A Comprehensive View of Microbial Communities in the Laundering Cycle Suggests a Preventive Effect of Soil Bacteria on Malodour Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Microbial Count on Contaminated Towels
2.3. Statistical Methods
- c is the sum of all countable agar plates;
- n1 is the number of cells taken into account in the lower dilution, i.e., 10−6;
- n2 is the number of cells taken into account in the higher dilution, i.e., 10−7; and
- d is the dilution factor corresponding to the lower dilution (10−6).
2.4. DNA Extraction and Metagenome Analysis
3. Results and Discussion
3.1. Laundry Routines and Malodour Formation
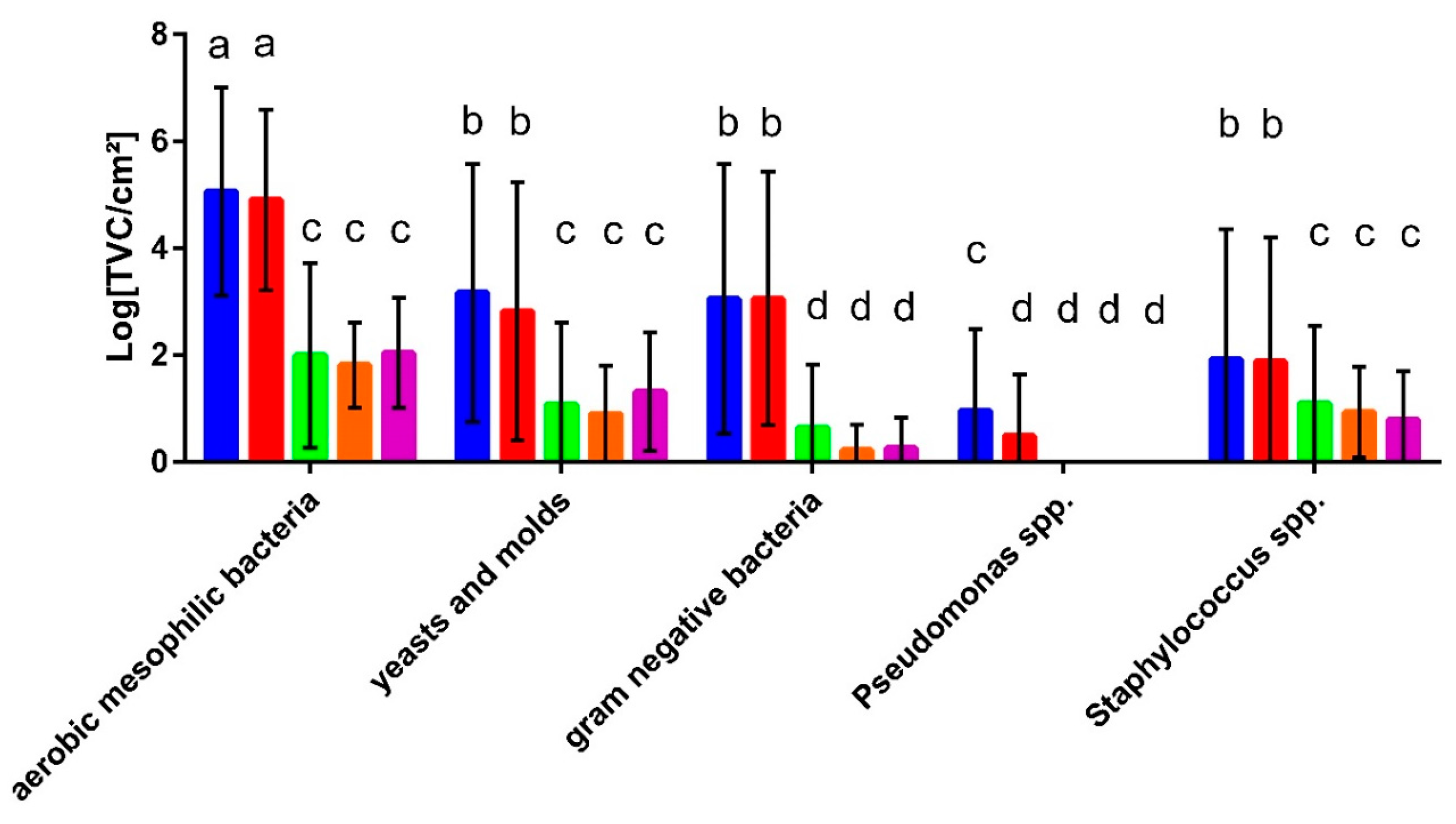
3.2. Microbial Contamination of Washing Machines and Textiles
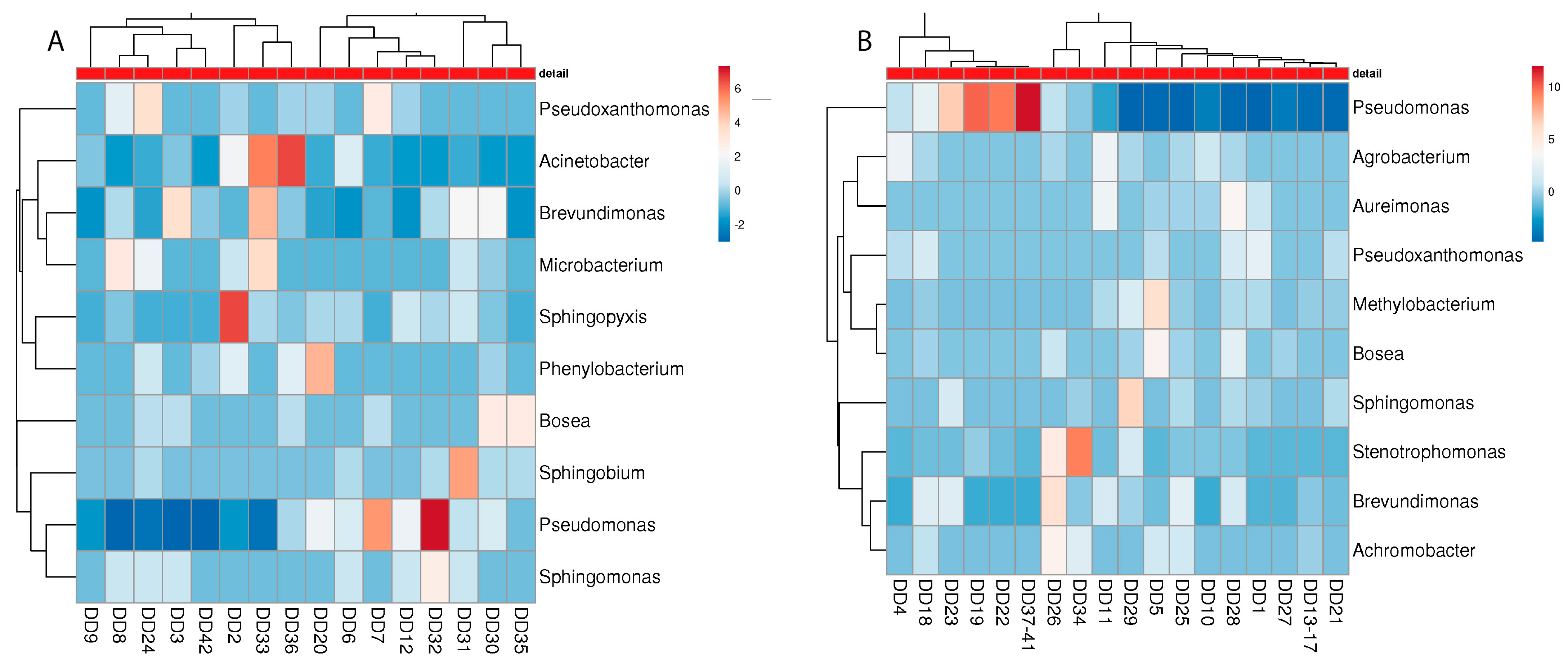
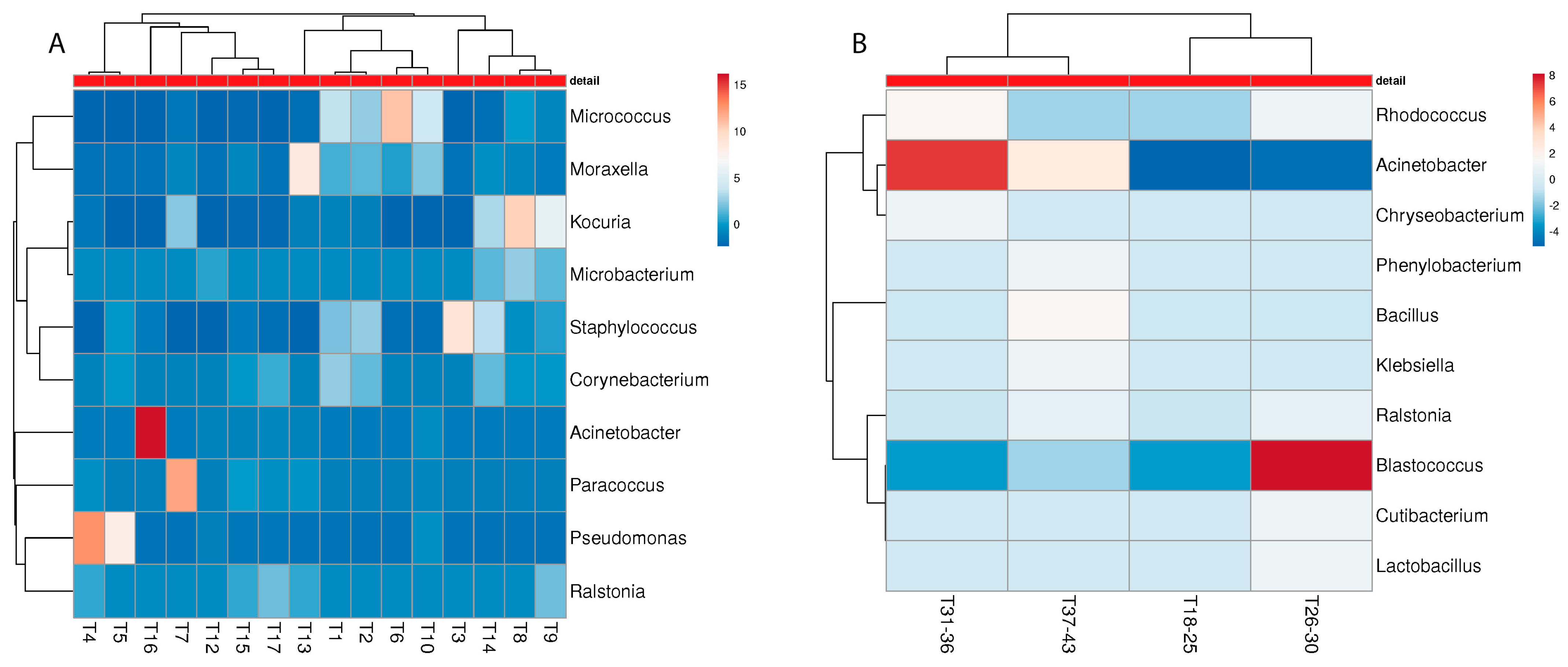
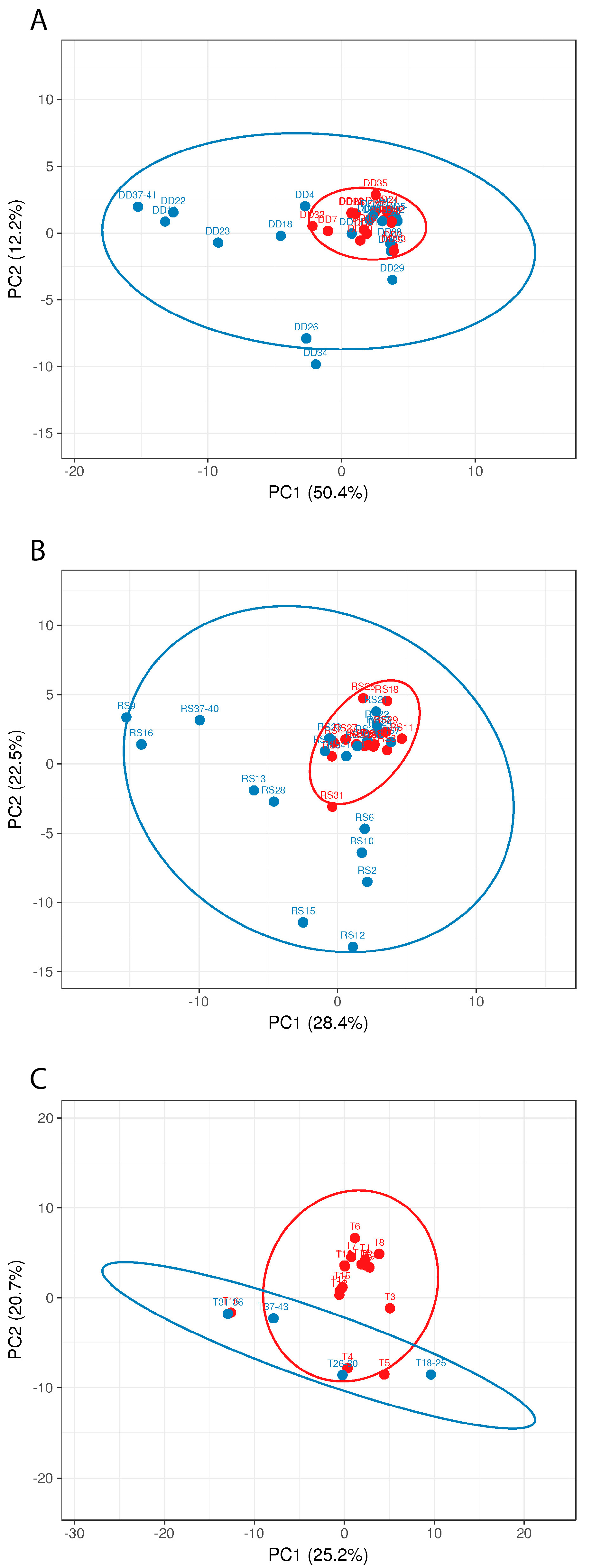
3.3. Metagenome Analysis
3.4. Malodour-Associated Microbial Contamination
3.5. Risk Assessment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Van Herreweghen, F.; Amberg, C.; Marques, R.; Callewaert, C. Biological and chemical processes that lead to textile malodour development. Microorganisms 2020, 8, 1709. [Google Scholar] [CrossRef]
- Bockmühl, D.P. Laundry hygiene—how to get more than clean. J. Appl. Microbiol. 2017, 122, 1124–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bockmühl, D.; Schages, J.; Rehberg, L. Laundry and textile hygiene in healthcare and beyond. Microb. Cell 2019, 6, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Munk, S.; Johansen, C.; Stahnke, L.H.; Adler-Nissen, J. Microbial survival and odor in laundry. J. Surfactants Deterg. 2001, 4, 385–394. [Google Scholar] [CrossRef]
- Callewaert, C.; Van Nevel, S.; Kerckhof, F.M.; Granitsiotis, M.S.; Boon, N. Bacterial exchange in household washing machines. Front. Microbiol. 2015, 6, 1381. [Google Scholar] [CrossRef] [Green Version]
