Abstract
By integrating phylogenomic and comparative analyses of 1104 high-quality genome sequences, we identify the core proteins and the lineage-specific fingerprint proteins of the various evolutionary clusters (clades/groups/species) of the Bacillus genus. As fingerprints, we denote those core proteins of a certain lineage that are present only in that particular lineage and absent in any other Bacillus lineage. Thus, these lineage-specific fingerprints are expected to be involved in particular adaptations of that lineage. Intriguingly, with a few notable exceptions, the majority of the Bacillus species demonstrate a rather low number of species-specific fingerprints, with the majority of them being of unknown function. Therefore, species-specific adaptations are mostly attributed to highly unstable (in evolutionary terms) accessory proteomes and possibly to changes at the gene regulation level. A series of comparative analyses consistently demonstrated that the progenitor of the Cereus Clade underwent an extensive genomic expansion of chromosomal protein-coding genes. In addition, the majority (76–82%) of the B. subtilis proteins that are essential or play a significant role in sporulation have close homologs in most species of both the Subtilis and the Cereus Clades. Finally, the identification of lineage-specific fingerprints by this study may allow for the future development of highly specific vaccines, therapeutic molecules, or rapid and low-cost molecular tests for species identification.
1. Introduction
Bacillus are rod-shaped, Gram-positive aerobic (or facultatively anaerobic) bacteria that form endospores and colonize many diverse habitats [1]. The type species of the genus, Bacillus subtilis, was first described by Ehrenberg in 1835 (as Vibrio subtilis), whereas the genus was established by Cohn and Koch in 1872 [2]. Members of the genus have been isolated from soil, water, and sediment, as well as from many diverse hosts, such as humans, animals, and plants [3,4], and they act as both human and/or animal pathogens [5]. Bacillus species have been exploited as plant growth-promoting factors [6], as pest controllers [7], and as bioreactors for the production of important enzymes, metabolites, antibiotics, food preservatives [8], and probiotics [9]. Bacillus subtilis is a very popular model organism for studying Gram-positive bacteria and sporulation as a developmental pathway [10]; it is also emerging as a synthetic biology “chassis” [11]. Moreover, many other Bacillus species are also focal points of research, including B. cereus (a cause of foodborne illnesses), B. thuringiensis (a pest control agent), and B. anthracis (a lethal pathogen of livestock and humans).
Taxonomically, the Bacillus genus belongs to the Firmicutes phylum and includes more than 104 species that display high diversity (https://lpsn.dsmz.de/genus/bacillus, accessed on 22 August 2022) [4,12,13]. Phylogenetic analyses based on 16S rRNA and on Multi-Locus Sequence Typing (MLST) have studied the evolution and taxonomy of this genus [14]. However, the advent of low-cost whole-genome sequencing technologies has played a critical role in resolving evolutionary relationships at an even finer scale (even at the species or strain level) [15]. This new phylogenomic approach utilizes the phylogenetic signal from hundreds or even thousands of orthologous genes/proteins [16,17,18]. Furthermore, phylogenomics is considered robust against phenomena, such as Horizontal Gene Transfer, that may scramble the evolutionary signal of certain gene families but not the majority of them [19,20,21]. The phylogenomic approach relies on the pangenome concept, where genes and proteins are characterized as core, dispensable/accessory or strain specific, based on their evolutionary/phylogenetic distribution [22,23]. As more genomes become available, the concept of core genes/proteins needs to become more relaxed (i.e., presence in 95% of the analyzed strains) so as to account for sequencing errors, among other things, [24]. In addition, comparisons of average nucleotide identity (ANI) between whole-genome sequences have been utilized in this new genomic era for delimiting species boundaries, usually with an implemented cut-off value of 94–96% identity [25,26].
The importance of the Bacillus genus has led to the sequencing of almost 5800 genomes (source: NCBI Assembly; Bethesda, MA, USA, May 2022), with at least 20% of them being annotated as whole-genome or chromosome-level assemblies (high quality). Thus, several recent phylogenomic studies have utilized the wealth of these genomic data to delineate, with much higher accuracy and confidence, the major and minor Bacillus lineages and their evolutionary relationships [3,27,28,29,30,31,32,33,34,35,36]. For example, [3] analyzed 79 representative Bacillus genomes and identified 196 core genes that were utilized for phylogenomic analyses. They identified 9 well-supported clades within the genus, with the B. cereus and B. subtilis clades being the most prominent. A recent phylogenomic study based on 352 genomes proposed that the genus should include the two major clades of B. subtilis and B. cereus and some additional Bacillus species, whereas several other previously classified Bacillus species should be re-assigned to new genera [4]. A later study of 303 genomes proposed that the Bacillus genus should only include the two major clades (termed the Subtilis Clade and Cereus Clade), whereas all the other previously classified Bacillus species should become new genera [37]. Based strictly on phylogenomics, the Cereus Clade should also be a distinct genus; however, it has been decided to retain the Bacillus name for health and safety reasons [38,39]. Recently, the NCBI taxonomy adopted some of these conclusions and removed several clades from the Bacillus genus.
The goal of this study is to utilize the publicly available complete sequences of genomes/chromosomes of Bacillus species to identify the distinct evolutionary lineages at the clade and species level based on phylogenomics and ANI values. We also determine the chromosomally encoded core and the fingerprint proteins of these lineages that characterize/define them at both relaxed and strict stringencies. This should reveal any molecular adaptations at the gene/protein content level. We define those fingerprint proteins that are present in all analyzed members of a lineage/group but are absent in all other analyzed Bacillus genomes. Therefore, these fingerprints constitute lineage-specific core proteins. In addition, another category of strict fingerprints will be erected that do not have a close homolog (>50% amino acid identity, over 50% of the protein’s length) in any other Bacillus lineage. For a more detailed definition of relaxed and strict fingerprints, see Section 2.4. We applied a similar approach in an analysis of the Pseudomonas genus and identified fingerprint proteins of Pseudomonas aeruginosa (present in all P. aeruginosa members, but absent in all other Pseudomonas groups) that are involved in its pathogenicity in humans [40].
2. Materials and Methods
2.1. Analyzed Genomes
A total of 1154 genomes of the Bacillus genus (NCBI taxonomy ID: 1386) were downloaded from the NCBI RefSeq database (latest download in April 2022), whose assembly level was annotated as being a complete genome or chromosome. Next, we filtered out genomes that were from confounding strains (i.e., those that have been artificially manipulated) or had less than 10× genome coverage, more than 1% unknown nucleotides, or many pseudogenes (≥10%) [34]. The final dataset contained 1104 genomes. The goal was to filter out all genomes whose assembly level was of lower quality and would result in a significant underestimation of the core proteomes and the accompanying fingerprints.
2.2. Orthology Detection and Phylogenomic Analysis
In order to identify the core proteome of this set of organisms, we implemented a series of Python scripts that we developed for studying the core proteome of the Pseudomonas genus [40]. In brief, these scripts rely on best reciprocal BLAST hits between a reference proteome of a defined evolutionary group, and all the other proteomes of that evolutionary group that are under investigation. In this way, a core set of orthologs present in them all was identified. Thus, each evolutionary group has its own reference proteome (see Supplementary Excel File S1, spreadsheet 1). For all reciprocal BLAST hits of the reference proteome against another proteome, the Python script gathers all the best reciprocal BLAST-result percent identities, estimates the mean value and standard deviation and then filters out all hits that have identities two standard deviations below the average value. This approach permits the definition of an adjustable orthology cut-off, depending on the genetic distance of the two genomes/proteomes undergoing reciprocal BLAST, instead of fixed cut-offs of sequence identity/similarity or defined BLAST score ratios, as implemented in many other pangenome analyses [22,41,42,43,44]. Afterwards, multiple alignments for all identified groups of core orthologs are generated with Muscle software [45]; they are concatenated in a super-alignment and then filtered with G-blocks software [46] for removing badly aligned regions (default parameters). A maximum likelihood (ML) phylogenomic tree is generated, using the IQTree2 software [47], which automatically calculates the best-fit model. In our study, tree visualization was performed using Treedyn [48] and iTOL [49].
Species boundary determination was based on Average Nucleotide identity with the FastANI [50] software and MUMmer/pyani software [51]. Functional category assignment of the identified core and fingerprint proteins was based on the EGGNOG database v5 [52] and the KEGG Orthologies with the KAAS tool [53] and COG [54].
2.3. Detection of Core Proteomes
A protein is considered to be a member of the core proteome of a certain lineage if it is present in all its members. However, the number of proteomes analyzed seriously affects the number of core proteins; more genomes result in fewer core proteins [40]. Given the imbalanced sequencing of the various lineages, we generated a normalized core proteome for each evolutionary lineage of interest by using a maximum number of only five randomly selected proteomes from that lineage. Such normalized datasets allow for meaningful comparisons between lineages of varying sampling coverage, in terms of numbers of genomes. They, nevertheless, remain within the concept of a soft-core proteome [24]. The list of normalized core proteins for each evolutionary lineage (Clade/group/species) is given in Supplementary Excel File S1, spreadsheet 2. We tested the statistical significance of enrichment of a certain functional category within the normalized core proteins of a certain species using the hypergeometric test. The results are summarized in Supplementary Excel File S1, spreadsheet 3.
