Long-Term Fertilization Strategy Impacts Rhizoctonia solani–Microbe Interactions in Soil and Rhizosphere and Defense Responses in Lettuce
Abstract
:1. Introduction
2. Materials and Methods
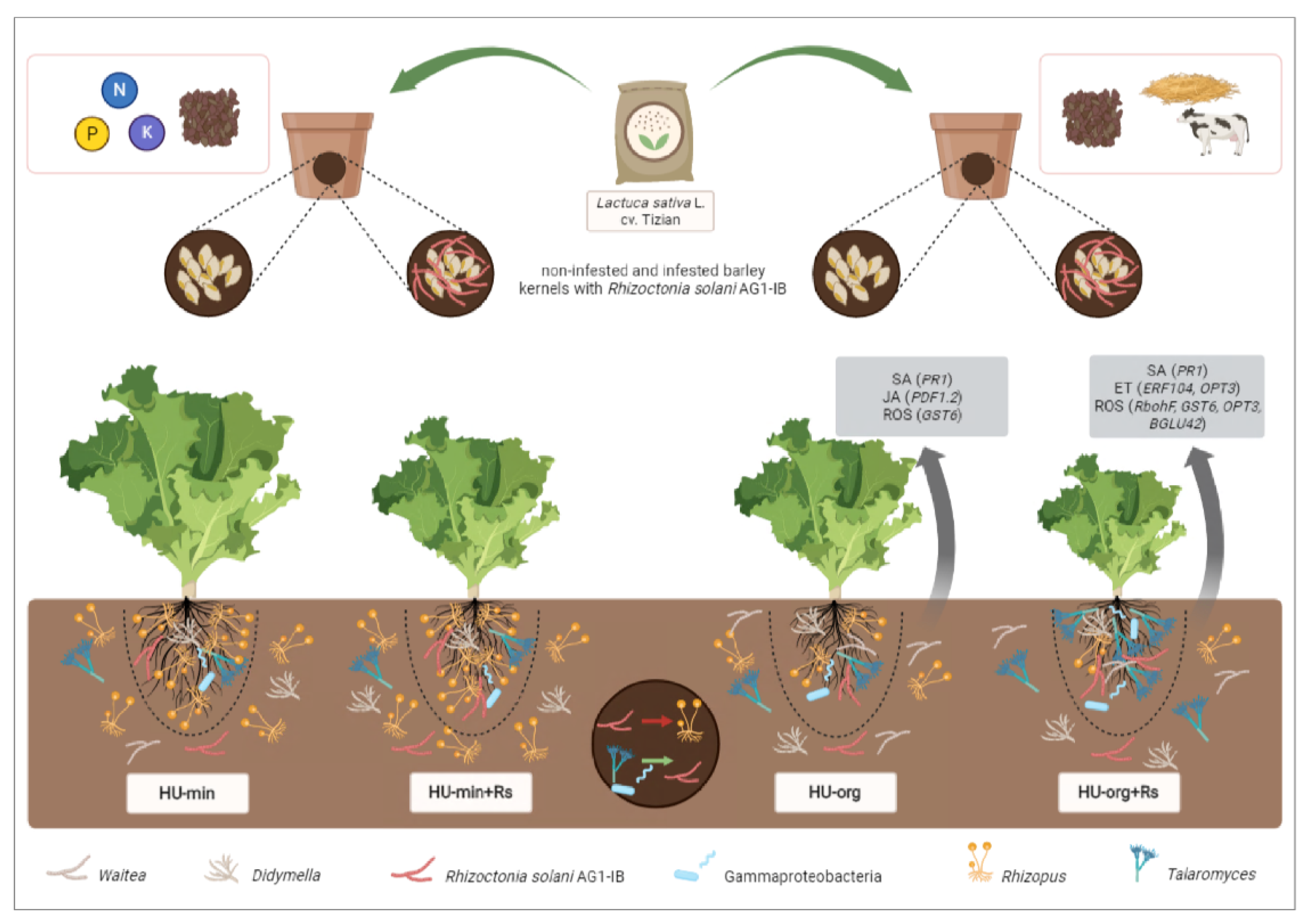
2.1. Field Site and Soil Sampling Strategy
2.2. Pathogens Used
2.3. Assessment of Soil Suppressiveness
2.4. Growth Chamber Experiments to Study Lettuce Health and Rhizosphere Microbiota
2.5. Analysis of Plant Gene Expression
2.6. Collection of Bulk Soils, Root-Associated Soils and Rhizosphere Samples and Total Community DNA Extraction
2.7. Microbial Community Analyses
2.8. Statistical Analysis
3. Results
3.1. Long-Term Mineral Fertilization Reduced the Spread of Rhizoctonia solani
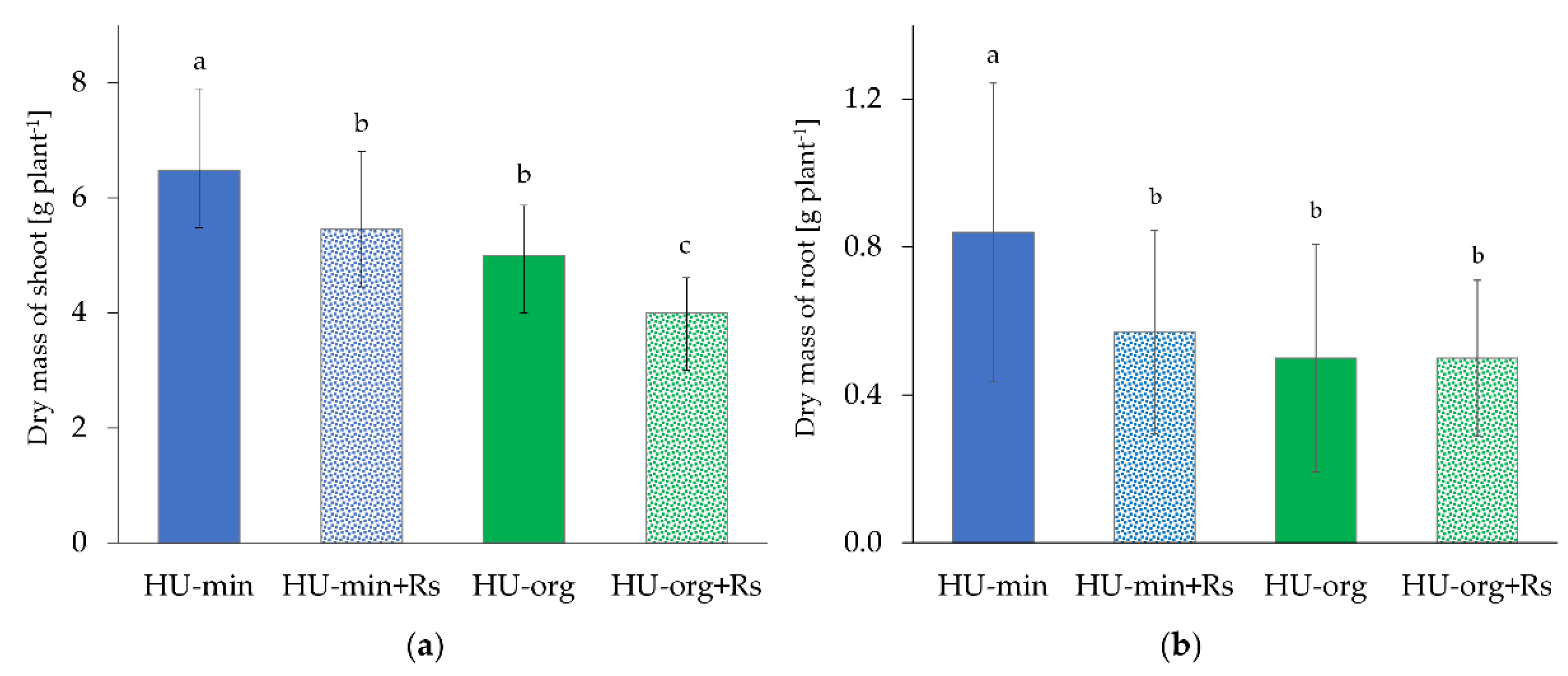
3.2. Fertilization Strategy and Presence of R. solani AG1-IB Limited Lettuce Growth
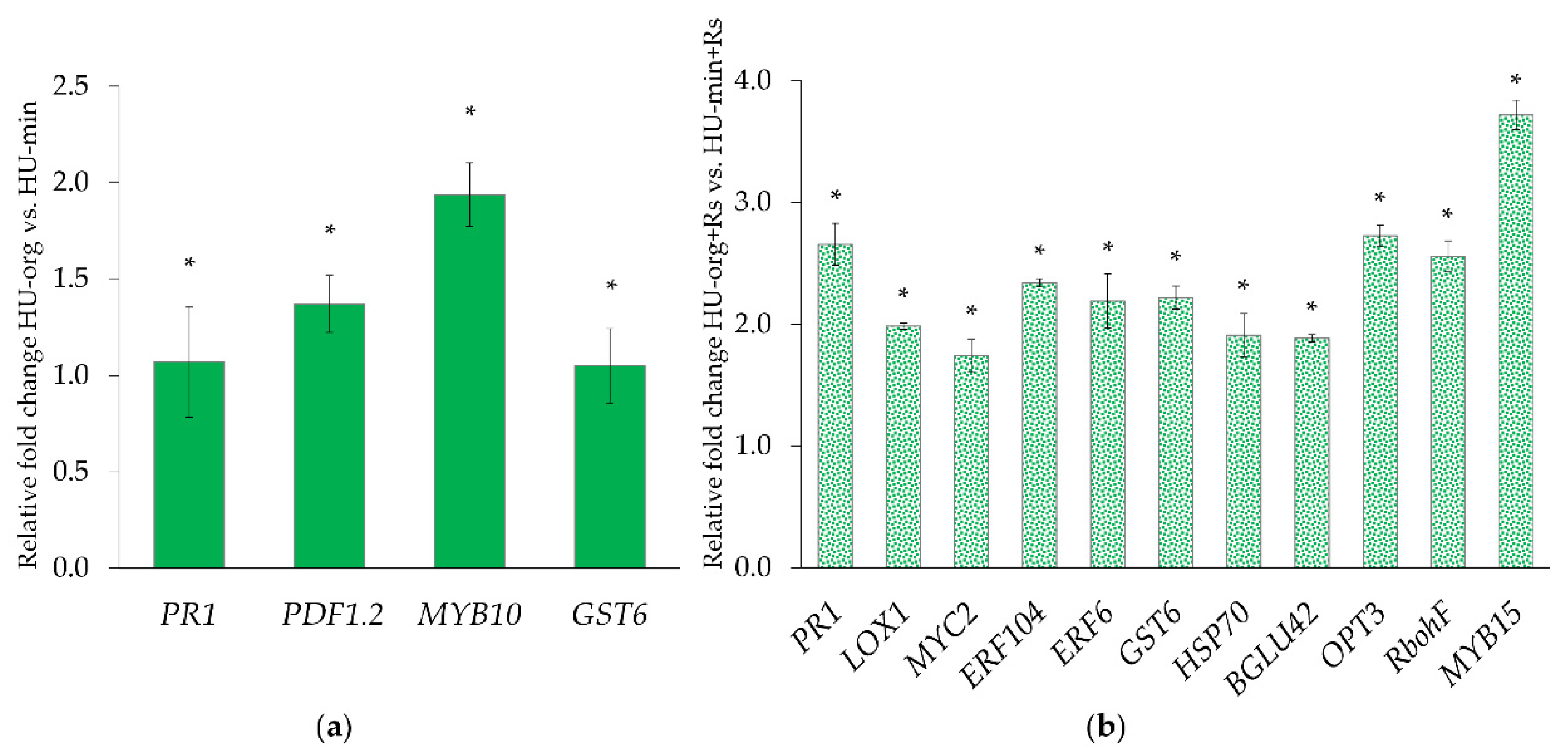
3.3. R. solani AG1-IB and Fertilization Strategy Influenced Gene Expression Profiles of Lettuce
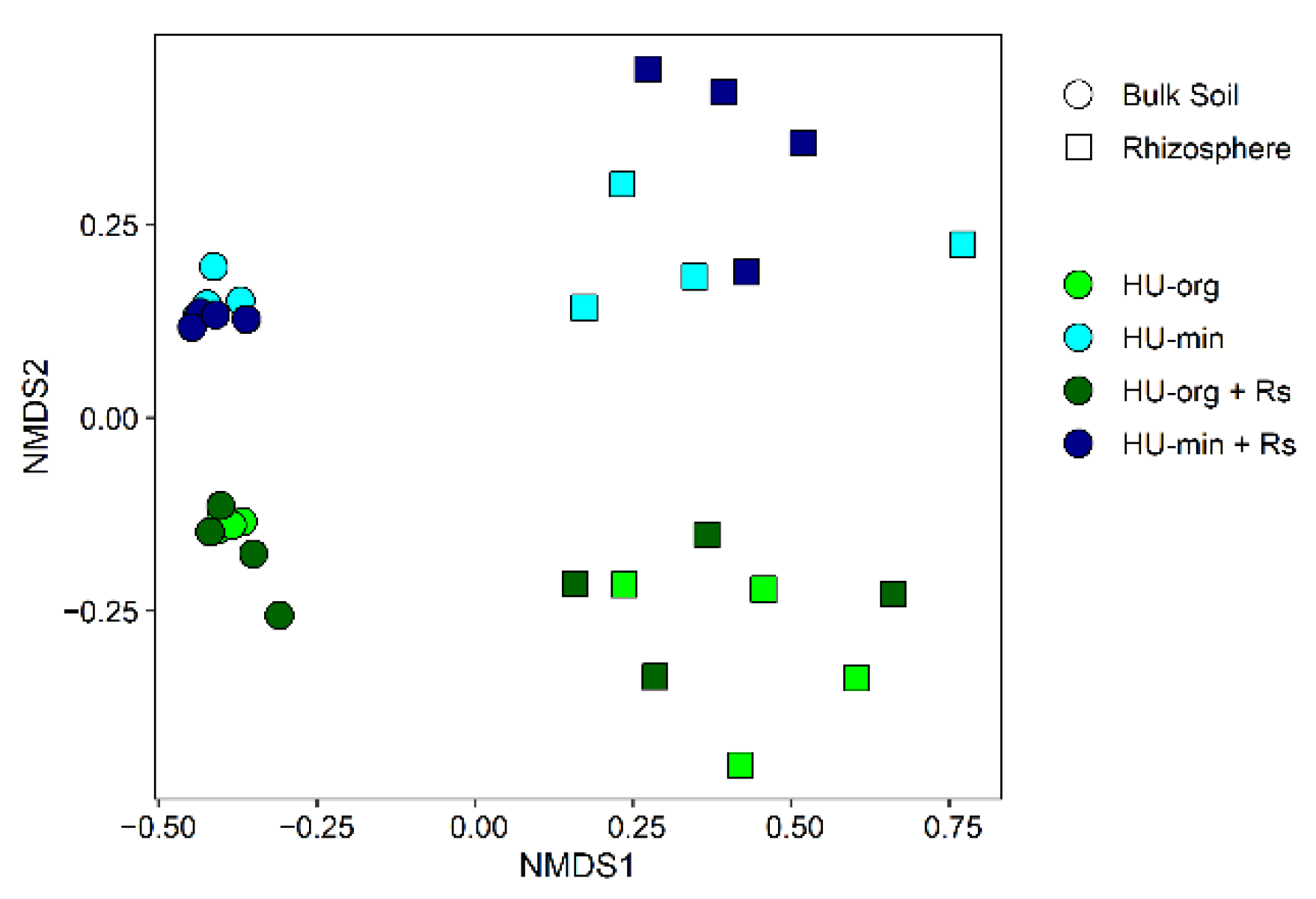
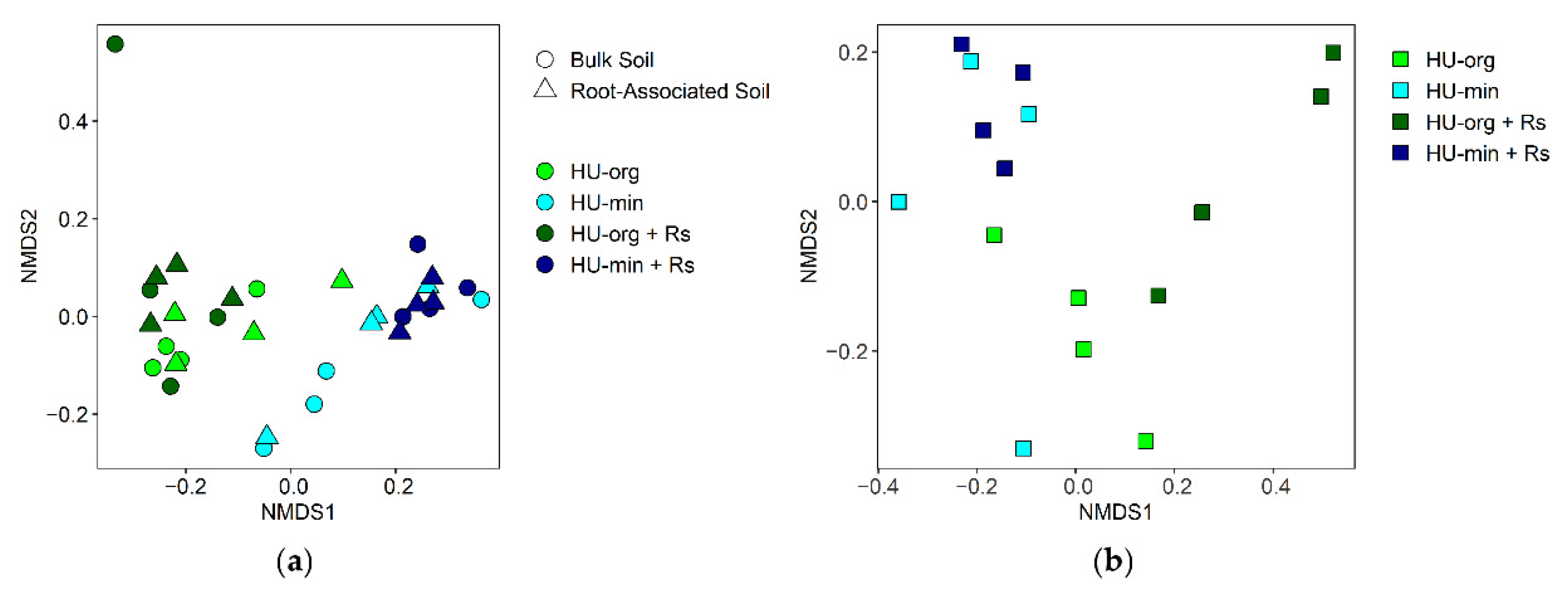
3.4. Microbial Communities in Soil and Lettuce Rhizosphere
3.4.1. Fertilization Strategy but Not R. solani AG1-IB Shifted Bacterial Community Composition
3.4.2. Fertilization Strategy and R. solani AG1-IB Shaped Fungal Community Composition
3.4.3. Rhizoctonia solani AG1-IB Affected Relative Abundance of Fungal Taxa in Organic-Fertilized Soils
3.4.4. No Clear Indication of R. solani AG1-IB Establishment in the Differently Fertilized Soils
4. Discussion
4.1. No Inhibition of the Spread of Rhizoctonia Pathogens in Organic-Fertilized Soil
4.2. Long-Term Organic Fertilization Impacted R. solani AG1-IB Interaction with Indigenous Soil Fungi
4.3. Bacterial Community Shifts in Response to Fertilization Practice but Not to Pathogen Inoculation
4.4. R. solani AG1-IB Induced Systemic Expression of Defense-Related Genes in Lettuce Plants Grown in Soils with Long-Term Organic Fertilization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meena, R.S.; Kumar, S.; Datta, R.; Lal, R.; Vijayakumar, V.; Brtnicky, M.; Sharma, M.P.; Yadav, G.S.; Jhariya, M.K.; Jangir, C.K.; et al. Impact of Agrochemicals on Soil Microbiota and Management: A Review. Land 2020, 9, 34. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil quality—A critical review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Sukhdev, P.; May, P.; Müller, A. Fix food metrics. Nature 2016, 540, 33–34. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection: A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Hoitink, H.A.J.; Boehm, M.J. Biocontrol within the Context of Soil Microbial Communities: A Substrate-Dependent Phenomenon. Annu. Rev. Phytopathol. 1999, 37, 427–446. [Google Scholar] [CrossRef]