- Gattlen, J.; Amberg, C.; Zinn, M.; Mauclaire, L. Biofilms isolated from washing machines from three continents and their tolerance to a standard detergent. Biofouling 2010, 26, 873–882. [Google Scholar] [CrossRef]
- Nix, I.D.; Frontzek, A.; Bockmühl, D.P. Characterization of microbial communities in household washing machines. Tenside Surfactants Deterg. 2015, 52, 432–440. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Neu, T.R.; Wingender, J. The Perfect Slime: Microbial Extracellular Polymeric Substances (EPS); IWA Publishing: London, UK, 2016; Volume 15, ISBN 9781780407418. [Google Scholar]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [Green Version]
- Costerton, J.W.; Cheng, K.; Geesey, G.G.; Ladd, T.; Nickel, J.; Dasgupta, M.; Marrie, T. Bacterial biofilms in nature and disease. Ann. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef]
- Zalar, P.; Novak, M.; De Hoog, G.S.; Gunde-Cimerman, N. Dishwashers—A man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 2011, 115, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Feazel, L.M.; Baumgartner, L.K.; Peterson, K.L.; Frank, D.N.; Harris, J.K.; Pace, N.R. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. USA 2009, 106, 16393–16398. [Google Scholar] [CrossRef] [Green Version]
- Schages, J.; Brands, B.; Heldt, P.; Kohlmann, R.; Pickel, A.; DP, B.; Bockmühl, D. The Influence of Technical and Microbiological Parameters on the Hygienic Quality of Coffee from Fully Automated Coffee Machines. J. Bioprocess. Biotech. 2018, 8, 322. [Google Scholar] [CrossRef]
- Rayner, J.; Veeh, R.; Flood, J. Prevalence of microbial biofilms on selected fresh produce and household surfaces. Int. J. Food Microbiol. 2004, 95, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Novak Babič, M.; Gostinčar, C.; Gunde-Cimerman, N. Microorganisms populating the water-related indoor biome. Appl. Microbiol. Biotechnol. 2020, 104, 6443–6462. [Google Scholar] [CrossRef]