We investigated if a normalized core proteome based on five genomes would be equivalent to a soft-core proteome at the 85%, 90%, or 95% level, assuming that many more genomes would be available. We thus performed permutation analyses for four different species (B. subtilis, B. velezensis, B. anthracis, and Cereus subclade 2) with more than 100 available genomes each. We randomly sampled twenty times each, genomes of that species for different genome numbers available and estimated how many of its proteins would be present in 85%, 90%, or 95% of the selected genomes. We plotted these permutated soft-cores together with the normalized core based on the five or less genomes for that species. Less than five complete genomes results in an even softer core. As it is evident from Supplementary Figure S1, the normalized core should correspond to a soft core of 85% for B. velezensis, whereas for the other three species it corresponds to a soft core of between 90–95%.
2.4. Detection of Lineage-Specific Relaxed and Strict Fingerprint Proteins
In order to identify fingerprint proteins of a particular evolutionary lineage, we applied two criteria, one relaxed and the other stringent. Based on our relaxed criteria, the orthologs of this protein (relaxed fingerprint) were present in the five (or less) analyzed members of this particular clade (that were used for the normalized core proteome) and absent in all other Bacillus proteomes (that were included in normalized datasets). Based on our more stringent criterion (strict fingerprints), the previously identified relaxed fingerprints should additionally not have any other close homolog in any of the other Bacillus proteomes with BLASTP percent identity above 50%, across 50% of the protein’s length. From this point on, we will refer to the fingerprint proteins with two numbers, one for the normalized relaxed fingerprints and the other for the normalized strict fingerprints. The list and table of relaxed/strict fingerprints for each evolutionary lineage (Clade/group/species) together with their functional category is given in Supplementary Excel File S1, spreadsheets 3 and 4.
3. Results and Discussion
3.1. Phylogenomic Analysis of the Bacillus Genus
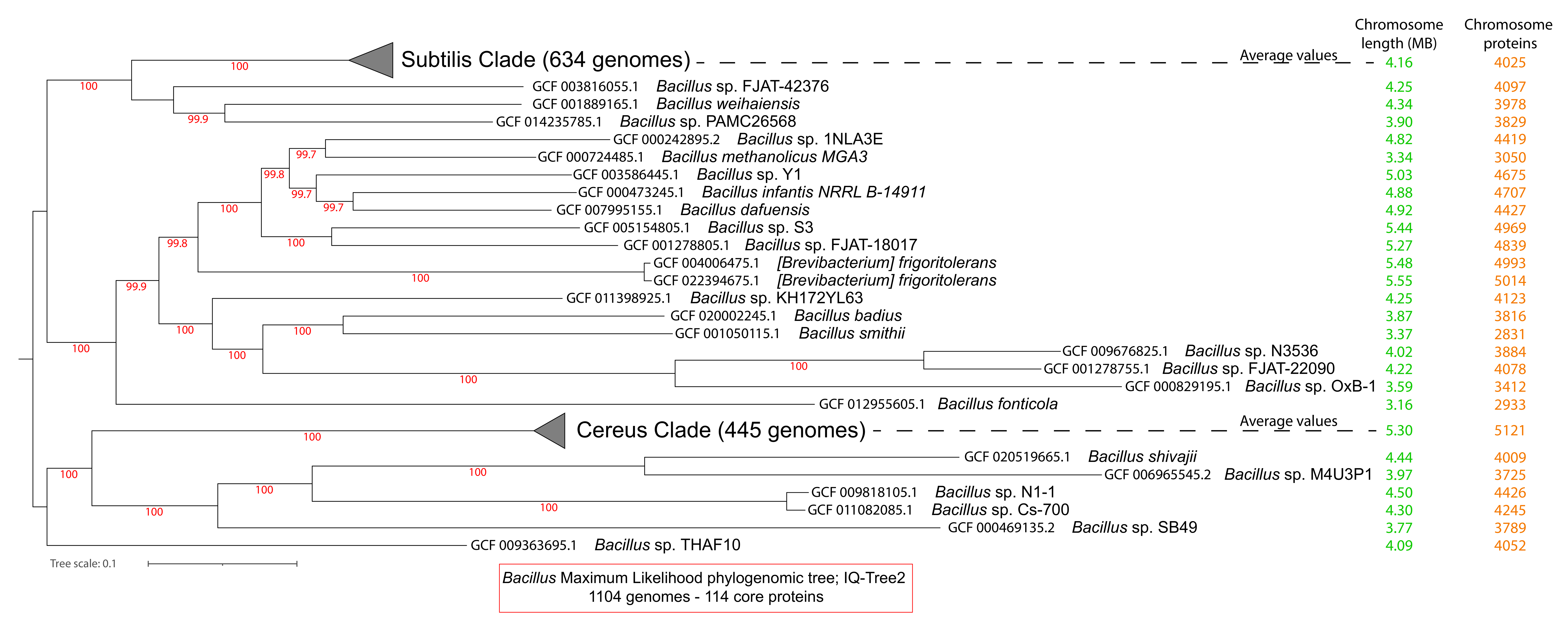
We analyzed all of the 1104 complete proteomes of the Bacillus genus, based on the latest NCBI Taxonomy [55]. B. subtilis strain 168 [10,56] was used as a reference proteome for the whole genus. This was the first Gram-positive bacterium to have its whole genome completely sequenced; moreover, this genome has a high quality annotation [57,58]. In this set of complete proteomes, the most numerous groups were B. subtilis strains (194), B. velezensis (202), B. cereus (131), B. anthracis (114), B. thuringiensis (81), and B. amyloliquefaciens (58). Our first analysis identified 114 core proteins for the whole Bacillus genus (see Supplementary Excel File S1, spreadsheet 2). The multiple alignment of these 114 core proteins contained 20,041 variable sites (after G-blocks filtering) that were used to generate a maximum likelihood phylogenomic tree in IQ-Tree2 (LG + I + F + G4 model-aLRT) [47] (see Figure 1).
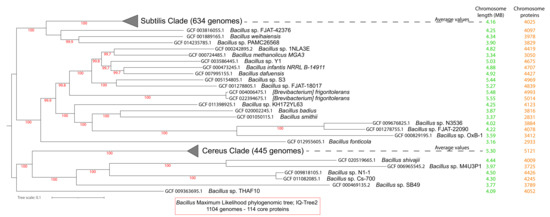
Figure 1.
The phylogenomic maximum likelihood tree (IQ-Tree2) of the 1104 Bacillus proteomes. The tree was based on 114 core proteins and 20,041 variable sites, using the LG + I + F + G4 model and aLRT. For ease of visualization, the entire Subtilis and Cereus Clades are collapsed. Next to each leaf of the tree, the chromosome size, and the number of all chromosomally encoded proteins are given.
We identified two major clades that are also focal points of research in this genus: one that includes B. subtilis and many other species and is now referred to in the literature as the Subtilis Clade; and another that includes B. cereus and many other species and is now referred to in the literature as the Cereus Clade [37].
3.2. Phylogenomic Analysis of the Subtilis Clade
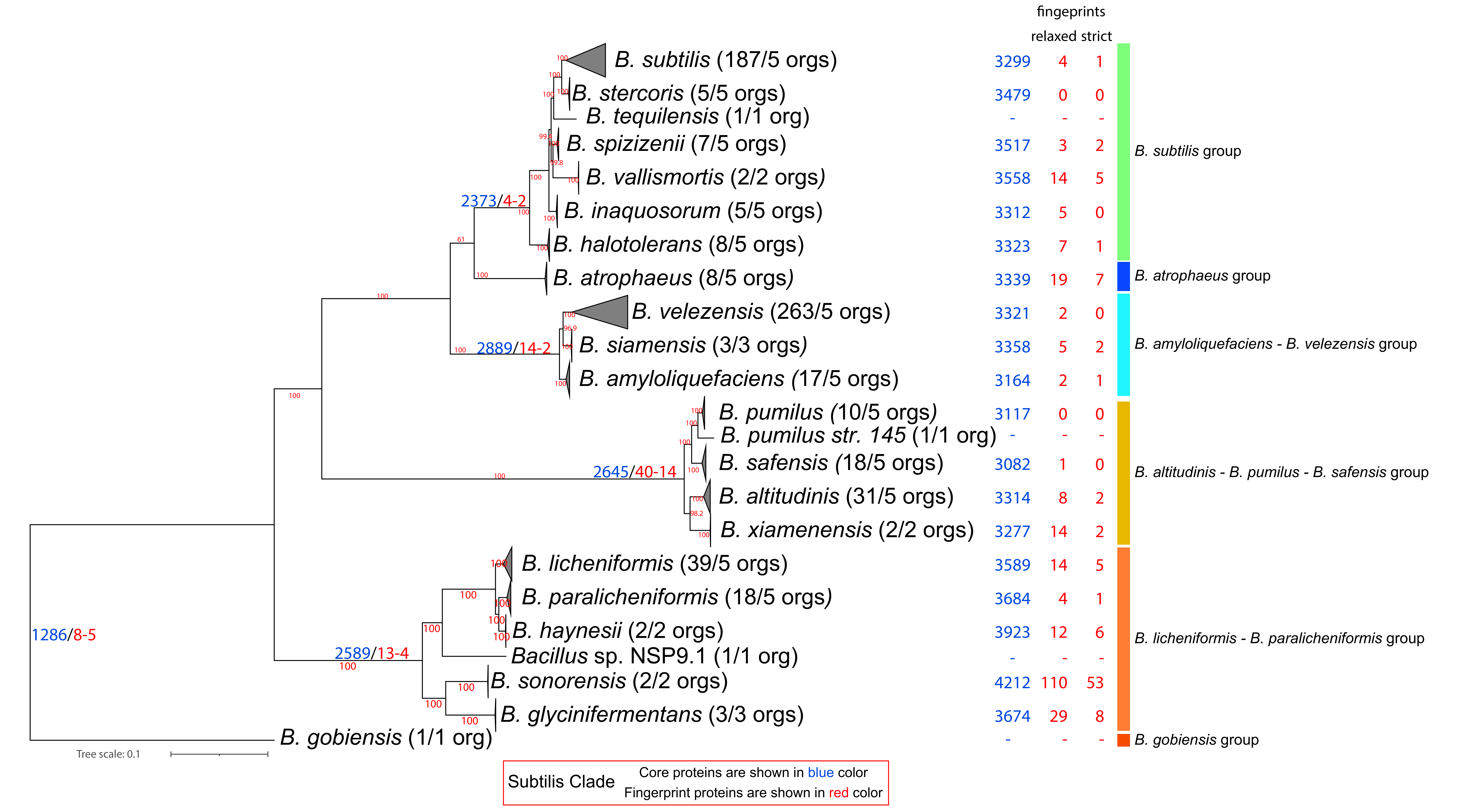
The Subtilis Clade [1,31,35,37] includes six major groups and 23 species (see Figure 2 and Supplementary Figure S2, for a complete tree): (i) B. subtilis (7 species), (ii) B. atrophaeus (1 species), (iii) B. amyloliquefaciens—B. velezensis (3 species), (iv) B. altitudinis—B. pumilus—B. safensis (5 species), (v) B. licheniformis—B. paralicheniformis (6 species), (vi) B. gobiensis (1 species). The members of this clade are hard to distinguish from each other based on phenotypic characteristics or the 16S rRNA phylogeny [59]. This clade has also been verified by our analysis and includes 634 genomes (see Figure 2). It is comprised of 1286 core proteins, with only 8/5 of them being Subtilis Clade relaxed/strict fingerprints, meaning that they are found only within this clade and in no other Bacillus proteome that we analyzed. Most of them are of unknown function, whereas one of them is involved in energy production and conversion and another is involved in nucleotide transport and metabolism.
