Talanta 2008, 77, 606–623. [Google Scholar] [CrossRef]
- Liu, G.; Wan, Y.; Gau, V.; Zhang, J.; Wang, L.; Song, S.; Fan, C. An Enzyme-Based E-DNA Sensor for Sequence-Specific Detection of Femtomolar DNA Targets. J. Am. Chem. Soc. 2008, 130, 6820–6825. [Google Scholar] [CrossRef]
- Volpe, G.; Compagnone, D.; Draisci, R.; Palleschi, G. 3,3′,5,5′-Tetramethylbenzidine as Electrochemical Substrate for Horseradish Peroxidase Based Enzyme Immunoassays. A Comparative Study. Analyst 1998, 123, 1303–1307. [Google Scholar] [CrossRef]
- Wang, L.J.; Wei, Q.S.; Wu, C.S.; Hu, Z.Y.; Ji, J.; Wang, P. The Escherichia Coli O157:H7 DNA Detection on a Gold Nanoparticle-Enhanced Piezoelectric Biosensor. Chin. Sci. Bull. 2008, 53, 1175–1184. [Google Scholar] [CrossRef] [Green Version]
- Eksin, E.; Muti, M.; Erdem, A. Chitosan/Ionic Liquid Composite Electrode for Electrochemical Monitoring of the Surface-Confined Interaction between Mitomycin C and DNA. Electroanalysis 2013, 25, 2321–2329. [Google Scholar] [CrossRef]
- Erdem, A.; Muti, M.; Mese, F.; Eksin, E. Chitosan-Ionic Liquid Modified Single-Use Sensor for Electrochemical Monitoring of Sequence-Selective DNA Hybridization. Colloids Surf. B. Biointerfaces 2014, 114, 261–268. [Google Scholar] [CrossRef]
- Congur, G.; Eksin, E.; Erdem, A. Chitosan Modified Graphite Electrodes Developed for Electrochemical Monitoring of Interaction between Daunorubicin and DNA. Sens. Bio-Sens. Res. 2019, 22, 100255. [Google Scholar] [CrossRef]
- Albareda-Sirvent, M.; Merkoçi, A.; Alegret, S. Configurations Used in the Design of Screen-Printed Enzymatic Biosensors. A Review. Sens. Actuators B Chem. 2000, 69, 153–163. [Google Scholar] [CrossRef]
- Metters, J.P.; Kadara, R.O.; Banks, C.E. New Directions in Screen Printed Electroanalytical Sensors: An Overview of Recent Developments. Analyst 2011, 136, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S.; Minotti, C.; Moscone, D.; Palleschi, G.; Arduini, F. Fully Integrated Ready-to-Use Paper-Based Electrochemical Biosensor to Detect Nerve Agents. Biosens. Bioelectron. 2017, 93, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S.; Basso, M.; Moscone, D.; Arduini, F. A Paper-Based Nanomodified Electrochemical Biosensor for Ethanol Detection in Beers. Anal. Chim. Acta 2017, 960, 123–130. [Google Scholar] [CrossRef]
- Cinti, S.; Neagu, D.; Carbone, M.; Cacciotti, I.; Moscone, D.; Arduini, F. Novel Carbon Black-Cobalt Phthalocyanine Nanocomposite as Sensing Platform to Detect Organophosphorus Pollutants at Screen-Printed Electrode. Electrochim. Acta 2016, 188, 574–581. [Google Scholar] [CrossRef]
- Mancini, F.; Villa, L.; Menegon, M.; Luca, M.D.; Toma, L.; De Liberato, C.; Magliano, A.; Romiti, F.; Carattoli, A.; Ciervo, A. First Evidence of BlaNDM-1 and BlaOXA-23 Carbapenemase Genes in Human Body Lice Infesting a Second-Hand T-Shirt in a Street Market in Italy. Ann. Ist. Super. Sanita 2021, 57, 33–36. [Google Scholar]