- Bailey, K.L.; Lazarovits, G. Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 2003, 72, 169–180. [Google Scholar] [CrossRef]
- van Bruggen, A.H.C.; Semenov, A.M. Soil Health and Soilborne Diseases in Organic Agriculture. In Plant Diseases and Their Management in Organic Agriculture; Finckh, M.R., van Bruggen, A.H.C., Tamm, L., Eds.; APS Press: St. Paul, MN, USA, 2017; pp. 67–89. [Google Scholar] [CrossRef]
- van Bruggen, A.H.C.; Gamliel, A.; Finckh, M.R. Plant disease management in organic farming systems. Pest Manag. Sci. 2016, 72, 30–44. [Google Scholar] [CrossRef]
- Postma, J.; Schilder, M.T.; Bloem, J.; van Leeuwen-Haagsma, W.K. Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biol. Biochem. 2008, 40, 2394–2406. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Garbeva, P.; Postma, J.; van Veen, J.A.; van Elsas, J.D. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ. Microbiol. 2006, 8, 233–246. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Sommermann, L.; Geistlinger, J.; Wibberg, D.; Deubel, A.; Zwanzig, J.; Babin, D.; Schlüter, A.; Schellenberg, I. Fungal community profiles in agricultural soils of a long-term field trial under different tillage, fertilization and crop rotation conditions analyzed by high-throughput ITS-amplicon sequencing. PLoS ONE 2018, 13, e0195345. [Google Scholar] [CrossRef] [PubMed]
- Babin, D.; Deubel, A.; Jacquiod, S.; Sørensen, S.J.; Geistlinger, J.; Grosch, R.; Smalla, K. Impact of long-term agricultural management practices on soil prokaryotic communities. Soil Biol. Biochem. 2019, 129, 17–28. [Google Scholar] [CrossRef]
- Allan, E.; Manning, P.; Alt, F.; Binkenstein, J.; Blaser, S.; Blüthgen, N.; Böhm, S.; Grassein, F.; Hölzel, N.; Klaus, V.H.; et al. Land use intensification alters ecosystem multifunctionality via loss of biodiversity and changes to functional composition. Ecol. Lett. 2015, 18, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Köberl, M.; Rybakova, D.; Müller, H.; Grosch, R.; Smalla, K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 2017, 93, fix050. [Google Scholar] [CrossRef]
- Hol, W.H.G.; De Boer, W.; De Hollander, M.; Kuramae, E.E.; Meisner, A.; Van Der Putten, W.H. Context dependency and saturating effects of loss of rare soil microbes on plant productivity. Front. Plant Sci. 2015, 6, 485. [Google Scholar] [CrossRef]
- Lapsansky, E.R.; Milroy, A.M.; Andales, M.J.; Vivanco, J.M. Soil memory as a potential mechanism for encouraging sustainable plant health and productivity. Curr. Opin. Biotechnol. 2016, 38, 137–142. [Google Scholar] [CrossRef]
