High Prevalence and Varied Distribution of Antibiotic-Resistant Bacteria in the Rhizosphere and Rhizoplane of Citrus medica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
2.2. DNA Extraction and Taxonomic Classification
2.3. Antibiotic Susceptibility Assay
2.4. Statistics Analysis
3. Results
3.1. Isolation and Identification of Bacterial Isolates from the Rhizosphere and Rhizoplane of C. medica
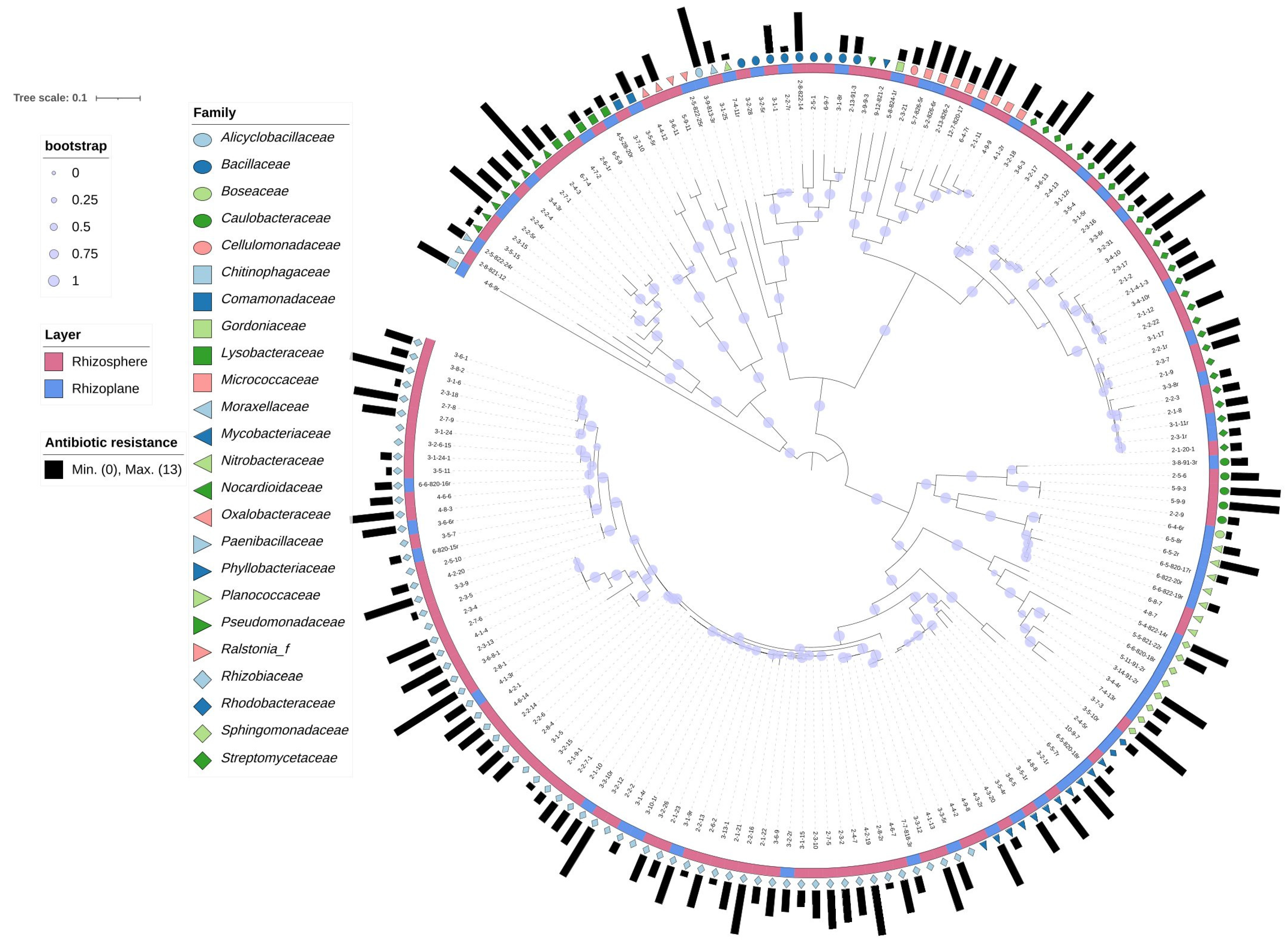
3.2. Comparison of Taxonomic Distribution between Isolates from the Rhizoplane and Rhizosphere
3.3. Comparison of Antibiotic Resistance Profiles between Isolates from the Rhizoplane and Rhizosphere
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, M.; Tang, J.C. Research of antibiotics pollution in soil environments and its ecological toxicity. J. Agro-Environ. Sci. 2010, 29, 261–266. [Google Scholar]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Ondon, B.S.; Li, S.; Zhou, Q.; Li, F. Sources of Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARGs) in the Soil: A Review of the Spreading Mechanism and Human Health Risks; Springer International Publishing: Cham, Switzerland, 2021; pp. 121–153. [Google Scholar]
- Zhang, Y.-J.; Hu, H.-W.; Chen, Q.-L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.-Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef]
- Chen, Q.-L.; Cui, H.-L.; Su, J.-Q.; Penuelas, J.; Zhu, Y.-G. Antibiotic Resistomes in Plant Microbiomes. Trends Plant Sci. 2019, 24, 530–541. [Google Scholar] [CrossRef]
- Brandt, K.K.; Amézquita, A.; Backhaus, T.; Boxall, A.; Coors, A.; Heberer, T.; Lawrence, J.R.; Lazorchak, J.; Schönfeld, J.; Snape, J.R.; et al. Ecotoxicological assessment of antibiotics: A call for improved consideration of microorganisms. Environ. Int. 2015, 85, 189–205. [Google Scholar] [CrossRef]
- Huang, R.; Ding, J.; Guo, Y.; Sun, B.; Liang, Y. Habitat determines the relationships among bacteria, resistance genes and mobile genetic elements in the soil–plant system. Eur. J. Soil Sci. 2022, 73, e13132. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, B.; Yu, G. Antibiotic resistance genes in China: Occurrence, risk, and correlation among different parameters. Environ. Sci. Pollut. Res. Int. 2018, 25, 21467–21482. [Google Scholar] [CrossRef]
- Sharma, R.; Bisaria, V.S.; Sharma, S. Rhizosphere: A Home for Human Pathogens; Springer International Publishing: Cham, Switzerland, 2019; pp. 113–127. [Google Scholar]