- Jacksch, S.; Kaiser, D.; Weis, S.; Weide, M.; Ratering, S.; Schnell, S.; Egert, M. Influence of sampling site and other environmental factors on the bacterial community composition of domestic washing machines. Microorganisms 2020, 8, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, H.; Mitani, A.; Niwano, Y.; Takeuchi, K.; Tanaka, A.; Yamaguchi, N.; Kawamura, Y.; Hitomi, J. Moraxella species are primarily responsible for generating malodor in laundry. Appl. Environ. Microbiol. 2012, 78, 3317–3324. [Google Scholar] [CrossRef] [Green Version]
- Leyden, J.J.; McGinley, K.J.; Holzle, E.; Labows, J.N.; Kligman, A.M. The microbiology of the human axilla and its relationship to axillary odor. J. Investig. Dermatol. 1981, 77, 413–416. [Google Scholar] [CrossRef] [Green Version]
- Klepp, I.G.; Buck, M.; Laitala, K.; Kjeldsberg, M. What’s the Problem? Odor-control and the Smell of Sweat in Sportswear. Fash. Pract. 2016, 8, 296–317. [Google Scholar] [CrossRef]
- Kanda, F.; Yagi, E.; Fukuda, M.; Nakajima, K.; Ohta, T.; Nakata, O. Elucidation of chemical compounds responsible for foot malodour. Br. J. Dermatol. 1990, 122, 771–776. [Google Scholar] [CrossRef]
- Caroprese, A.; Gabbanini, S.; Beltramini, C.; Lucchi, E.; Valgimigli, L. HS-SPME-GC-MS analysis of body odor to test the efficacy of foot deodorant formulations. Ski. Res. Technol. 2009, 15, 503–510. [Google Scholar] [CrossRef]
- Nagoh, Y.; Tobe, S.; Watanabe, T.; Mukaiyama, T. Analysis of odorants produced from Indoor drying laundries and effects of enzyme for preventing malodor generation. Tenside Surfactants Deterg. 2005, 42, 7–12. [Google Scholar] [CrossRef]
- Takeuchi, K.; Hasegawa, Y.; Ishida, H.; Kashiwagi, M. Identification of novel malodour compounds in laundry. Flavour Fragr. J. 2012, 27, 89–94. [Google Scholar] [CrossRef]
- Abney, S.E.; Ijaz, M.K.; McKinney, J.; Gerba, C.P. Laundry Hygiene and Odor Control—State of the Science. Appl. Environ. Microbiol. 2021, 87, e03002-20. [Google Scholar] [CrossRef]