- Widder, S.; Allen, R.J.; Pfeiffer, T.; Curtis, T.P.; Wiuf, C.; Sloan, W.T.; Cordero, O.X.; Brown, S.P.; Momeni, B.; Shou, W.; et al. Challenges in microbial ecology: Building predictive understanding of community function and dynamics. ISME J. 2016, 10, 2557–2568. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, X.; Gong, J.; Xu, T.; Ding, G.-C.; Li, J. Long-Term Organic Farming Manipulated Rhizospheric Microbiome and Bacillus Antagonism Against Pepper Blight (Phytophthora capsici). Front. Microbiol. 2019, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Dennert, F.; Imperiali, N.; Staub, C.; Schneider, J.; Laessle, T.; Zhang, T.; Wittwer, R.; Van Der Heijden, M.G.A.; Smits, T.H.M.; Schlaeppi, K.; et al. Conservation tillage and organic farming induce minor variations in Pseudomonas abundance, their antimicrobial function and soil disease resistance. FEMS Microbiol. Ecol. 2018, 94, fiy075. [Google Scholar] [CrossRef] [PubMed]
- van Elsas, J.D.; Postma, J. Suppression of Soil-Borne Phytopathogens by Compost. In Compost Science and Technology; Diaz, L.F., de Bertoldi, M., Bidlingmaier, W., Stentiford, E., Eds.; Waste Management Series; Elsevier: Boston, MA, USA, 2007; Volume 8, pp. 201–214. [Google Scholar]
- Bonanomi, G.; Antignani, V.; Pane, C.; Scala, F. Suppression of Soilborne Fungal Diseases with Organic Amendments. J. Plant Pathol. 2007, 89, 311–324. [Google Scholar]
- Yogev, A.; Raviv, M.; Hadar, Y.; Cohen, R.; Katan, J. Plant waste-based composts suppressive to diseases caused by pathogenic Fusarium oxysporum. Eur. J. Plant Pathol. 2006, 116, 267–278. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Soil immune responses: Soil microbiomes may be harnessed for plant health. Science 2016, 352, 1392–1393. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Berendsen, R.L.; Van Pelt, J.A.; Vismans, G.; Yu, K.; Li, E.; Van Bentum, S.; Poppeliers, S.W.M.; Sanchez Gil, J.J.; Zhang, H.; et al. The Soil-Borne Identity and Microbiome-Assisted Agriculture: Looking Back to the Future. Mol. Plant 2020, 13, 1394–1401. [Google Scholar] [CrossRef]
- Semenov, M.V.; Krasnov, G.S.; Semenov, V.M.; van Bruggen, A. Mineral and Organic Fertilizers Distinctly Affect Fungal Communities in the Crop Rhizosphere. J. Fungi 2022, 8, 251. [Google Scholar] [CrossRef]
- Bonanomi, G.; Antignani, V.; Capodilupo, M.; Scala, F. Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol. Biochem. 2010, 42, 136–144. [Google Scholar] [CrossRef]
- van Bruggen, A.H.C.; Sharma, K.; Kaku, E.; Karfopoulos, S.; Zelenev, V.V.; Blok, W.J. Soil health indicators and Fusarium wilt suppression in organically and conventionally managed greenhouse soils. Appl. Soil Ecol. 2015, 86, 192–201. [Google Scholar] [CrossRef]
- Antoniou, A.; Tsolakidou, M.-D.; Stringlis, I.A.; Pantelides, I.S. Rhizosphere Microbiome Recruited from a Suppressive Compost Improves Plant Fitness and Increases Protection against Vascular Wilt Pathogens of Tomato. Front. Plant Sci. 2017, 8, 2022. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Babin, D.; Sandmann, M.; Jacquiod, S.; Sommermann, L.; Sørensen, S.J.; Fliessbach, A.; Mäder, P.; Geistlinger, J.; Smalla, K.; et al. Effect of long-term organic and mineral fertilization strategies on rhizosphere microbiota assemblage and performance of lettuce. Environ. Microbiol. 2019, 21, 2426–2439. [Google Scholar] [CrossRef] [PubMed]