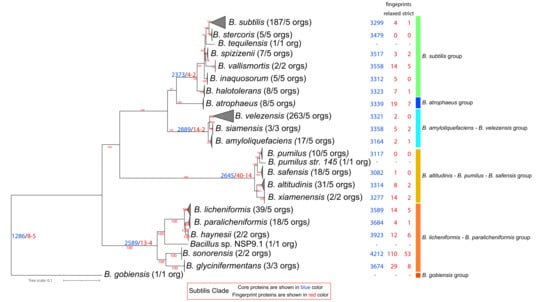
Figure 2.
Phylogenomic ML tree (IQ-Tree2-Q.Plant + I + F + G4-aLRT) of the Subtilis Clade based on 457 core protein-orthologous groups from 634 proteomes. For ease of visualization, certain evolutionary clusters have been collapsed. The full tree is available as Supplementary Figure S2. Next to the species name, in parentheses, is the number of complete genomes that are available and, on their right, is the number of genomes used in the normalized dataset. Further to the right of the species names and at the common ancestor of a lineage, with blue and red colors we denote the number of core and relaxed/strict fingerprint proteins for each lineage (based on the normalized dataset).
3.3. Phylogenomic Analysis of the Cereus Clade
This large phylogenetic clade is organized into three major groups or else subclades [27,28,29,30,60,61,62,63,64,65]. It consists of approximately 30 evolutionary clusters, representing 11 known species and 19–20 putative novel species [29,61]. They are mostly soil bacteria, with some of them being opportunistic pathogens and some being recently emerged pathogens in humans and other organisms [65]. Accordingly, they have been characterized as “hopeful monsters” that can be transformed into pathogens, under the right conditions and circumstances [27,66]. Thus, insights into their evolution are important for understanding basic mechanisms of pathogen emergence. The type species of this large clade is B. cereus, a common soil bacterium that is frequently involved in food poisoning [67]. It has also been implicated in skin infection, pneumonia, bacteremia, and meningitis (mostly in immunocompromised individuals) [30]. Its pathogenicity has been attributed to an emetic toxin (cereulide), to enterotoxins/hemolysins, phospholipases, and proteases that function as tissue-destructive exoenzymes. Another prominent member of this clade, B. thuringiensis, is an insect pathogen that is characterized by the production of parasporal crystals that contain the insect toxins cry, cyt, and vip, encoded by plasmids [67]. If these plasmids are lost, then B. thuringiensis cannot be distinguished from B. cereus. Accordingly, B. thuringiensis has also been reported as an opportunistic human pathogen [67]. B. anthracis is another prominent member of this clade; it was identified by Koch and Pasteur as the etiological agent of anthrax and is pathogenic for both humans and herbivores [67]. Its pathogenicity is mostly attributed to two large plasmids, pXO1 (that encodes three toxin peptides) and pXO2 (that produces the poly-γ-d-glutamic acid antiphagocytic capsule via the capBCADE operon) [68]. The phylogenetic distribution of the pathogenic plasmids and genes has shown that a classification that is mostly based on phenotype and virulence is improper [61,62]. A consistently observed scattered phylogenetic distribution of B. cereus, B. thuringiensis, and B. anthracis has been the focus of many previous studies. Several studies have proposed that B. cereus, B. thuringiensis, and B. anthracis should be treated as one species, based on high levels of chromosome synteny and protein similarity [69,70]. The components that differentiate them are mostly attributed to the plasticity of the accessory genomes, with plasmids playing a key role [71]. However, adaptive mutations, recombination events, and re-organization of the gene regulatory network also contribute to this phenotypic heterogeneity. In addition, the impact of positive selection on the core genome shapes the evolution of this lineage [72].
Early comparative analyses of two representative genomes from subclades 1 and 2 vs. B. subtilis revealed an under-representation of genes for the degradation of carbohydrate polymers, an abundance of genes encoding proteolytic enzymes, peptide and amino-acid transporters, and a variety of amino-acid degradation pathways [73,74]. Thus, they and others [75] supported the view that the common ancestor of subclades 1 and 2 inhabited the intestine of insects as an opportunistic pathogen, instead of being a benign soil bacterium.
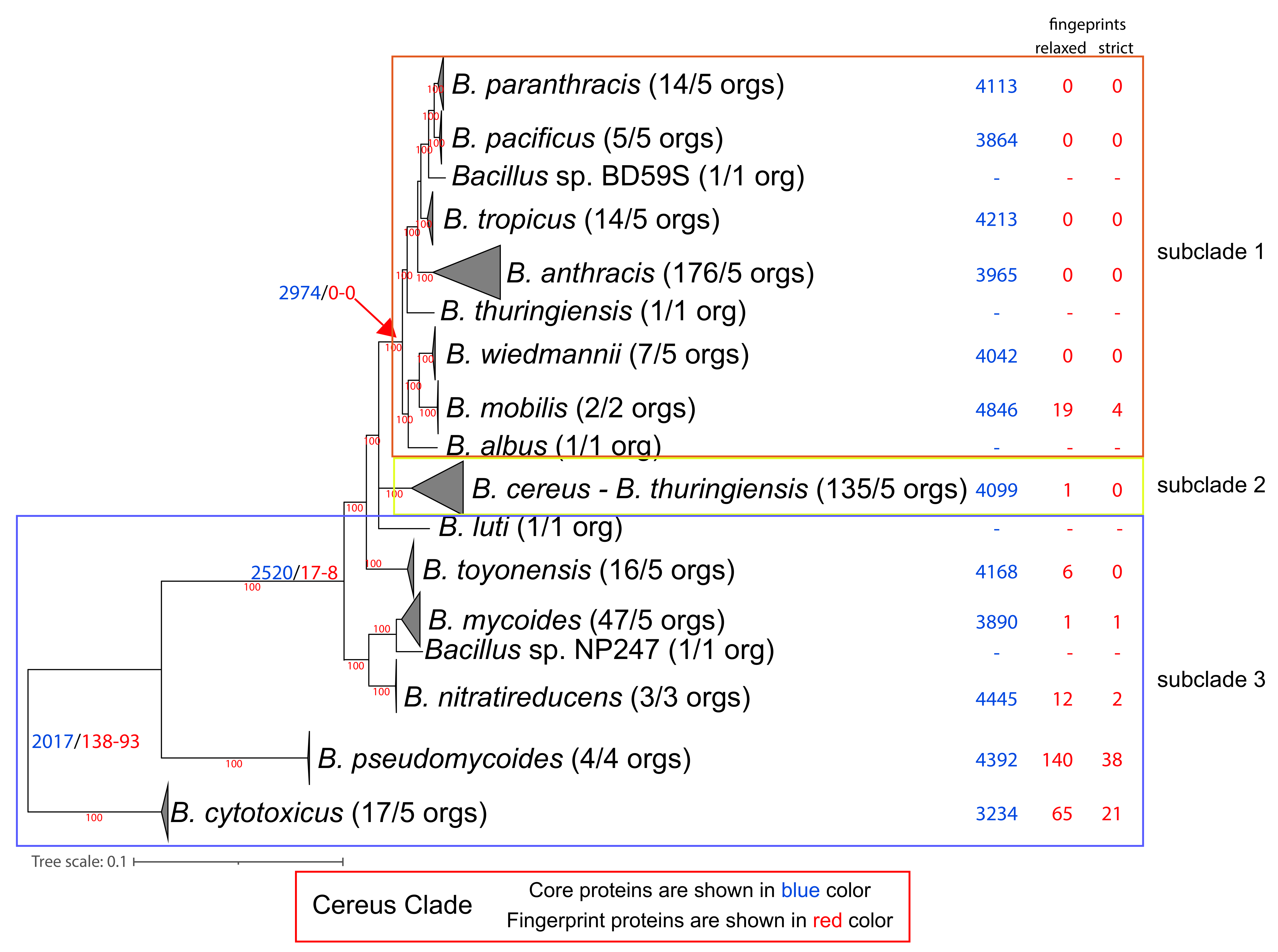
Based on our phylogenomic analyses (see Figure 3 and Supplementary Figure S3), we partitioned the Clade into 3 major evolutionary groups or else subclades in accordance with previous evolutionary analyses [27,28,29,30,60,61,62,63,64,65]. Subclade 1 includes 9 species, such as B. anthracis; moreover, several B. cereus and B. thuringiensis strains are also within this subclade. Of note, the B. anthracis species (based on the phylogenomic tree and ANI values) includes the clonal lineage as well as several B. anthracis Biovars and several strains annotated as B. cereus and B. thuringiensis. Subclade 2 is organized as a single species (based on the phylogenomic tree and the ANI values) and includes most B. cereus (with the reference strain) and B. thuringiensis strains. Subclades 1 and 2 are two monophyletic sister groups, whereas subclade 3 is basal and paraphyletic, consisting of seven species. Of note, a few strains annotated as B. cereus and B. thuringiensis are also found within subclade 3.