- Zinn, M.; Singer, M.; Bockmühl, D. Smells Like Teen Spirit—A Model to Generate Laundry-Associated Malodour In Vitro. Microorganisms 2021, 9, 974. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Stapleton, K.; Hill, K.; Day, K.; Perry, J.D.; Dean, J.R. The potential impact of washing machines on laundry malodour generation. Lett. Appl. Microbiol. 2013, 56, 299–306. [Google Scholar] [CrossRef]
- Lucassen, R.; Blümke, H.; Born, L.; Fritz, A.; Geurtz, P.; Hofmann, N.; Hoffmann, L.; Steiner, R.; Merettig, N.; Bockmühl, D. The washing machine as a source of microbial contamination of domestic laundry—A case study. Househ. Pers. Care Today 2014, 9, 54–57. [Google Scholar]
- Howe, C.; Silva, T.; Marston, A.; Woo, D. Staphylococcal contamination of mattresses and blankets on a surgical ward under nonepidemic conditions. Med. Sci. Sports Exerc. 1961, 264, 625–632. [Google Scholar] [CrossRef]
- Smith, J.A.; Neil, K.R.; Davidson, C.G.; Davidson, R.W. Effect of water temperature on bacterial killing in laundry. Infect. Control 1987, 8, 204–209. [Google Scholar] [CrossRef]
- Bloomfield, S.F.; Exner, M.; Signorelli, C.; Nath, K.J.; Scott, E.A. The Infection Risks Associated with Clothing and Household Linens in Home and Everyday Life Settings, and the Role of Laundry; International Scientific Forum on Home Hygiene (IFH): Somerset, UK, 2011; pp. 1–43. [Google Scholar]
- Blaser, M.J.; Smith, P.F.; Cody, H.J.; Wang, W.L.L.; LaForce, F.M. Killing of fabric-associated bacteria in hospital laundry by low temperature washing. J. Infect. Dis. 1984, 149, 48–57. [Google Scholar] [CrossRef]
- Honisch, M.; Brands, B.; Weide, M.; Speckmann, H.D.; Stamminger, R.; Bockmühl, D. Antimicrobial efficacy of laundry detergents with regard to time and temperature in domestic washing machines. Tenside Surfactants Deterg. 2016, 53, 547–552. [Google Scholar] [CrossRef]