- Beckers, G.J.M.; Conrath, U. Priming for stress resistance: From the lab to the field. Curr. Opin. Plant Biol. 2007, 10, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense Priming: An Adaptive Part of Induced Resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef]
- Trost, B.; Prochnow, A.; Baumecker, M.; Meyer-Aurich, A.; Drastig, K.; Ellmer, F. Effects of nitrogen fertilization and irrigation on N2O emissions from a sandy soil in Germany. Arch. Agron. Soil Sci. 2015, 61, 569–580. [Google Scholar] [CrossRef]
- Windisch, S.; Sommermann, L.; Babin, D.; Chowdhury, S.P.; Grosch, R.; Moradtalab, N.; Walker, F.; Höglinger, B.; El-Hasan, A.; Armbruster, W.; et al. Impact of Long-Term Organic and Mineral Fertilization on Rhizosphere Metabolites, Root–Microbial Interactions and Plant Health of Lettuce. Front. Microbiol. 2021, 11, 597745. [Google Scholar] [CrossRef]
- Schneider, J.; Schilder, M.; Dijst, G. Characterization of Rhizoctonia solani AG 2 isolates causing bare patch in field grown tulips in the Netherlands. Eur. J. Plant Pathol. 1997, 103, 265–279. [Google Scholar] [CrossRef]
- Verwaaijen, B.; Wibberg, D.; Nelkner, J.; Gordin, M.; Rupp, O.; Winkler, A.; Bremges, A.; Blom, J.; Grosch, R.; Pühler, A.; et al. Assembly of the Lactuca sativa, L. cv. Tizian draft genome sequence reveals differences within major resistance complex 1 as compared to the cv. Salinas reference genome. J. Biotechnol. 2018, 267, 12–18. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. 2019. Available online: https://cran.r-project.org/package=vegan (accessed on 14 June 2022).
- Schreiter, S.; Ding, G.-C.; Heuer, H.; Neumann, G.; Sandmann, M.; Grosch, R.; Kropf, S.; Smalla, K. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 2014, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Babin, D.; Sommermann, L.; Chowdhury, S.P.; Behr, J.H.; Sandmann, M.; Neumann, G.; Nesme, J.; Sørensen, S.J.; Schellenberg, I.; Rothballer, M.; et al. Distinct rhizomicrobiota assemblages and plant performance in lettuce grown in soils with different agricultural management histories. FEMS Microbiol. Ecol. 2021, 97, fiab027. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and Validation of Some ITS Primer Pairs Useful for Fungal Metabarcoding Studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Hannon, G. FASTX-Toolkit. FASTQ/A Short-Reads Pre-Processing Tools. 2010. Available online: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 23 June 2022).