- Berg, G.; Eberl, L.; Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 2005, 7, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Cernava, T.; Erlacher, A.; Soh, J.; Sensen, C.W.; Grube, M.; Berg, G. Enterobacteriaceae dominate the core microbiome and contribute to the resistome of arugula (Eruca sativa Mill.). Microbiome 2019, 7, 13. [Google Scholar] [CrossRef]
- Nygård, K.; Lassen, J.; Vold, L.; Andersson, Y.; Fisher, I.; Löfdahl, S.; Threlfall, J.; Luzzi, I.; Peters, T.; Hampton, M.; et al. Outbreak of Salmonella Thompson infections linked to imported rucola lettuce. Foodborne Pathog. Dis. 2008, 5, 165–173. [Google Scholar]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Riera, N.; Jin, T.; Li, J.; Wang, N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 2017, 5, 97. [Google Scholar] [CrossRef]
- Zhang, Y.; Trivedi, P.; Xu, J.; Roper, M.C.; Wang, N. The Citrus Microbiome: From Structure and Function to Microbiome Engineering and Beyond. Phytobiomes J. 2021, 5, 249–262. [Google Scholar] [CrossRef]
- Kawasaki, K.; Kamagata, Y. Phosphate-Catalyzed Hydrogen Peroxide Formation from Agar, Gellan, and kappa-Carrageenan and Recovery of Microbial Cultivability via Catalase and Pyruvate. Appl. Environ. Microbiol. 2017, 83, e01366-17. [Google Scholar] [CrossRef]
- Kato, S.; Yamagishi, A.; Daimon, S.; Kawasaki, K.; Tamaki, H.; Kitagawa, W.; Abe, A.; Tanaka, M.; Sone, T.; Asano, K.; et al. Isolation of Previously Uncultured Slow-Growing Bacteria by Using a Simple Modification in the Preparation of Agar Media. Appl. Environ. Microbiol. 2018, 84, e00807-18. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, 9–15. [Google Scholar]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Szczesny, G.; Leszczynski, P.; Sokol-Leszczynska, B.; Maldyk, P. Identification of human-dependent routes of pathogen’s transmission in a tertiary care hospital. Jt. Dis. Relat. Surg. 2022, 33, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Balmer, L.; Seth-Smith, H.M.B.; Egli, A.; Casanova, C.; Kronenberg, A.; Schrenzel, J.; Marschall, J.; Sommerstein, R. Agrobacterium species bacteraemia, Switzerland, 2008 to 2019: A molecular epidemiological study. Antimicrob. Resist. Infect. Control 2022, 11, 47. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas spp: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Wang, E.; Wang, N. Mechanisms Underlying the Rhizosphere-To-Rhizoplane Enrichment of Cellvibrio Unveiled by Genome-Centric Metagenomics and Metatranscriptomics. Microorganisms 2020, 8, 583. [Google Scholar] [CrossRef]