- Lucassen, R.; Merettig, N.; Bockmühl, D.P. Antimicrobial efficacy of hygiene rinsers under consumer-related conditions. Tenside Surfactants Deterg. 2013, 50, 259–262. [Google Scholar] [CrossRef]
- Linke, S.; Gemein, S.; Koch, S.; Gebel, J.; Exner, M. Orientating investigation of the inactivation of Staphylococcus aureus in the laundry process. Hyg. Medizin 2011, 36, 25–29. [Google Scholar]
- Bockmühl, D. Hygiene aspects in domestic laundry. Hyg. Medizin 2011, 36, 280. [Google Scholar]
- Deutsches Institut für Normung. DIN EN 16616 Chemische Desinfektionsmittel und Antiseptika—Chemothermische Wäschedesinfektion—Prüfverfahren und Anforderungen (Phase 2, Stufe 2); Beuth Verlag GmbH: Berlin, Germany, 2015. [Google Scholar]
- Gebel, J.; Werner, H.; Kirsch-Altena, A.; Bansemir, K. Standardmethoden der DGHM zur Prüfung chemsicher Desinfektionsverfahren; mhp Verlag: Wiesbaden, Germany, 2001. [Google Scholar]
- European Committee for Standardization. prEN 17658:2021; Chemical Disinfectants and Antisepticschemical Textile Disinfection for the Domestic area—Test Method and Requirements (Phase 2, Step 2). European Committee: Brussels, Belgium, 2021. [Google Scholar]
- Technischen, D.; Arbeitsstoffe, B.; Der, D.S. Technische Regeln für Biologische Arbeitsstoffe Einstufung von Viren in Risikogruppen Anwendungsbereich Allgemeines Liste der Einstufungen; Bundesanstalt für Arbeitsschutz und Arbeitsmedizin: Dortmund, Germany, 1998; pp. 1–8. [Google Scholar]
- Arbeitsstoffe, T.R. für B. Technische Regeln für BiologischeArbeitsstoffe Einstufung von Pilzen in Risikogruppen Anwendungsbereich Allgemeines Liste der Einstufungen; Wolters Kluwer Deutschland GmbH: Alphen aan den Rijn, The Netherlands, 2016. [Google Scholar]
- Schages, L.; Wichern, F.; Geisen, S.; Kalscheuer, R.; Bockmühl, D. Distinct resistomes and microbial communities of soils, wastewater treatment plants and households suggest development of antibiotic resistances due to distinct environmental conditions in each environment. Antibiotics 2021, 10, 514. [Google Scholar] [CrossRef]