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Antweiler, K.; Schreiter, S.; Keilwagen, J.; Baldrian, P.; Kropf, S.; Smalla, K.; Grosch, R.; Heuer, H. Statistical test for tolerability of effects of an antifungal biocontrol strain on fungal communities in three arable soils. Microb. Biotechnol. 2017, 10, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Larsson, K.-H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- UNITE Community. UNITE General FASTA Release for Fungi: Version 18.11.2018. 2019. Available online: https://unite.ut.ee/repository.php (accessed on 14 June 2022). [CrossRef]
- Wallon, T.; Sauvageau, A.; Van Der Heyden, H. Detection and Quantification of Rhizoctonia solani and Rhizoctonia solani AG1-IB Causing the Bottom Rot of Lettuce in Tissues and Soils by Multiplex qPCR. Plants 2021, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2020. Available online: http://www.r-project.org/index.html (accessed on 5 July 2022).
- Kolde, R. pheatmap: Pretty Heatmaps. 2019. Available online: https://cran.r-project.org/package=pheatmap (accessed on 14 June 2022).
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage Publications: Thousand Oaks, CA, USA, 2019; Available online: https://socialsciences.mcmaster.ca/jfox/Books/Companion/ (accessed on 5 July 2022).
- Mangiafico, S. rcompanion: Functions to Support Extension Education Program. 2020. Available online: https://cran.r-project.org/package=rcompanion (accessed on 14 June 2022).
- de Mendiburu, F. agricolae: Statistical Procedures for Agricultural Research. 2020. Available online: https://cran.r-project.org/package=agricolae (accessed on 14 June 2022).
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 5 July 2022).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes. 2014. Available online: https://cran.r-project.org/package=RColorBrewer (accessed on 14 June 2022).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; Available online: http://www.stats.ox.ac.uk/pub/MASS4/ (accessed on 5 July 2022).
- Wang, Y.; Naumann, U.; Eddelbuettel, D.; Wilshire, J.; Warton, D. mvabund: Statistical Methods for Analysing Multivariate Abundance Data. 2020. Available online: https://cran.r-project.org/package=mvabund (accessed on 14 June 2022).
- Grünwald, N.; Hu, S.; Van Bruggen, A. Short-term Cover Crop Decomposition in Organic and Conventional Soils: Soil Microbial and Nutrient Cycling Indicator Variables Associated with Different Levels of Soil Suppressiveness to Pythium aphanidermatum. Eur. J. Plant Pathol. 2000, 106, 51–65. [Google Scholar] [CrossRef]
- Wibberg, D.; Jelonek, L.; Rupp, O.; Kröber, M.; Goesmann, A.; Grosch, R.; Pühler, A.; Schlüter, A. Transcriptome analysis of the phytopathogenic fungus Rhizoctonia solani AG1-IB 7/3/14 applying high-throughput sequencing of expressed sequence tags (ESTs). Fungal Biol. 2014, 118, 800–813. [Google Scholar] [CrossRef]
- Ehrmann, J.; Ritz, K. Plant: Soil interactions in temperate multi-cropping production systems. Plant Soil 2014, 376, 1–29. [Google Scholar] [CrossRef]
- Qiu, W.; Su, H.; Yan, L.; Ji, K.; Liu, Q.; Jian, H. Organic Fertilization Assembles Fungal Communities of Wheat Rhizosphere Soil and Suppresses the Population Growth of Heterodera avenae in the Field. Front. Plant Sci. 2020, 11, 1225. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Yeoh, Y.K.; Kasinadhuni, N.R.P.; Lonhienne, T.G.A.; Robinson, N.; Hugenholtz, P.; Ragan, M.A.; Schmidt, S. Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci. Rep. 2015, 5, 8678. [Google Scholar] [CrossRef] [PubMed]
- Suleiman, A.K.A.; Harkes, P.; van den Elsen, S.; Holterman, M.; Korthals, G.W.; Helder, J.; Kuramae, E.E. Organic amendment strengthens interkingdom associations in the soil and rhizosphere of barley (Hordeum vulgare). Sci. Total Environ. 2019, 695, 133885. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ling, N.; Guo, J.; Wang, M.; Guo, S.; Shen, Q. Impacts of Fertilization Regimes on Arbuscular Mycorrhizal Fungal (AMF) Community Composition Were Correlated with Organic Matter Composition in Maize Rhizosphere Soil. Front. Microbiol. 2016, 7, 1840. [Google Scholar] [CrossRef] [PubMed]
- Marois, J.J.; Fravel, D.R.; Papavizas, G.C. Ability of Talaromyces flavus to occupy the rhizosphere and its interaction with Verticillium dahliae. Soil Biol. Biochem. 1984, 16, 387–390. [Google Scholar] [CrossRef]
- Madi, L.; Katan, T.; Katan, J.; Henis, Y. Biological Control of Sclerotium rolfsii and Verticillium dahliae by Talaromyces flavus Is Mediated by Different Mechanisms. Phytopathology 1997, 87, 1054–1060. [Google Scholar] [CrossRef]
- Kakvan, N.; Heydari, A.; Zamanizadeh, H.R.; Rezaee, S.; Naraghi, L. Development of new bioformulations using Trichoderma and Talaromyces fungal antagonists for biological control of sugar beet damping-off disease. Crop Prot. 2013, 53, 80–84. [Google Scholar] [CrossRef]
- Kazerooni, E.A.; Rethinasamy, V.; Al-Sadi, A.M. Talaromyces pinophilus inhibits Pythium and Rhizoctonia-induced damping-off of cucumber. J. Plant Pathol. 2019, 101, 377–383. [Google Scholar] [CrossRef]
- Kim, K.K.; Fravel, D.R.; Papavizas, G.C. Identification of a Metabolite Produced by Talaromyces flavus as Glucose Oxidase and its Role in the Biocontrol of Verticillium dahliae. Phytopathology 1988, 78, 488–492. [Google Scholar] [CrossRef]