- Xie, W.; Shen, Q.; Zhao, F.J. Antibiotics and antibiotic resistance from animal manures to soil: A review. Eur. J. Soil Sci. 2018, 69, 181–195. [Google Scholar] [CrossRef]
- Liao, X.; Ma, Y.; Daliri, E.B.-M.; Koseki, S.; Wei, S.; Liu, D.; Ye, X.; Chen, S.; Ding, T. Interplay of antibiotic resistance and food-associated stress tolerance in foodborne pathogens. Trends Food Sci. Technol. 2020, 95, 97–106. [Google Scholar] [CrossRef]
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Bradford, Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Ngoi, S.T.; Muhamad, A.N.; The, C.S.J.; Chong, C.W.; Abdul Jabar, K.; Chai, L.C.; Leong, K.C.; Tee, L.H.; AbuBakar, S. beta-Lactam Resistance in Upper Respiratory Tract Pathogens Isolated from a Tertiary Hospital in Malaysia. Pathogens 2021, 10, 1602. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Azad, M.A.K.; Velkov, T.; Zhou, Q.; Li, J. Rescuing the Last-Line Polymyxins: Achievements and Challenges. Pharmacol. Rev. 2021, 73, 679–728. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-T.; Li, X.; Zhang, Q.; Shan, H.; Zou, M.; Song, F.-J. Colistin-Resistant mcr-Positive Enterobacteriaceae in Fresh Vegetables, an Increasing Infectious Threat in China. Int. J. Antimicrob. Agents 2019, 54, 89–94. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Wang, Y.; Liu, Q.; Xu, B.; Chen, H.; Li, Y.; Wang, K.; Liang, G.; Zhang, R.; Jiao, X.; et al. High Prevalence and Varied Distribution of Antibiotic-Resistant Bacteria in the Rhizosphere and Rhizoplane of Citrus medica. Microorganisms 2022, 10, 1708. https://doi.org/10.3390/microorganisms10091708
Yang F, Wang Y, Liu Q, Xu B, Chen H, Li Y, Wang K, Liang G, Zhang R, Jiao X, et al. High Prevalence and Varied Distribution of Antibiotic-Resistant Bacteria in the Rhizosphere and Rhizoplane of Citrus medica. Microorganisms. 2022; 10(9):1708. https://doi.org/10.3390/microorganisms10091708
Chicago/Turabian StyleYang, Fang, Yu Wang, Qianwen Liu, Bo Xu, Huan Chen, Yaomen Li, Kun Wang, Guixin Liang, Ruiqi Zhang, Xin’an Jiao, and et al. 2022. "High Prevalence and Varied Distribution of Antibiotic-Resistant Bacteria in the Rhizosphere and Rhizoplane of Citrus medica" Microorganisms 10, no. 9: 1708. https://doi.org/10.3390/microorganisms10091708
APA StyleYang, F., Wang, Y., Liu, Q., Xu, B., Chen, H., Li, Y., Wang, K., Liang, G., Zhang, R., Jiao, X., & Zhang, Y. (2022). High Prevalence and Varied Distribution of Antibiotic-Resistant Bacteria in the Rhizosphere and Rhizoplane of Citrus medica. Microorganisms, 10(9), 1708. https://doi.org/10.3390/microorganisms10091708