- Takeuchi, K.; Yabuki, M.; Hasegawa, Y. Review of odorants in human axillary odour and laundry malodour: The importance of branched C7 chain analogues in malodours perceived by humans. Flavour Fragr. J. 2013, 28, 223–230. [Google Scholar] [CrossRef]
- Madsen, A.M.; Moslehi-Jenabian, S.; Islam, M.Z.; Frankel, M.; Spilak, M.; Frederiksen, M.W. Concentrations of Staphylococcus species in indoor air as associated with other bacteria, season, relative humidity, air change rate, and S. aureus-positive occupants. Environ. Res. 2018, 160, 282–291. [Google Scholar] [CrossRef]
- Kooken, J.M.; Fox, K.F.; Fox, A. Characterization of micrococcus strains isolated from indoor air. Mol. Cell. Probes 2012, 26, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Weisskopf, L.; Schulz, S.; Garbeva, P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 2021, 19, 391–404. [Google Scholar] [CrossRef]




| Species | Risk Group (According to [41,42]) | Detergent Drawer (in %) | Rubber Sealant (in %) | Hand Towel (in %) | Body Towel (in %) | Kitchen Cloth (in %) |
|---|---|---|---|---|---|---|
| Actinomycetospora succinea | 1 | 0.21 | 0.02 | 0.02 | 0.00 | 0.00 |
| Agrobacterium tumefaciens | 1 | 0.51 | 0.36 | 0.02 | 0.00 | 0.00 |
| Aspergillus glaucus | 1 | 0.00 | 0.00 | 0.73 | 0.00 | 0.00 |
| Aureimonas altamirensis | 1 | 0.56 | 0.03 | 0.01 | 0.00 | 0.00 |
| Bacillus subtilis | 1 | 0.01 | 0.00 | 2.89 | 0.00 | 0.00 |
| Brevundimonas bullata | 1 | 0.22 | 0.27 | 0.00 | 0.00 | 0.00 |
| Brevundimonas diminuta | 1 | 0.17 | 0.30 | 0.01 | 0.00 | 0.00 |
| Brevundimonas sp DS20 | 1 | 0.13 | 0.23 | 0.00 | 0.00 | 0.00 |
| Brevundimonas sp SH203 | 1 | 0.35 | 0.31 | 0.01 | 0.00 | 0.01 |
| Flavobacterium lindanitolerans | 1 | 0.05 | 0.26 | 0.00 | 0.00 | 0.00 |
| Homo sapiens | 1 | 0.03 | 0.24 | 0.24 | 2.17 | 0.15 |
| Kocuria rhizophila | 1 | 0.01 | 0.07 | 1.60 | 0.07 | 0.01 |