- Ababneh, Q.; Aldaken, N.; Jaradat, Z.; Al Sbei, S.; Alawneh, D.; Al-zoubi, E.; Alhomsi, T.; Saadoun, I. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Isolated from Three Major Hospitals in Jordan. Int. J. Clin. Pract. 2021, 75, e14998. [Google Scholar] [CrossRef]
- Al-ahmer, S.D.; Moslim, A.M.; Al-asady, Z.H.A. Molecular Detection of Acientobacter Baumannii Isolated from Nosocomial Infections in Baghdad Hospitals. Ann. Rom. Soc. Cell Biol. 2021, 25, 2–7. [Google Scholar]
- Cisneros, J.M.; Reyes, M.J.; Pachón, J.; Becerril, B.; Caballero, F.J.; García-Garmendía, J.L.; Ortiz, C.; Cobacho, A.R. Bacteremia Due to Acinetobacter baumannii: Epidemiology, Clinical Findings, and Prognostic Features. Clin. Infect. Dis. 1996, 22, 1026–1032. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases from a Prospective Nationwide Surveillance Study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef] [Green Version]
- Al-Soud, W.A.; Rådström, P. Purification and Characterization of PCR-Inhibitory Components in Blood Cells. J. Clin. Microbiol. 2001, 39, 485–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedoux, A.; Paccou, L.; Achir, S.; Guinet, Y. Mechanism of Protein Stabilization by Trehalose during Freeze-Drying Analyzed by in Situ Micro-Raman Spectroscopy. J. Pharm. Sci. 2013, 102, 2484–2494. [Google Scholar] [CrossRef]
- Dash, S.K.; Sharma, M.; Khare, S.; Kumar, A. Omp85 Genosensor for Detection of Human Brain Bacterial Meningitis. Biotechnol. Lett. 2013, 35, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Rafique, B.; Iqbal, M.; Mehmood, T.; Shaheen, M.A. Electrochemical DNA Biosensors: A Review. Sens. Rev. 2019, 39, 34–50. [Google Scholar] [CrossRef]
- Vigneshvar, S.; Sudhakumari, C.C.; Senthilkumaran, B.; Prakash, H. Recent Advances in Biosensor Technology for Potential Applications—An Overview. Front. Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil Rosa, B.; Akingbade, O.E.; Guo, X.; Gonzalez-Macia, L.; Crone, M.A.; Cameron, L.P.; Freemont, P.; Choy, K.L.; Güder, F.; Yeatman, E.; et al. Multiplexed Immunosensors for Point-of-Care Diagnostic Applications. Biosens. Bioelectron. 2022, 203, 114050. [Google Scholar] [CrossRef]
- Xi, H.; Jiang, H.; Juhas, M.; Zhang, Y. Multiplex Biosensing for Simultaneous Detection of Mutations in SARS-CoV-2. ACS Omega 2021, 6, 25846–25859. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]


| Reference Strain | Quantity | Source |
|---|---|---|
| Acinetobacter baumannii ATCC 19606 | 1 | American Type Culture Collection (ATCC) |
| Shigella sonnei ATCC 25931 | 1 | |
| Acinetobacter baumannii NCTC13301 | 1 | National Collection Type Cultures (NCTC) |
| Acinetobacter baumannii NCTC13302 | 1 | |
| Acinetobacter baumannii NCTC13303 | 1 | |
| Acinetobacter baumannii NCTC13304 | 1 | |
| Acinetobacter baumannii NCTC13305 | 1 | |