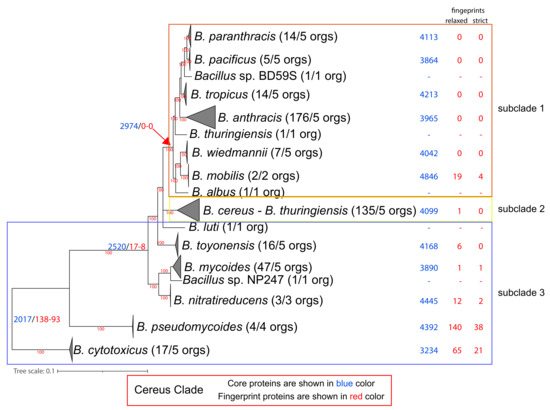
Figure 3.
Phylogenomic ML tree (QTree2-Q.Plant + I + F + G4-aLRT) of the Cereus Clade, based on 812 core protein-orthologous groups from 445 proteomes. For ease of visualization, certain evolutionary clusters have been collapsed. The full tree is available as Supplementary Figure S3. Next to the species name, in parentheses, is the number of complete genomes that are available and, on their right, is the number of genomes used in the normalized dataset. Further to the right of the species names and at the common ancestor of a lineage, we denote with blue and red colors the number of core and (relaxed/strict) fingerprint proteins for each lineage (based on the normalized dataset).
Our analysis identified 2017 normalized core proteins for the entire Cereus Clade. We also identified 138/93 (relaxed/strict) fingerprints for the Cereus Clade (as an entire lineage), meaning that these fingerprints are found only within the Cereus clade and in no other Bacillus proteome that we analyzed. Notably, the entire Subtilis Clade has 1286 normalized core proteins and only 8/5 (relaxed/strict) fingerprints. This is a strong indication that the common ancestor of the Cereus Clade underwent an extensive series of genomic adaptations in terms of gene content that were most probably shaped by its lifestyle. Alternatively, the common ancestor of the Subtilis Clade could have undergone extensive gene losses. However, the chromosome size as well as the number of chromosomally encoded proteins in the other Bacillus species (excluding the Subtilis and Cereus Clades) are very similar to those of the species in the Subtilis Clade (no statistically significant difference) and significantly smaller than the species of the Cereus Clade (t-test p-value < 0.05). This is a strong indication that the ancestor of the Cereus Clade underwent extensive genomic expansion, rather than the Subtilis Clade experiencing a major loss of genes.
The vast majority of Cereus Clade fingerprints (102/86) are of unknown function. Nevertheless, the other three most numerous functional categories are amino acid transport and metabolism (8/1), cell wall/membrane/envelope biogenesis (5/3), and transcription (4/0).
3.4. The Core Proteome and the General Genomic Characteristics of the Genus and Its Species
The general genomic characteristics, the core proteome and the fingerprints of the various lineages, are summarized in Table 1. In addition, the specific normalized core and fingerprint proteins of each lineage and their annotations are available in Supplementary Excel File S1, spreadsheets 2, 3, 4, and 5.

Table 1.
The genomic characteristics of the evolutionary lineages and species of the Subtilis and Cereus Clades. The number of core proteins and fingerprints for each clade/group/species are based on the normalized dataset. As normalized, we denote a dataset where the maximum number of genomes used per species is five (or less, if not available). Thus, sampling biases for certain lineages with very high numbers of available genomes are mitigated and the results between different lineages become comparable. The stars indicate species with only one or two available complete genomes.
We calculated, using the hypergeometric test, the statistical significance of the fold enrichment/depletion of certain functional categories in the core proteomes of 31 species from the Subtilis and Cereus Clades (see Supplementary Excel File S1, spreadsheet 3). Consistently, in all 31 species, the category of unknown function is significantly depleted, as might be expected. Although their absolute numbers are substantial, the proportion of proteins of unknown function is low. This contrasts with the situation in eukaryotes, where 75% of the proteins of unknown function encoded by the genome of the fission yeast, Schizosaccharomyces pombe, are conserved in other fungi and fully 23% are also found in humans [76]. Furthermore, the functional categories of nucleotide transport and metabolism (F); translation, ribosomal structure, and biogenesis (J); energy production and conversion (C); inorganic ion transport and metabolism (P); amino acid transport and metabolism (E); and coenzyme transport and metabolism (H) are consistently enriched in the vast majority (28–30/31) of the species. A recent pangenome analysis focused on 238 strains (ranging between 20–58 per species) of 5 Bacillus species and identified their core genes (indicated in the parentheses), namely those of B. amyloliquefaciens (2870), B. subtilis (1022), B. anthracis (3972), B. cereus (1656) and B. thuringiensis (2299) [77]. For every species, the core gene-set was consistently enriched for functions related to energy production and conversion (C); amino acid transport and metabolism (E); coenzyme transport and metabolism (H); and inorganic ion transport and metabolism (P). Thus, independent studies that utilize different approaches and datasets consistently observe enrichment of the same functional categories.
We also observed that, at the species level, the 31 species of the Subtilis and Cereus Clades had a total of 497 and 162 relaxed and strict fingerprints. Barring three outlier species with very high numbers of fingerprints (B. sonoresnsis, B. pseudomycoides, and B. cytotoxicus), the average number of relaxed and strict fingerprints for the other species were 7 and 2, respectively. The vast majority of relaxed fingerprints (411/497—83%) were of unknown function, whereas the second and third largest functional categories were cell wall/membrane/envelope biogenesis (M: 16/497—3%) and amino acid transport and metabolism (E: 12/497—2%). Notably, 158/162 (98%) of the species-level strict fingerprints were of unknown function.
We compared several genomic characteristics among the species belonging to the Cereus and Subtilis Clades (see Table 1). Interestingly, we observed that (i) the length of the chromosome was on average 28% higher (5.3 vs. 4.13 Mbp; t-test p-value < 0.05) in the species of the Cereus Clade; (ii) the number of chromosomally encoded proteins was on average 28% higher (5124 vs. 3989; t-test p-value < 0.05) in the species of the Cereus Clade; (iii) the number of core proteins was on average 19% higher (4106 vs. 3450; t-test p-value < 0.05) in the species of the Cereus Clade; and (iv) the number of accessory proteins was on average 89% higher (1018 vs. 539; t-test p-value < 0.05) in the species of the Cereus Clade. However, when we tested for differences at the level of relaxed and strict fingerprint proteins, no statistically significant difference was observed. Nevertheless, these findings are compatible with our previous findings (see Section 3.3.) that the Cereus Clade has been through an expansion of its genome.