| Methylorubrum extorquens | 1 | 0.23 | 0.00 | 0.01 | 0.00 | 0.00 |
| Micrococcus luteus | 1 | 0.22 | 0.16 | 2.54 | 0.02 | 0.01 |
| Mycolicibacterium tusciae | 1 | 0.19 | 0.05 | 0.01 | 0.00 | 0.00 |
| Pseudomonas fluorescens | 1 | 0.20 | 0.15 | 0.14 | 0.03 | 0.02 |
| Pseudomonas fragi | 1 | 0.01 | 0.12 | 0.01 | 6.57 | 4.49 |
| Pseudomonas oleovorans | 1 | 0.04 | 0.27 | 0.00 | 0.00 | 0.00 |
| Pseudomonas veronii | 1 | 0.29 | 0.05 | 0.00 | 0.00 | 0.00 |
| Pseudoxanthomonas spadix | 1 | 0.02 | 0.58 | 0.00 | 0.00 | 0.00 |
| Pseudoxanthomonas suwonensis | 1 | 0.11 | 0.39 | 0.00 | 0.00 | 0.00 |
| Rhizobiales bacterium | 1 | 0.16 | 0.21 | 0.01 | 0.00 | 0.01 |
| Rhizobium sp ACO-34A | 1 | 0.00 | 0.34 | 0.00 | 0.00 | 0.00 |
| Rhodococcus erythropolis | 1 | 0.04 | 0.10 | 0.18 | 0.17 | 0.20 |
| Skermanella aerolata | 1 | 0.22 | 0.10 | 0.00 | 0.00 | 0.00 |
| Staphylococcus warneri | 1 | 0.00 | 0.00 | 0.66 | 0.02 | 0.00 |
| Stenotrophomonas rhizophila | 1 | 0.06 | 0.20 | 0.00 | 0.00 | 0.07 |
| Xanthobacter autotrophicus | 1 | 0.37 | 0.08 | 0.00 | 0.00 | 0.00 |
| Xanthobacter tagetidis | 1 | 0.04 | 0.26 | 0.00 | 0.00 | 0.00 |
| Acinetobacter johnsonii | 2 | 0.17 | 0.47 | 0.16 | 0.00 | 3.00 |
| Acinetobacter ursingii | 2 | 0.26 | 0.32 | 0.03 | 0.00 | 0.55 |
| Brevibacterium casei | 2 | 0.57 | 0.03 | 0.11 | 0.01 | 0.00 |
| Cutibacterium acnes | 2 | 0.02 | 0.15 | 0.54 | 12.26 | 0.21 |
| Moraxella osloensis | 2 | 0.42 | 2.75 | 3.26 | 0.05 | 0.60 |
| Paracoccus yeei | 2 | 0.11 | 0.37 | 0.74 | 0.00 | 0.03 |
| Pseudomonas aeruginosa | 2 | 1.04 | 0.35 | 2.84 | 0.07 | 0.05 |
| Pseudomonas alcaligenes | 2 | 0.20 | 0.07 | 0.00 | 0.00 | 0.00 |
| Pseudomonas stutzeri | 2 | 0.64 | 0.50 | 0.08 | 0.00 | 0.01 |
| Roseomonas gilardii | 2 | 0.21 | 0.03 | 0.05 | 0.00 | 0.00 |
| Staphylococcus aureus | 2 | 0.00 | 0.02 | 1.40 | 0.16 | 0.04 |
| Staphylococcus epidermidis | 2 | 0.02 | 0.01 | 0.47 | 0.44 | 0.02 |
| Stenotrophomonas maltophilia | 2 | 0.70 | 1.54 | 0.04 | 0.01 | 0.02 |