| Acinetobacter calcoaceticus LMG 1046 | 1 | Belgian Coordinated Collections of Microorganisms (BCCM™/LMG) |
| Acinetobacter nosocomialis LMG 993 | 1 | |
| Acinetobacter haemolyticus LMG 996 | 1 | |
| Acinetobacter junii LMG 998 | 1 | |
| Acinetobacter genospecies 3 LMG 1035 | 1 | |
| Total | 12 |
| Clinical Strain | Source | |
|---|---|---|
| Quantity | Department of Medical Microbiology and Parasitology, Universiti Sains Malaysia | |
| Acinetobacter baumannii | 42 | |
| Other Bacteria: | Quantity | |
| Gram-Positive Bacteria | ||
| Methicillin-Resistant Staphylococcus aureus (MRSA) | 1 | |
| Staphylococcus aureus | 2 | |
| Streptococcus spp. | 1 | |
| Gram-Negative Bacteria | Quantity | |
| Aeromonas hydrophila | 3 | |
| Citrobacter spp. | 1 | |
| Coagulase-negative Staphylococcus (CoNS) | 1 | |
| Escherichia coli | 1 | |
| Enteropathogenic E. coli (EPEC) | 2 | |
| Enterohaemorrhagic E. coli (EHEC) | 1 | |
| Enterobacter cloacae | 1 | |
| Enterobacter spp. | 1 | |
| Klebsiella pneumoniae | 2 | |
| Pseudomonas aeruginosa | 2 | |
| Proteus mirabilis | 1 | |
| Proteus vulgaris | 1 | |
| Proteus spp. | 1 | |
| Salmonella spp. | 1 | |
| Salmonella Paratyphi B | 1 | |
| Salmonella Typhi | 3 | |
| Serratia spp. | 1 | |
| Shigella flexneri | 2 | |
| Shigella sonnei | 1 | |
| Vibrio cholerae | 1 | |
| Yersinia enterocolitica | 2 | |
| Total | 76 | |
| Oligomers | Sequence | Gene | Amplicon (bp) |
|---|---|---|---|
| blaOXA-51_F (forward primer) | 5′-TTT AGC TCG TCG TAT TGG ACT TGA-3′ | blaOXA-51-like | 135 |
| blaOXA-51_R (reverse primer) | 5′-/56-FAM/GCC TCT TGC TGA GGA GTA ATT TTT-3′ | ||
| Capture probe | 5′-/5Bio/TGG CAA TGC AGA TAT CGG TAC CCA AGT C-3′ | ||
| Synthetic blaOXA-51-like | 5′/56FAM/ GCCTCTTGCTGAGGAGTAATTTTTAAAGAATTATCGACTTGGGTACCGATATCTGCATTGCCATAACGAGTTCAAGTCCAATACGACGAGCTAAA | ||
| IC forward primer | 5′-/5Bio/AGG CGG TTG CTC CTG CGT CTT TT -3′ | zot (IC) | 245 |
| IC reverse primer | 5′-/56-FAM/CGG TAA CGG TAG CAC CTT GTA G -3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanapathy, S.; Obande, G.A.; Chuah, C.; Shueb, R.H.; Yean, C.Y.; Banga Singh, K.K. Sequence-Specific Electrochemical Genosensor for Rapid Detection of blaOXA-51-like Gene in Acinetobacter baumannii. Microorganisms 2022, 10, 1413. https://doi.org/10.3390/microorganisms10071413
Kanapathy S, Obande GA, Chuah C, Shueb RH, Yean CY, Banga Singh KK. Sequence-Specific Electrochemical Genosensor for Rapid Detection of blaOXA-51-like Gene in Acinetobacter baumannii. Microorganisms. 2022; 10(7):1413. https://doi.org/10.3390/microorganisms10071413
Chicago/Turabian StyleKanapathy, Swarnaletchumi, Godwin Attah Obande, Candy Chuah, Rafidah Hanim Shueb, Chan Yean Yean, and Kirnpal Kaur Banga Singh. 2022. "Sequence-Specific Electrochemical Genosensor for Rapid Detection of blaOXA-51-like Gene in Acinetobacter baumannii" Microorganisms 10, no. 7: 1413. https://doi.org/10.3390/microorganisms10071413