- Duo-Chuan, L.I.; Chen, S.; Jing, L.U. Purification and partial characterization of two chitinases from the mycoparasitic fungus Talaromyces flavus. Mycopathologia 2005, 159, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Lan, D.; Wu, B. Chemistry and Bioactivities of Secondary Metabolites from the Genus Talaromyces. Chem. Biodivers. 2020, 17, e2000229. [Google Scholar] [CrossRef] [PubMed]
- Zhai, M.-M.; Li, J.; Jiang, C.-X.; Shi, Y.-P.; Di, D.-L.; Crews, P.; Wu, Q.-X. The Bioactive Secondary Metabolites from Talaromyces species. Nat. Prod. Bioprospect. 2016, 6, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, E.; Benoit, I.; van den Brink, J.; Wiebenga, A.; Coutinho, P.M.; Henrissat, B.; De Vries, R.P. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: A highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genom. 2011, 12, 38. [Google Scholar] [CrossRef]
- Celestino, K.R.S.; Cunha, R.B.; Felix, C.R. Characterization of a β-glucanase produced by Rhizopus microsporus var. microsporus, and its potential for application in the brewing industry. BMC Biochem. 2006, 7, 23. [Google Scholar] [CrossRef]
- Miyake, T.; Kato, A.; Tateishi, H.; Teraoka, T.; Arie, T. Mode of action of Talaromyces sp. KNB422, a biocontrol agent against rice seedling diseases. J. Pestic. Sci. 2012, 37, 56–61. [Google Scholar] [CrossRef]
- Butler, E.E. Rhizoctonia solani as a Parasite of Fungi. Mycologia 1957, 49, 354–373. [Google Scholar] [CrossRef]
- Davis, R.M.; Subbarao, K.V.; Raid, R.N. (Eds.) Compendium of Lettuce Diseases; Disease Compendium Series; APS Press: St. Paul, MN, USA, 1997; 79p. [Google Scholar]
- Schreiter, S.; Babin, D.; Smalla, K.; Grosch, R. Rhizosphere Competence and Biocontrol Effect of Pseudomonas sp. RU47 Independent from Plant Species and Soil Type at the Field Scale. Front. Microbiol. 2018, 9, 97. [Google Scholar] [CrossRef]
- Iorio, M.; Tocchetti, A.; Cruz, J.C.S.; Del Gatto, G.; Brunati, C.; Maffioli, S.I.; Sosio, M.; Donadio, S. Novel Polyethers from Screening Actinoallomurus spp. Antibiotics 2018, 7, 47. [Google Scholar] [CrossRef]
- Windisch, S.; Bott, S.; Ohler, M.-A.; Mock, H.-P.; Lippmann, R.; Grosch, R.; Smalla, K.; Ludewig, U.; Neumann, G. Rhizoctonia solani and Bacterial Inoculants Stimulate Root Exudation of Antifungal Compounds in Lettuce in a Soil-Type Specific Manner. Agronomy 2017, 7, 44. [Google Scholar] [CrossRef]
- Chapelle, E.; Mendes, R.; Bakker, P.A.H.M.; Raaijmakers, J.M. Fungal invasion of the rhizosphere microbiome. ISME J. 2016, 10, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Köberl, M.; Dita, M.; Martinuz, A.; Staver, C.; Berg, G. Members of Gammaproteobacteria as indicator species of healthy banana plants on Fusarium wilt-infested fields in Central America. Sci. Rep. 2017, 7, 45318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ménard, R.; Li, Y.; Coruzzi, G.M.; Heitz, T.; Shen, W.-H.; Berr, A. Arabidopsis SDG8 Potentiates the Sustainable Transcriptional Induction of the Pathogenesis-Related Genes PR1 and PR2 During Plant Defense Response. Front. Plant Sci. 2020, 11, 277. [Google Scholar] [CrossRef]
- Kumar, S.; Trivedi, P.K. Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef]
- Verbon, E.H.; Trapet, P.L.; Stringlis, I.A.; Kruijs, S.; Bakker, P.A.H.M.; Pieterse, C.M.J. Iron and Immunity. Annu. Rev. Phytopathol. 2017, 55, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Baba, S.A.; Vishwakarma, R.A.; Ashraf, N. Functional Characterization of CsBGlu12, a β-Glucosidase from Crocus sativus, Provides Insights into Its Role in Abiotic Stress through Accumulation of Antioxidant Flavonols. J. Biol. Chem. 2017, 292, 4700–4713. [Google Scholar] [CrossRef] [Green Version]
- Otulak-Kozieł, K.; Kozieł, E.; Bujarski, J.J.; Frankowska-Łukawska, J.; Torres, M.A. Respiratory Burst Oxidase Homologs RBOHD and RBOHF as Key Modulating Components of Response in Turnip Mosaic Virus—Arabidopsis thaliana (L.) Heyhn System. Int. J. Mol. Sci. 2020, 21, 8510. [Google Scholar] [CrossRef]
- Bethke, G.; Unthan, T.; Uhrig, J.F.; Pöschl, Y.; Gust, A.A.; Scheel, D.; Lee, J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 8067–8072. [Google Scholar] [CrossRef]
- Garcia, A.; Martinez, M.; Diaz, I.; Santamaria, M.E. The Price of the Induced Defense Against Pests: A Meta-Analysis. Front. Plant Sci. 2021, 11, 615122. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Van den Broeck, L.; Claeys, H.; Van Vlierberghe, K.; Matsui, M.; Inzé, D. The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 Antagonistically Regulate Mannitol-Induced Growth Inhibition in Arabidopsis. Plant Physiol. 2015, 169, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Kurt, F. An Insight into Oligopeptide Transporter 3 (OPT3) Family Proteins. Protein Pept. Lett. 2021, 28, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Wendehenne, D.; Gao, Q.-M.; Kachroo, A.; Kachroo, P. Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 2014, 20, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Yamagiwa, Y.; Inagaki, Y.; Ichinose, Y.; Toyoda, K.; Hyakumachi, M.; Shiraishi, T. Talaromyces wortmannii FS2 emits β-caryphyllene, which promotes plant growth and induces resistance. J. Gen. Plant Pathol. 2011, 77, 336–341. [Google Scholar] [CrossRef]