| Aquabacterium sp SJQ9 | (nd) | 0.64 | 0.03 | 0.00 | 0.00 | 0.00 |
| Blastococcus sp CCUG 61487 | (nd) | 0.02 | 0.20 | 0.05 | 0.00 | 0.00 |
| Janibacter indicus | (nd) | 0.14 | 0.23 | 0.04 | 0.00 | 0.00 |
| Micavibrio aeruginosavorus | (nd) | 0.01 | 0.63 | 0.00 | 0.00 | 0.00 |
| Paracoccus salipaludis | (nd) | 0.00 | 0.02 | 0.52 | 0.00 | 0.00 |
| Phenylobacterium sp Root700 | (nd) | 0.36 | 0.19 | 0.25 | 0.00 | 0.00 |
| Rahnella inusitata | (nd) | 0.00 | 0.00 | 0.01 | 3.03 | 1.28 |
| Sample | Shannon Diversity | Standard Deviation |
|---|---|---|
| M detergent drawer | 15.03 | 7.28 |
| NM detergent drawer | 10.56 | 9.38 |
| M rubber sealant | 10.57 | 8.02 |
| NM rubber sealant | 5.03 | 4.09 |
| M towel | 7.25 | 5.56 |
| NM towel | 2.75 | 1.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zinn, M.-K.; Flemming, H.-C.; Bockmühl, D. A Comprehensive View of Microbial Communities in the Laundering Cycle Suggests a Preventive Effect of Soil Bacteria on Malodour Formation. Microorganisms 2022, 10, 1465. https://doi.org/10.3390/microorganisms10071465
Zinn M-K, Flemming H-C, Bockmühl D. A Comprehensive View of Microbial Communities in the Laundering Cycle Suggests a Preventive Effect of Soil Bacteria on Malodour Formation. Microorganisms. 2022; 10(7):1465. https://doi.org/10.3390/microorganisms10071465
Chicago/Turabian StyleZinn, Marc-Kevin, Hans-Curt Flemming, and Dirk Bockmühl. 2022. "A Comprehensive View of Microbial Communities in the Laundering Cycle Suggests a Preventive Effect of Soil Bacteria on Malodour Formation" Microorganisms 10, no. 7: 1465. https://doi.org/10.3390/microorganisms10071465
APA StyleZinn, M.-K., Flemming, H.-C., & Bockmühl, D. (2022). A Comprehensive View of Microbial Communities in the Laundering Cycle Suggests a Preventive Effect of Soil Bacteria on Malodour Formation. Microorganisms, 10(7), 1465. https://doi.org/10.3390/microorganisms10071465