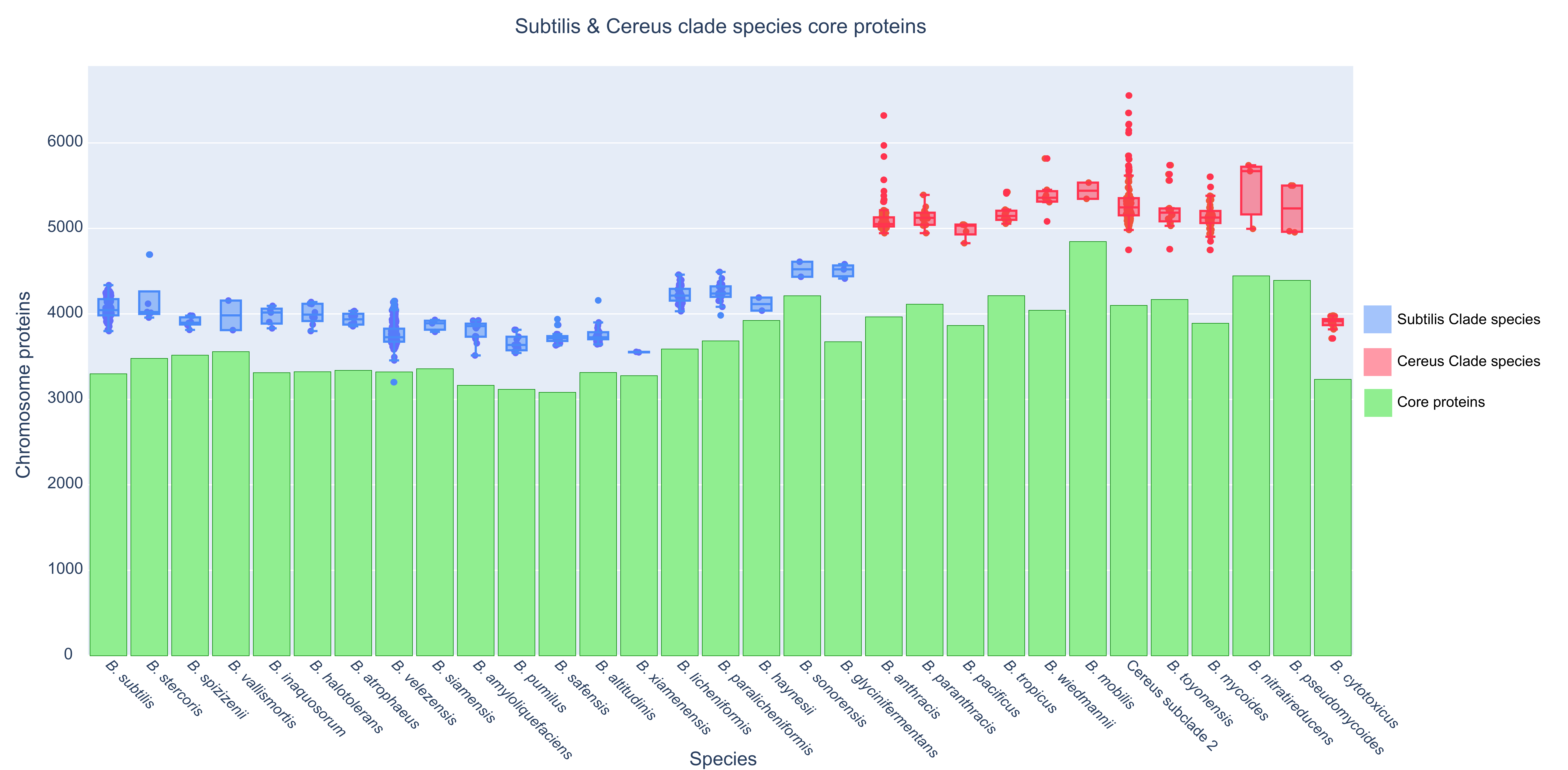
For every species of the Subtilis and Cereus Clades, we also plotted the total number of chromosomally encoded proteins per strain (for all available strains with complete genomes) together with the normalized core proteome of that species (see Figure 4). Thus, the extent of variability of the accessory proteome for every species is visualized. Notably, the B. anthracis species (including the Anthracis clonal lineage and several other strains—see Supplementary Figure S2), the Cereus subclade 2 species, and the B. mycoides species demonstrate an elevated variability in terms of chromosomally encoded accessory proteins.
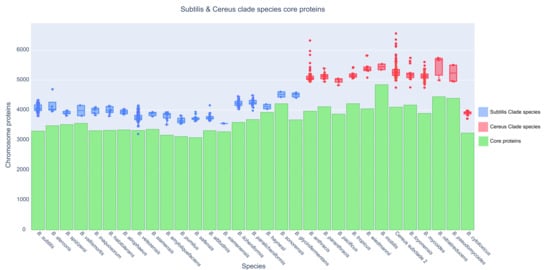
Figure 4.
Boxplot of the total number of proteins (y-axis) for every available strain (dot—each genome) of a species (x-axis) and its normalized core proteome (green bar).
3.5. The Phylogenetic Distribution Profile of the Core and Accessory Protein Components of the Subtilis and Cereus Clades
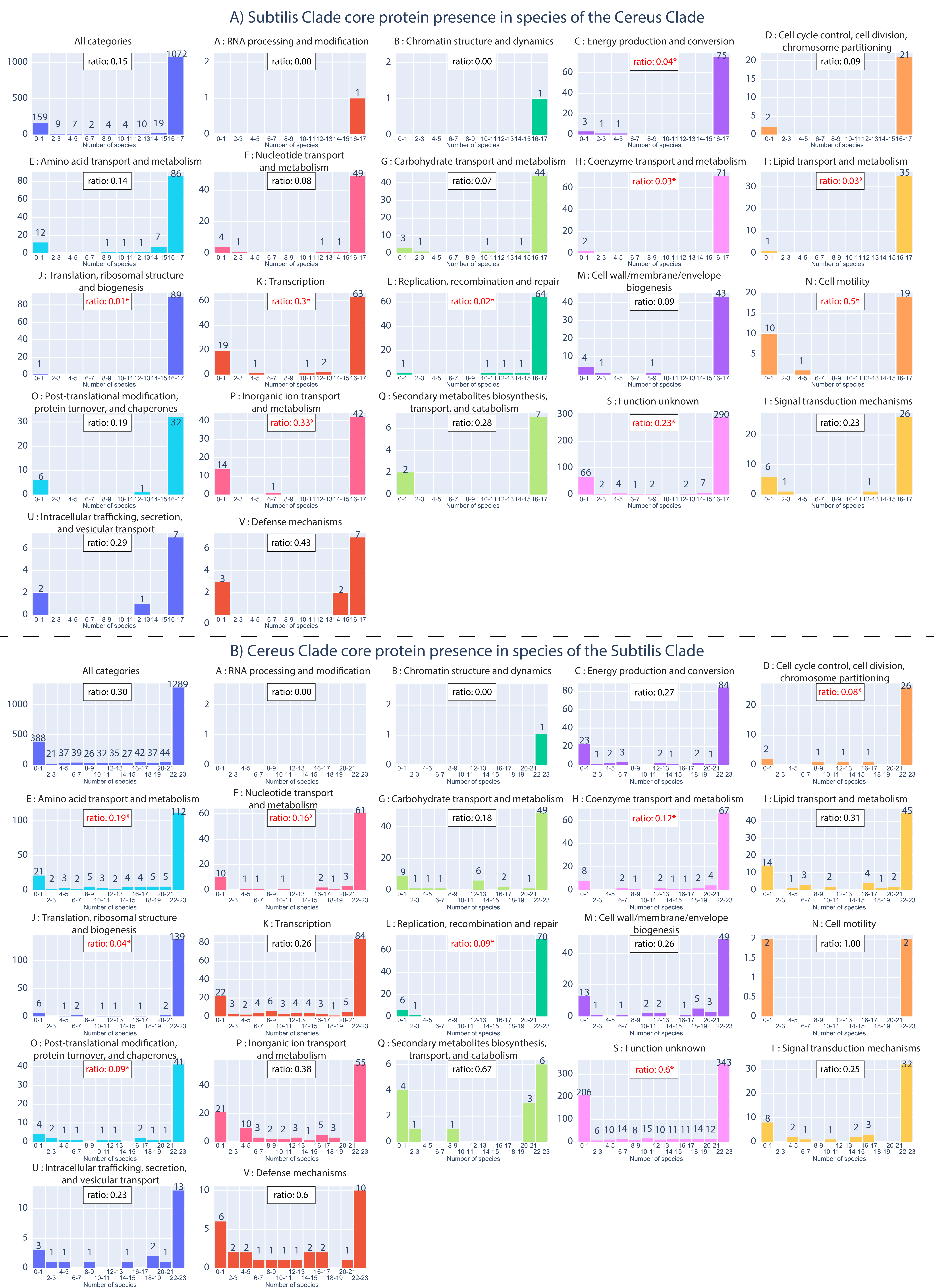
We investigated what proportion of core proteins of the Subtilis Clade (as an entire lineage) were also present (and how often) in the various species of the Cereus Clade and vice versa (see Supplementary Excel File S1, spreadsheets 6 and 7). In this way, it is possible to understand the similarities of the core genomic components of these two Clades, and determine where they diverge from each other. The results are summarized in Figure 5, for all the core proteins and for each functional category of the core proteins, separately. Overall, the vast majority (1072/1286–83%) of the Subtilis Clade core proteins have a presence in most species (16 or 17 species) of the Cereus Clade; the second largest bin consists of 159 (12%) Subtilis Clade core proteins that have a very low presence (in 0 or 1 species) in the Cereus Clade. This unbalanced bimodal distribution or even unimodal distribution in favor of presence in most species is observed for all individual functional categories as well. Next, we calculated the ratio of the low-presence bin (0–1 species) to the high presence bin (16–17 species) for the entire core proteins (background) and for each functional category separately. In addition, we performed a hypergeometric test to identify any categories that have a significantly different ratio of low-presence core proteins compared to the background (all core proteins). The highest and statistically significant ratio was observed for proteins of unknown function. However, it is noteworthy that Subtilis Clade core proteins belonging to functional categories, such as secondary metabolism, signal transduction, intracellular trafficking/secretion, and defense mechanisms, also have a very low presence in the species of the Cereus Clade, though this is not statistically significant. Therefore, these categories of core proteins may be involved in fundamental adaptations that differentiate the Subtilis Clade from the Cereus Clade.
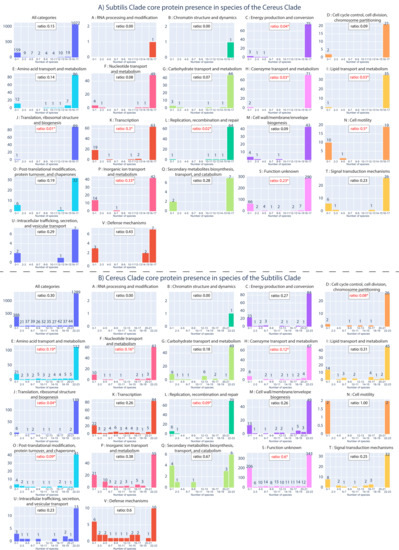
Figure 5.