- Grosch, R.; Dietel, K.; Junge, H.; Chowdhury, S.P.; Hartmann, A.; Borriss, R. Interaktion von Bacillus amyloliquefaciens FZB42 mit dem Salatfäuleerreger und der mikrobiellen Rhizosphärengemeinschaft von Salat. In 58. Deutsche Pflanzenschutztagung: 10.–14. September 2012, Technische Universität Braunschweig; Kurzfassungen der Beiträge; Julius-Kühn-Archiv, Julius Kühn-Institut, Eds.; Julius Kühn-Institut, Bundesforschungsinstitut für Kulturpflanzen: Braunschweig, Germany, 2012; Volume 438, pp. 255–256. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on Lettuce Growth and Health under Pathogen Pressure and Its Impact on the Rhizosphere Bacterial Community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, W. (Ed.) Ernährungsstörungen bei Kulturpflanzen: Entstehung, visuelle u. analytische Diagnose, 2nd ed.; Gustav Fischer: Jena, Germany, 1988; 762p. [Google Scholar]


| Treatment | Spread of AG1-IB in Soils with Lettuce Plants [cm day−1] | Spread of AG2-2IIIb in Soils with Sugar Beet Plants [cm day−1] | ||||
|---|---|---|---|---|---|---|
| 2015 | 2016 | 2017 | 2015 | 2016 | 2017 | |
| HU-min | 0.09 ± 0.06 b | 0.25 ± 0.12 b | 0.34 ± 0.09 b | 0.49 ± 0.07 b | 0.17 ± 0.17 b | 0.59 ± 0.16 a |
| HU-org | 0.36 ± 0.09 a | 0.50 ± 0.21 a | 0.41 ± 0.01 a | 0.99 ± 0.13 a | 0.67 ± 0.16 a | 0.64 ± 0.12 a |
| Factor | Ev [%] | p-Value |
|---|---|---|
| Fertl | 11.6 | <0.05 |
| R. solani (Rs) | 5.7 | n.s. |
| Fertl × Rs | 43.7 | <0.001 |
| Residuals | 38.9 |
| (A) | Bulk Soil | |||
|---|---|---|---|---|
| Phylum | Family | Genus | HU-org | HU-min |
| Acidobacteria | Gp4# | Gp4# | 2.3 ± 1.2 | 0.4 ± 0.1 |
| Firmicutes | Paenibacillaceae_1 | Fontibacillus | 0.5 ± 0.7 | 0.1 ± 0.0 |
| Acidobacteria | Gp1# | Gp1# | 3.0 ± 0.7 | 5.4 ± 0.8 |
| Acidobacteria | Occallatibacter# | Occallatibacter# | 0.0 ± 0.0 | 0.5 ± 0.1 |
| Actinobacteria | Solirubrobacterales# | Solirubrobacterales# | 0.5 ± 0.1 | 0.8 ± 0.1 |
| Actinobacteria | Thermomonosporaceae | Thermomonosporaceae# | 0.2 ± 0.0 | 0.5 ± 0.1 |
| Actinobacteria | Thermomonosporaceae | Actinoallomurus | 0.1 ± 0.0 | 0.7 ± 1.3 |
| Candidatus_Saccharibacteria | Candidatus_Saccharibacteria# | Candidatus_Saccharibacteria# | 0.9 ± 0.4 | 3.2 ± 1.8 |
| Chloroflexi | Ktedonobacterales# | Ktedonobacterales# | 0.2 ± 0.0 | 0.6 ± 0.2 |
| Firmicutes | Alicyclobacillaceae | Tumebacillus | 0.5 ± 0.1 | 1.1 ± 0.3 |
| Firmicutes | Bacillaceae_1 | Neobacillus | 1.2 ± 0.2 | 2.2 ± 0.1 |
| Proteobacteria | Bradyrhizobiaceae | Bradyrhizobium | 1.1 ± 0.3 | 1.9 ± 0.6 |
| Proteobacteria | Acetobacteraceae | Acetobacteraceae# | 0.3 ± 0.1 | 0.5 ± 0.1 |
| Proteobacteria | Gammaproteobacteria _incertae_sedis | Acidibacter | 0.4 ± 0 | 0.8 ± 0.5 |
| Verrucomicrobia | Subdivision3# | Subdivision3# | 0.2 ± 0 | 0.5 ± 0.2 |
| (B) | Bulk Soil | |||
| Phylum | Family | Genus | HU-org + Rs | HU-min + Rs |
| Acidobacteria | Gp4# | Gp4# | 1.7 ± 0.6 | 0.5 ± 0.1 |
| Actinobacteria | Micrococcaceae | Micrococcaceae # | 1.1 ± 1.9 | 0.0 ± 0.0 |
| Acidobacteria | Gp1# | Gp1# | 2.4 ± 0.9 | 4.7 ± 1.1 |
| Acidobacteria | Gp2# | Gp2# | 0.3 ± 0.1 | 0.7 ± 0.2 |
| Chloroflexi | Ktedonobacterales# | Ktedonobacterales# | 0.2 ± 0.1 | 0.6 ± 0.2 |
| Firmicutes | Alicyclobacillaceae | Tumebacillus | 0.5 ± 0.2 | 1.2 ± 0.3 |
| Firmicutes | Bacillaceae_2 | Bacillaceae_2# | 0.2 ± 0.1 | 0.7 ± 0.6 |
| Proteobacteria | Proteobacteria# | Proteobacteria# | 0.3 ± 0.1 | 0.5 ± 0.1 |
| Factor | BS Ev [%] | p-Value | RA Ev [%] | p-Value | RH Ev [%] | p-Value |
|---|---|---|---|---|---|---|
| Fertl | 36.9 | <0.001 | 46.7 | <0.001 | 30.1 | <0.001 |
| R. solani (Rs) | 10.8 | <0.05 | 5.8 | n.s. | 11.8 | <0.05 |
| Fertl × Rs | 5.6 | n.s. | 6.4 | n.s. | 11.6 | <0.05 |
| Residuals | 46.6 | 41.1 | 46.5 |
| (A) | Bulk Soil | Root-Associated Soil | Rhizosphere | |||||
|---|---|---|---|---|---|---|---|---|
| Genus | HU-org | HU-min | Genus | HU-org | HU-min | Genus | HU-org | HU-min |
| Didymella | 10.3 ± 2.0 | 0.6 ± 0.3 | Didymella | 8.8 ± 1.6 | 0.5 ± 0.3 | Didymella | 3.7 ± 1.7 | 0.2 ± 0.1 |
| Ceratobasidiaceae# | 1.4 ± 2.8 | 0.1 ± 0.1 | Metarhizium | 3.5 ± 4.9 | 0.6 ± 0.3 | Humicola | 3.2 ± 5.9 | 0.1 ± 0.1 |
| Sordariales# | 1.2 ± 0.4 | 0.3 ± 0.3 | Actinomucor | 2.4 ± 1.5 | 0.0 ± 0.0 | Ilyonectria | 1.2 ± 0.9 | 0.1 ± 0.0 |
| Humicola | 1.0 ± 0.2 | 0.3 ± 0.1 | Powellomycetaceae# | 1.0 ± 1.2 | 0.1 ± 0.1 | Arthrobotrys | 1.0 ± 0.4 | 0.1 ± 0.1 |
| Retroconis | 0.9 ± 0.4 | 0.3 ± 0.2 | Laetisaria | 0.8 ± 1.0 | 0.0 ± 0.0 | Plenodomus | 0.8 ± 0.3 | 0.1 ± 0.1 |
| Herpotrichiellaceae# | 0.8 ± 0.1 | 0.2 ± 0.1 | Herpotrichiellaceae# | 0.7 ± 0.1 | 0.2 ± 0.1 | Volucrispora | 0.8 ± 0.3 | 0.1 ± 0.1 |
| Volucrispora | 0.5 ± 0.1 | 0.1 ± 0.1 | Humicola | 0.6 ± 0.2 | 0.2 ± 0.1 | Rhizophagus | 0.7 ± 0.8 | 0.0 ± 0.0 |
| Laetisaria | 0.5 ± 0.6 | 0.0 ± 0.0 | Fungi# | 0.6 ± 0.3 | 0.1 ± 0.0 | Actinomucor | 0.7 ± 0.7 | 0.0 ± 0.0 |
| Ilyonectria | 0.5 ± 0.3 | 0.0 ± 0.0 | Sordariales# | 0.5 ± 0.3 | 0.1 ± 0.0 | Funneliformis | 0.5 ± 0.3 | 0.0 ± 0.0 |
| Rhizopus | 5.7 ± 9.8 | 29.0 ± 30.5 | Apiotrichum | 0.1 ± 0.1 | 2.7 ± 4.6 | Herpotrichiellaceae# | 0.5 ± 0.2 | 0.1 ± 0.1 |