(A) The phylogenetic distribution of core proteins of the Subtilis Clade in the species of the Cereus Clade. (B) The phylogenetic distribution of core proteins of the Cereus Clade in the species of the Subtilis Clade. The bins on the x-axis correspond to the number of species (in the other Clade), while the y-axis corresponds to the absolute number of core proteins (for that bin). For example, the first graph of Figure 5A shows that 1072 of the core proteins of the Subtilis Clade are also present in 16–17 species of the Cereus Clade. The ratio of the low-presence to high presence bin is shown in the box at the top of the graph. Stars identify any ratio whose difference from the background (in all categories) is statistically significant (based on the hypergeometric test; p-value < 0.05).
Analysis of the equivalent phylogenetic distribution profiles of the Cereus Clade core proteins also demonstrated a bimodal-like pattern; the majority of them (1289/2017–64%) have a high presence (22–23) in most species of the Subtilis Clade, whereas 388 (19%) of them have a very low presence (0–1) in the species of the Subtilis Clade. Interestingly, the ratio (0.3) of low/high presence is significantly higher in the Cereus Clade, compared to the equivalent ratio (0.15) in the Subtilis Clade. This is another clear indication that the Cereus Clade is much more specialized in terms of the proteins it encodes, compared to the Subtilis Clade. For the individual functional categories, the highest and statistically significant ratio (0.6) was once again observed for unknown function. Cereus Clade core proteins that belong to functional categories, such as secondary metabolism and defense mechanisms, also have a very low presence in the species of the Subtilis Clade, though this is not statistically significant. Therefore, these core proteins may be involved in fundamental adaptations that differentiate the Cereus Clade from the Subtilis Clade.
We performed a similar analysis for the accessory proteins (i.e., not members of the core set) of each species (we used the reference strain) from one of the two Clades, that have a very low presence (in 0–1 species) in the other Clade. We observed that accessory proteins from species of the Subtilis Clade that had a very low presence in the Cereus Clade were consistently enriched for the category of unknown function (see Supplementary Excel File S1, spreadsheet 8). The same (consistent enrichment of unknown function) applied for accessory proteins from species of the Cereus Clade that had a very low presence in the Subtilis Clade.
3.6. The Phylogenetic Distribution Profile of Sporulation and Essential Proteins of the Model Organism Bacillus Subtilis
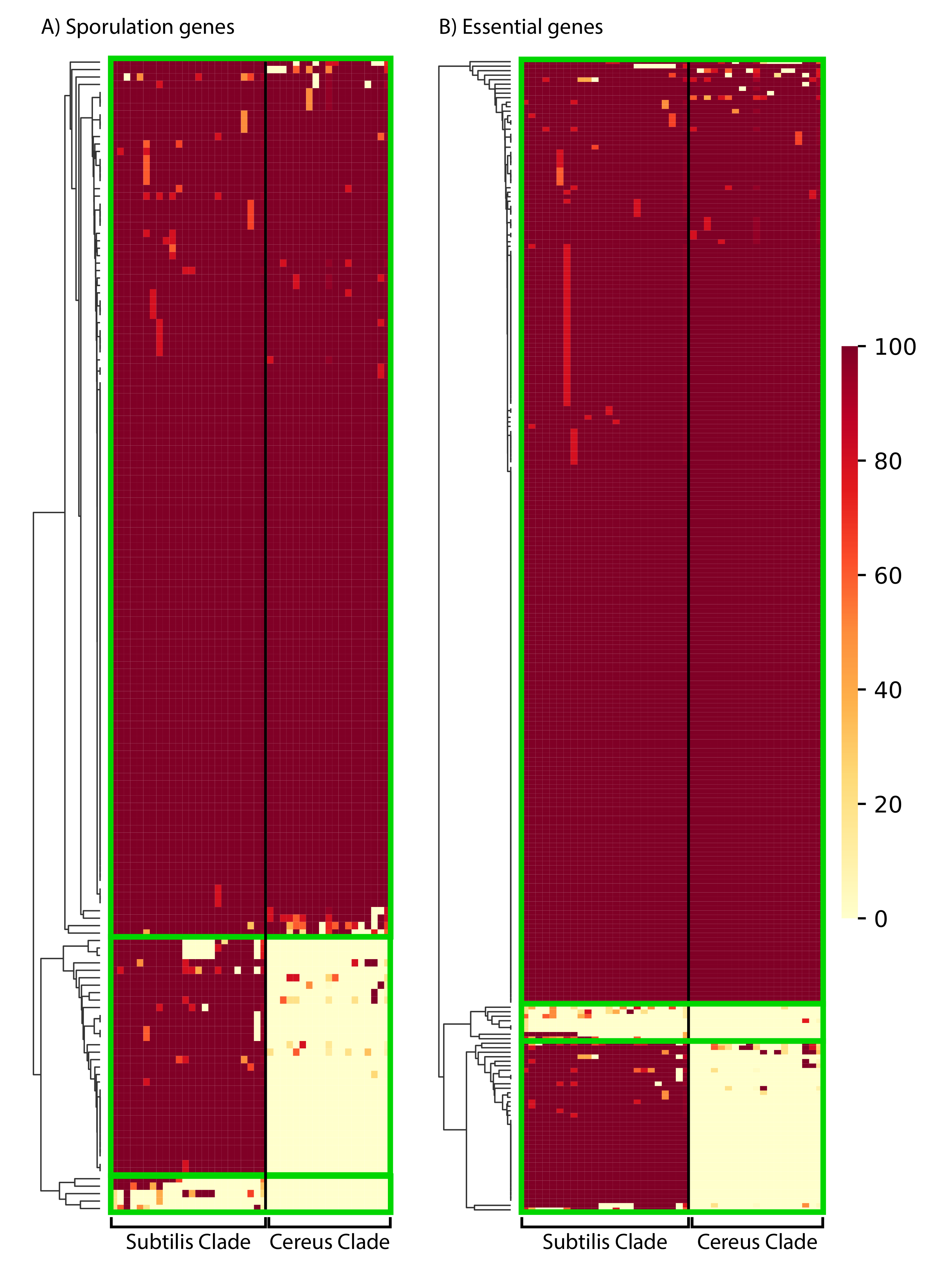
A major characteristic of Bacilli is their ability to form very resistant spores under harsh conditions [10,78,79]. Accordingly, B. subtilis has been widely used as a model organism for understanding bacterial developmental biology [80,81,82,83,84,85]. Although a large number of genes are involved in sporulation [80,81,82,86], a study exploiting a transposon mutagenesis screen identified 155 protein-coding genes whose disruption showed sporulation defects [87]. Thus, we investigated the phylogenetic distribution profile (presence of a homolog with 50% aa identity over 50% of the protein’s length) of each of these 155 important proteins in the various species of the Subtilis and Cereus Clades (see Figure 6, Supplementary Figure S4, and Supplementary Excel File S1, spreadsheet 9 for details—gene names and distribution patterns). The vast majority (118/155–76%) of these important sporulation proteins have a close homolog in the majority of species, in both the Subtilis and the Cereus Clades. This is a clear indication that most of the crucial genetic components of sporulation are highly conserved across the entire genus. However, we also identified a significant number of key sporulation proteins with very limited phylogenetic distribution, or even absence, from a given lineage. For example, 33 of the 155 sporulation proteins (21%) have a close homolog in the majority of species within the Subtilis Clade, but not in the majority of species within the Cereus Clade. Such sporulation proteins may either be missing from the species of the Cereus Clade, or they have undergone rapid divergence and did not pass our identity criteria. Furthermore, an even smaller number (4/155–3%) of these important sporulation proteins have a very limited presence, even within the Subtilis Clade. Most probably, these proteins are missing from the other species of the Subtilis and Cereus Clades, because there would not have been sufficient evolutionary time to allow their divergence in close relatives to a level below the threshold (50% identity).
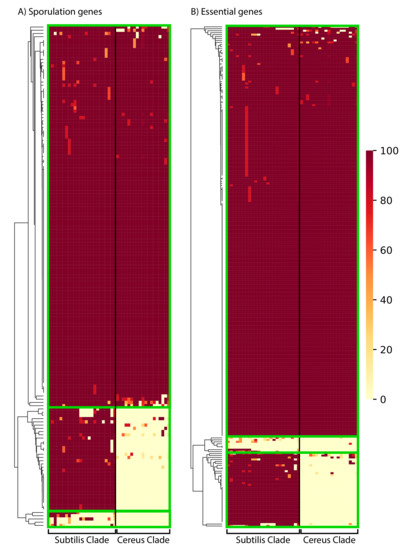
Figure 6.
The phylogenetic distribution pattern of: (A) 155 B. subtilis proteins important for sporulation; (B) 256 proteins that are essential in B. subtilis. Presence of a close homolog in a given species of the Subtilis and Cereus Clades was determined based on 50% amino acid identity over 50% of the protein’s length. The clustering of proteins (based on their distribution) was performed with the average Euclidean distance, within the seaborn.clustermap python package. A more detailed view of the cluster-heatmaps (including the individual gene names and species) is available in Supplementary Figures S4 and S5. Each row corresponds to a gene and each column corresponds to a certain species. The color in the heatmap corresponds to the % presence (how many strains of the species) of that gene in that certain species.
A very similar phylogenetic distribution pattern was observed for the 256 proteins of B. subtilis 168 model strain that have been experimentally determined to be essential (see Figure 6, Supplementary Figure S5, and Supplementary Excel File S1, spreadsheet 10 for details—gene names, and distribution patterns). This list of essential genes was based on the data in Subtiwiki [86]. Again, the majority of these proteins (210/256–82%) have a wide distribution (presence of a homolog with 50% aa identity over 50% of the protein’s length) in the majority of species of both the Subtilis and Cereus Clades. A rather limited number (39/256–15%) have no close homologs in most species of the Cereus Clade. Such B. subtilis essential proteins may either be missing from the species of the Cereus Clade, or they have undergone rapid divergence and did not pass our identity criteria. Furthermore, a very small number (7/256–3%) have a very limited distribution even within the Subtilis Clade. Most probably, these proteins are missing from the other species of the Subtilis and Cereus Clade because they would not have sufficient evolutionary time to allow their divergence in close relatives to a level below the threshold (50% identity).
4. Conclusions
All of the analyses in our study clearly and consistently demonstrate that the Cereus Clade is considerably more complex and diverse, in terms of its content of chromosomally encoded proteins, compared to the Subtilis Clade. A very significant proportion of proteins that distinguish the Cereus Clade are still of an unknown function, whereas other functional categories are related to secondary metabolism/transport/catabolism and defense mechanisms. B. subtilis is the well-established model organism for Bacilli and even for Gram-positive bacteria. However, several of the important components of the human/animal pathogens of the Cereus Clade are not present in B. subtilis. Therefore, this study demonstrates the strengths and the limitations of B. subtilis as a model organism for certain functions, including pathogenesis. A remarkable observation was that many species in both the Subtilis and the Cereus Clades have very low numbers of fingerprint proteins, with a few notable exceptions. Thus, it emerges that many of these species are much more homogeneous in terms of core protein content than was originally thought and that the species concept is much more relaxed; it is probably based on phenotypic characteristics whose molecular background is very unstable/dynamic. It is also plausible that many species adaptations could be related to gene-regulation, which would not be detected by our approach. Our observations concerning the phylogeny and fingerprints of the various species within the Cereus Clade suggest that subclades 1 and 2, together with several other species from subclade 3, should form one rather homogeneous monophyletic group. In contrast, both B. pseudomycoides and B. cytotoxicus are so divergent in terms of phylogenomics and fingerprints, that they should form two distinct monophyletic groups within the Clade. Finally, the identification of lineage-specific fingerprints may allow for the future development of highly specific vaccines, therapeutic molecules, or rapid and low-cost molecular tests for species identification.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10091720/s1. Figure S1: Saturation curves of soft core proteins at 85%, 90% and 95% depending on the number of genomes sampled; Figure S2: Subtilis clade tree based on 457 core proteins, IQ-Tree2; Figure S3: Cereus Clade tree based on 812 core proteins, IQ-Tree2; Figure S4: B. subtilis sporulation gene homologues; Figure S5: B. subtilis essential gene homologues. Supplementary Excel File S1: Information on Core and Fingerprint Genes for the various Bacillus lineages.
Author Contributions
Conceptualization, G.D.A. and S.G.O.; methodology, M.N., A.H., D.M., I.I., S.G.O. and G.D.A.; formal analysis, M.N. and G.D.A.; writing—original draft preparation, M.N., A.H., D.M., I.I., S.G.O. and G.D.A.; supervision, G.D.A. All authors have read and agreed to the published version of the manuscript.
Funding
M.N. thanks the University of Thessaly (Ph.D. studentship: DEKA-UTH-259) for financial support.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Valerie Wood, Cambridge University, UK, for helpful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fritze, D. Taxonomy of the Genus Bacillus and Related Genera: The Aerobic Endospore-Forming Bacteria. Phytopathology 2004, 94, 1245–1248. [Google Scholar] [CrossRef]
- Harwood, C.R. Introduction to the Biotechnology of Bacillus. In Bacillus; Harwood, C.R., Ed.; Springer US: Boston, MA, USA, 1989; pp. 1–4. ISBN 978-1-4899-3504-5. [Google Scholar]
- Hernández-González, I.L.; Moreno-Hagelsieb, G.; Olmedo-Álvarez, G. Environmentally-Driven Gene Content Convergence and the Bacillus Phylogeny. BMC Evol. Biol. 2018, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Gupta, R.S. A Phylogenomic and Comparative Genomic Framework for Resolving the Polyphyly of the Genus Bacillus: Proposal for Six New Genera of Bacillus Species, Peribacillus Gen. Nov., Cytobacillus Gen. Nov., Mesobacillus Gen. Nov., Neobacillus Gen. Nov., Metabacillus Gen. Nov. and Alkalihalobacillus Gen. Nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar] [CrossRef] [PubMed]
- Ehling-Schulz, M.; Koehler, T.M.; Lereclus, D. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Penha, R.O.; Vandenberghe, L.P.S.; Faulds, C.; Soccol, V.T.; Soccol, C.R. Bacillus lipopeptides as Powerful Pest Control Agents for a More Sustainable and Healthy Agriculture: Recent Studies and Innovations. Planta 2020, 251, 70. [Google Scholar] [CrossRef]
- Sumi, C.D.; Yang, B.W.; Yeo, I.-C.; Hahm, Y.T. Antimicrobial Peptides of the Genus Bacillus: A New Era for Antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef]
- Mingmongkolchai, S.; Panbangred, W. Bacillus Probiotics: An Alternative to Antibiotics for Livestock Production. J. Appl. Microbiol. 2018, 124, 1334–1346. [Google Scholar] [CrossRef]
- Barbe, V.; Cruveiller, S.; Kunst, F.; Lenoble, P.; Meurice, G.; Sekowska, A.; Vallenet, D.; Wang, T.; Moszer, I.; Médigue, C.; et al. From a Consortium Sequence to a Unified Sequence: The Bacillus subtilis 168 Reference Genome a Decade Later. Microbiology 2009, 155, 1758–1775. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, L.; Li, J.; Du, G.; Chen, J. Synthetic Biology Toolbox and Chassis Development in Bacillus subtilis. Trends Biotechnol. 2019, 37, 548–562. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of Prokaryotic Names with Standing in Nomenclature (Bacterio.Net), 20 Years On. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Logan, N.A.; Berge, O.; Bishop, A.H.; Busse, H.-J.; De Vos, P.; Fritze, D.; Heyndrickx, M.; Kämpfer, P.; Rabinovitch, L.; Salkinoja-Salonen, M.S.; et al. Proposed Minimal Standards for Describing New Taxa of Aerobic, Endospore-Forming Bacteria. Int. J. Syst. Evol. Microbiol. 2009, 59, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Côté, J.-C. Phylogenetic Relationships between Bacillus Species and Related Genera Inferred from Comparison of 3’ End 16S RDNA and 5’ End 16S-23S ITS Nucleotide Sequences. Int. J. Syst. Evol. Microbiol. 2003, 53, 695–704. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Nikolaidis, M.; Hesketh, A. The Notable Achievements and the Prospects of Bacterial Pathogen Genomics. Microorganisms 2022, 10, 1040. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A Phylogeny-Driven Genomic Encyclopaedia of Bacteria and Archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef]
- Kyrpides, N.C.; Hugenholtz, P.; Eisen, J.A.; Woyke, T.; Göker, M.; Parker, C.T.; Amann, R.; Beck, B.J.; Chain, P.S.G.; Chun, J.; et al. Genomic Encyclopedia of Bacteria and Archaea: Sequencing a Myriad of Type Strains. PLoS Biol. 2014, 12, e1001920. [Google Scholar] [CrossRef]
- Whitman, W.B.; Woyke, T.; Klenk, H.-P.; Zhou, Y.; Lilburn, T.G.; Beck, B.J.; De Vos, P.; Vandamme, P.; Eisen, J.A.; Garrity, G.; et al. Genomic Encyclopedia of Bacterial and Archaeal Type Strains, Phase III: The Genomes of Soil and Plant-Associated and Newly Described Type Strains. Stand. Genom. Sci. 2015, 10, 26. [Google Scholar] [CrossRef]
- Kunin, V.; Goldovsky, L.; Darzentas, N.; Ouzounis, C.A. The Net of Life: Reconstructing the Microbial Phylogenetic Network. Genome Res. 2005, 15, 954–959. [Google Scholar] [CrossRef]
- Kunin, V.; Ouzounis, C.A. The Balance of Driving Forces during Genome Evolution in Prokaryotes. Genome Res. 2003, 13, 1589–1594. [Google Scholar] [CrossRef]
- Gogarten, J.P.; Townsend, J.P. Horizontal Gene Transfer, Genome Innovation and Evolution. Nat. Rev. Microbiol. 2005, 3, 679–687. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus Agalactiae: Implications for the Microbial “Pan-Genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Vernikos, G.; Medini, D.; Riley, D.R.; Tettelin, H. Ten Years of Pan-Genome Analyses. Curr. Opin. Microbiol. 2015, 23, 148–154. [Google Scholar] [CrossRef]