| Umbelopsis | 0.6 ± 0.1 | 2.4 ± 1.0 | Umbelopsis | 0.4 ± 0.1 | 1.4 ± 0.7 | Apiotrichum | 0.1 ± 0.1 | 7.3 ± 14.0 |
| Oculimacula | 0.2 ± 0.1 | 1.2 ± 0.8 | Paraconiothyrium | 0.0 ± 0.0 | 0.7 ± 0.3 | Talaromyces | 0.4 ± 0.5 | 3.5 ± 4.4 |
| Paraconiothyrium | 0.0 ± 0.0 | 0.9 ± 0.2 | Fusicolla | 0.0 ± 0.0 | 0.6 ± 0.7 | Bionectriaceae# | 0.1 ± 0.1 | 2.3 ± 4.5 |
| Apiotrichum | 0.2 ± 0.1 | 0.8 ± 0.6 | Oculimacula | 0.2 ± 0.1 | 0.5 ± 0.3 | Saitozyma | 0.8 ± 0.5 | 2.0 ± 1.5 |
| Gibberella | 0.3 ± 0.1 | 0.8 ± 0.9 | Fusicolla | 0.0 ± 0.0 | 1.7 ± 1.8 | |||
| Cladophialophora | 0.2 ± 0.1 | 0.5 ± 0.3 | Umbelopsis | 0.2 ± 0.1 | 1.0 ± 0.7 | |||
| Paraconiothyrium | 0.0 ± 0.0 | 0.5 ± 0.5 | ||||||
| (B) | Bulk Soil | Root-Associated Soil | Rhizosphere | |||||
| Genus | HU-org + Rs | HU-min + Rs | Genus | HU-org + Rs | HU-min + Rs | Genus | HU-org + Rs | HU-min + Rs |
| Didymella | 8.5 ± 4.8 | 0.3 ± 0.0 | Didymella | 12.3 ± 1.3 | 0.4 ± 0.1 | Talaromyces | 51.2 ± 33.4 | 3.6 ± 4.7 |
| Waitea | 1.3 ± 2.5 | 0.0 ± 0.0 | Talaromyces | 11.8 ± 9.9 | 0.8 ± 0.4 | Didymella | 3.7 ± 3.7 | 0.1 ± 0.0 |
| Rhizopus | 3.0 ± 4.7 | 57.3 ± 11.6 | Actinomucor | 1.5 ± 2.1 | 0.1 ± 0.0 | Rhizophagus | 0.9 ± 1.6 | 0.0 ± 0.0 |
| Umbelopsis | 0.3 ± 0.2 | 1.2 ± 0.3 | Ascobolus | 0.9 ± 0.6 | 0.1 ± 0.0 | Ilyonectria | 0.9 ± 1.0 | 0.0 ± 0.0 |
| Herpotrichiellaceae# | 0.9 ± 0.4 | 0.2 ± 0.0 | Rhizopus | 0.6 ± 0.3 | 65.0 ± 14.9 | |||
| Agaricales# | 0.9 ± 0.5 | 0.0 ± 0.0 | Saitozyma | 0.4 ± 0.2 | 1.5 ± 1.3 | |||
| Tremellales# | 0.7 ± 1.1 | 0.0 ± 0.0 | Umbelopsis | 0.0 ± 0.0 | 0.5 ± 0.3 | |||
| Retroconis | 0.5 ± 0.1 | 0.1 ± 0.0 | ||||||
| Rhizopus | 4.3 ± 6.8 | 54.7 ± 7.7 | ||||||
| Umbelopsis | 0.5 ± 0.4 | 1.1 ± 0.3 | ||||||
| Sistotrema | 0.0 ± 0.0 | 0.9 ± 1.6 | ||||||
| (A) | Bulk Soil | Root-Associated Soil | Rhizosphere | |||||
|---|---|---|---|---|---|---|---|---|
| Genus | HU-org | HU-org + Rs | Genus | HU-org | HU-org + Rs | Genus | HU-org | HU-org + Rs |
| Ceratobasidiaceae# | 1.4 ± 2.8 | 0.0 ± 0.0 | Powellomycetaceae# | 1.0 ± 1.2 | 0.2 ± 0.0 | Rhizopus | 18.3 ± 19.2 | 0.6 ± 0.3 |
| Volucrispora | 0.5 ± 0.1 | 0.0 ± 0.0 | Talaromyces | 0.5 ± 0.6 | 11.8 ± 9.9 | Humicola | 3.2 ± 5.9 | 0.1 ± 0.1 |
| Talaromyces | 0.2 ± 0.2 | 24.7 ± 34.6 | Agaricales# | 0.1 ± 0.0 | 0.9 ± 0.5 | Arthrobotrys | 1.0 ± 0.4 | 0.2 ± 0.1 |
| Waitea | 0.0 ± 0.0 | 1.3 ± 2.5 | Tremellales# | 0.0 ± 0.0 | 0.7 ± 1.1 | Volucrispora | 0.8 ± 0.3 | 0.0 ± 0.0 |
| Actinomucor | 0.7 ± 0.7 | 0.0 ± 0.0 | ||||||
| Gibellulopsis | 0.6 ± 1.0 | 0.0 ± 0.0 | ||||||
| Talaromyces | 0.4 ± 0.5 | 51.2 ± 33.4 | ||||||
| Tremellales# | 0.1 ± 0.1 | 4.1 ± 7.9 | ||||||
| Apiosporaceae# | 0.0 ± 0.0 | 0.5 ± 1.0 | ||||||
| (B) | Bulk Soil | Root-Associated Soil | Rhizosphere | |||||
| Genus | HU-min | HU-min + Rs | Genus | HU-min | HU-min + Rs | Genus | HU-min | HU-min + Rs |
| Devriesia | 0.5 ± 0.2 | 0.0 ± 0.0 | Sistotrema | 0.0 ± 0.0 | 0.9 ± 1.6 | Apiotrichum | 7.3 ± 14.0 | 0.0 ± 0.0 |
| Talaromyces | 0.4 ± 0.4 | 5.9 ± 9.7 | Bionectriaceae# | 2.3 ± 4.5 | 0.0 ± 0.0 | |||
| Cantharellales# | 0.0 ± 0.0 | 1.0 ± 1.9 | Fusicolla | 1.7 ± 1.8 | 0.0 ± 0.0 | |||
| Plenodomus | 0.1 ± 0.1 | 0.7 ± 0.3 | ||||||
| Tremellales# | 0.0 ± 0.0 | 0.7 ± 1.0 | ||||||
| Treatment | BS [pg/gram Soil] | RA [pg/gram Soil] | RH [pg/gram Soil] |
|---|---|---|---|
| HU-min | 0.03 ± 0.01 ab | 0.02 ± 0.01 a | 0.08 ± 0.05 b |
| HU-min + Rs | 0.09 ± 0.07 a | 0.06 ± 0.09 a | 1.71 ± 1.98 a |
| HU-org | 0.04 ± 0.01 ab | 0.02 ± 0.01 a | 0.05 ± 0.02 b |
| HU-org + Rs | 0.04 ± 0.04 b | 0.02 ± 0.01 a | 0.90 ± 1.28 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sommermann, L.; Babin, D.; Behr, J.H.; Chowdhury, S.P.; Sandmann, M.; Windisch, S.; Neumann, G.; Nesme, J.; Sørensen, S.J.; Schellenberg, I.; et al. Long-Term Fertilization Strategy Impacts Rhizoctonia solani–Microbe Interactions in Soil and Rhizosphere and Defense Responses in Lettuce. Microorganisms 2022, 10, 1717. https://doi.org/10.3390/microorganisms10091717
Sommermann L, Babin D, Behr JH, Chowdhury SP, Sandmann M, Windisch S, Neumann G, Nesme J, Sørensen SJ, Schellenberg I, et al. Long-Term Fertilization Strategy Impacts Rhizoctonia solani–Microbe Interactions in Soil and Rhizosphere and Defense Responses in Lettuce. Microorganisms. 2022; 10(9):1717. https://doi.org/10.3390/microorganisms10091717
Chicago/Turabian StyleSommermann, Loreen, Doreen Babin, Jan Helge Behr, Soumitra Paul Chowdhury, Martin Sandmann, Saskia Windisch, Günter Neumann, Joseph Nesme, Søren J. Sørensen, Ingo Schellenberg, and et al. 2022. "Long-Term Fertilization Strategy Impacts Rhizoctonia solani–Microbe Interactions in Soil and Rhizosphere and Defense Responses in Lettuce" Microorganisms 10, no. 9: 1717. https://doi.org/10.3390/microorganisms10091717
APA StyleSommermann, L., Babin, D., Behr, J. H., Chowdhury, S. P., Sandmann, M., Windisch, S., Neumann, G., Nesme, J., Sørensen, S. J., Schellenberg, I., Rothballer, M., Geistlinger, J., Smalla, K., & Grosch, R. (2022). Long-Term Fertilization Strategy Impacts Rhizoctonia solani–Microbe Interactions in Soil and Rhizosphere and Defense Responses in Lettuce. Microorganisms, 10(9), 1717. https://doi.org/10.3390/microorganisms10091717