- Jun, S.-R.; Wassenaar, T.M.; Nookaew, I.; Hauser, L.; Wanchai, V.; Land, M.; Timm, C.M.; Lu, T.-Y.S.; Schadt, C.W.; Doktycz, M.J.; et al. Diversity of Pseudomonas Genomes, Including Populus-Associated Isolates, as Revealed by Comparative Genome Analysis. Appl. Environ. Microbiol. 2016, 82, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic Insights That Advance the Species Definition for Prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Zwick, M.E.; Joseph, S.J.; Didelot, X.; Chen, P.E.; Bishop-Lilly, K.A.; Stewart, A.C.; Willner, K.; Nolan, N.; Lentz, S.; Thomason, M.K.; et al. Genomic Characterization of the Bacillus cereus Sensu Lato Species: Backdrop to the Evolution of Bacillus anthracis. Genome Res. 2012, 22, 1512–1524. [Google Scholar] [CrossRef]
- Bazinet, A.L. Pan-Genome and Phylogeny of Bacillus Cereus Sensu Lato. BMC Evol. Biol. 2017, 17, 176. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, J.; Lai, Q.; Zeng, R.; Ye, D.; Xu, J.; Shao, Z. Proposal of Nine Novel Species of the Bacillus Cereus Group. Int. J. Syst. Evol. Microbiol. 2017, 67, 2499–2508. [Google Scholar] [CrossRef]
- Chang, T.; Rosch, J.W.; Gu, Z.; Hakim, H.; Hewitt, C.; Gaur, A.; Wu, G.; Hayden, R.T. Whole-Genome Characterization of Bacillus cereus Associated with Specific Disease Manifestations. Infect. Immun. 2018, 86, e00574-17. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Blom, J.; Klenk, H.-P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. Amyloliquefaciens” within the B. Subtilis Species Complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef]
- Tirumalai, M.R.; Stepanov, V.G.; Wünsche, A.; Montazari, S.; Gonzalez, R.O.; Venkateswaran, K.; Fox, G.E. Bacillus safensis FO-36b and Bacillus pumilus SAFR-032: A Whole Genome Comparison of Two Spacecraft Assembly Facility Isolates. BMC Microbiol. 2018, 18, 57. [Google Scholar] [CrossRef]
- Du, Y.; Ma, J.; Yin, Z.; Liu, K.; Yao, G.; Xu, W.; Fan, L.; Du, B.; Ding, Y.; Wang, C. Comparative Genomic Analysis of Bacillus paralicheniformis MDJK30 with Its Closely Related Species Reveals an Evolutionary Relationship between B. paralicheniformis and B. licheniformis. BMC Genom. 2019, 20, 283. [Google Scholar] [CrossRef]
- Wu, H.; Wang, D.; Gao, F. Toward a High-Quality Pan-Genome Landscape of Bacillus subtilis by Removal of Confounding Strains. Brief. Bioinform. 2021, 22, 1951–1971. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Bowman, M.J.; Zeigler, D.R. Promotion of Bacillus Subtilis Subsp. Inaquosorum, Bacillus subtilis Subsp. spizizenii and Bacillus subtilis Subsp. stercoris to Species Status. Antonie Van Leeuwenhoek 2020, 113, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.J.A.; Tasara, T.; Klumpp, J.; Stephan, R.; Ehling-Schulz, M.; Johler, S. Whole-Genome-Based Phylogeny of Bacillus cytotoxicus Reveals Different Clades within the Species and Provides Clues on Ecology and Evolution. Sci. Rep. 2019, 9, 1984. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust Demarcation of 17 Distinct Bacillus Species Clades, Proposed as Novel Bacillaceae Genera, by Phylogenomics and Comparative Genomic Analyses: Description of Robertmurraya kyonggiensis sp. Nov. and Proposal for an Emended Genus Bacillus Limiting It Only to the Members of the Subtilis and Cereus Clades of Species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef]
- Bhandari, V.; Ahmod, N.Z.; Shah, H.N.; Gupta, R.S. Molecular Signatures for Bacillus Species: Demarcation of the Bacillus subtilis and Bacillus cereus Clades in Molecular Terms and Proposal to Limit the Placement of New Species into the Genus Bacillus. Int. J. Syst. Evol. Microbiol. 2013, 63, 2712–2726. [Google Scholar] [CrossRef]
- Parker, C.T.; Tindall, B.J.; Garrity, G. International Code of Nomenclature of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2019, 69, S1–S111. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Mossialos, D.; Oliver, S.G.; Amoutzias, G.D. Comparative Analysis of the Core Proteomes among the Pseudomonas Major Evolutionary Groups Reveals Species-Specific Adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis. Diversity 2020, 12, 289. [Google Scholar] [CrossRef]
- Bentley, S.D.; Vernikos, G.S.; Snyder, L.A.S.; Churcher, C.; Arrowsmith, C.; Chillingworth, T.; Cronin, A.; Davis, P.H.; Holroyd, N.E.; Jagels, K.; et al. Meningococcal Genetic Variation Mechanisms Viewed through Comparative Analysis of Serogroup C Strain FAM18. PLoS Genet. 2007, 3, e23. [Google Scholar] [CrossRef]
- Hiller, N.L.; Janto, B.; Hogg, J.S.; Boissy, R.; Yu, S.; Powell, E.; Keefe, R.; Ehrlich, N.E.; Shen, K.; Hayes, J.; et al. Comparative Genomic Analyses of Seventeen Streptococcus pneumoniae Strains: Insights into the Pneumococcal Supragenome. J. Bacteriol. 2007, 189, 8186–8195. [Google Scholar] [CrossRef] [PubMed]
- Méric, G.; Yahara, K.; Mageiros, L.; Pascoe, B.; Maiden, M.C.J.; Jolley, K.A.; Sheppard, S.K. A Reference Pan-Genome Approach to Comparative Bacterial Genomics: Identification of Novel Epidemiological Markers in Pathogenic Campylobacter. PLoS ONE 2014, 9, e92798. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Rosovitz, M.J.; Myers, G.S.A.; Mongodin, E.F.; Fricke, W.F.; Gajer, P.; Crabtree, J.; Sebaihia, M.; Thomson, N.R.; Chaudhuri, R.; et al. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J. Bacteriol. 2008, 190, 6881–6893. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Chevenet, F.; Brun, C.; Bañuls, A.-L.; Jacq, B.; Christen, R. TreeDyn: Towards Dynamic Graphics and Annotations for Analyses of Trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded Microbial Genome Coverage and Improved Protein Family Annotation in the COG Database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef]
- Federhen, S. Type Material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015, 43, D1086–D1098. [Google Scholar] [CrossRef]
- Anagnostopoulos, C.; Spizizen, J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 1961, 81, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.; Azevedo, V.; Bertero, M.G.; Bessières, P.; Bolotin, A.; Borchert, S.; et al. The Complete Genome Sequence of the Gram-Positive Bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Borriss, R.; Danchin, A.; Harwood, C.R.; Médigue, C.; Rocha, E.P.C.; Sekowska, A.; Vallenet, D. Bacillus subtilis, the Model Gram-Positive Bacterium: 20 Years of Annotation Refinement. Microb. Biotechnol. 2018, 11, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Rooney, A.P.; Price, N.P.J.; Ehrhardt, C.; Swezey, J.L.; Bannan, J.D. Phylogeny and Molecular Taxonomy of the Bacillus subtilis Species Complex and Description of Bacillus subtilis Subsp. inaquosorum Subsp. Nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2429–2436. [Google Scholar] [CrossRef]
- Guinebretière, M.-H.; Auger, S.; Galleron, N.; Contzen, M.; De Sarrau, B.; De Buyser, M.-L.; Lamberet, G.; Fagerlund, A.; Granum, P.E.; Lereclus, D.; et al. Bacillus cytotoxicus Sp. Nov. Is a Novel Thermotolerant Species of the Bacillus cereus Group Occasionally Associated with Food Poisoning. Int. J. Syst. Evol. Microbiol. 2013, 63, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lai, Q.; Göker, M.; Meier-Kolthoff, J.P.; Wang, M.; Sun, Y.; Wang, L.; Shao, Z. Genomic Insights into the Taxonomic Status of the Bacillus cereus Group. Sci. Rep. 2015, 5, 14082. [Google Scholar] [CrossRef]
- Baek, I.; Lee, K.; Goodfellow, M.; Chun, J. Comparative Genomic and Phylogenomic Analyses Clarify Relationships within and Between Bacillus cereus and Bacillus thuringiensis: Proposal for the Recognition of Two Bacillus thuringiensis Genomovars. Front. Microbiol. 2019, 10, 1978. [Google Scholar] [CrossRef]
- Sorokin, A.; Candelon, B.; Guilloux, K.; Galleron, N.; Wackerow-Kouzova, N.; Ehrlich, S.D.; Bourguet, D.; Sanchis, V. Multiple-Locus Sequence Typing Analysis of Bacillus cereus and Bacillus thuringiensis Reveals Separate Clustering and a Distinct Population Structure of Psychrotrophic Strains. Appl. Environ. Microbiol. 2006, 72, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Barker, M.; Falush, D.; Priest, F.G. Evolution of Pathogenicity in the Bacillus cereus Group. Syst. Appl. Microbiol. 2009, 32, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Okinaka, R.T.; Keim, P. The Phylogeny of Bacillus cereus sensu lato. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, R. Some aspects of evolution. Science 1933, 78, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Kolstø, A.-B.; Tourasse, N.J.; Økstad, O.A. What Sets Bacillus anthracis Apart from Other Bacillus Species? Annu. Rev. Microbiol. 2009, 63, 451–476. [Google Scholar] [CrossRef]
- Moayeri, M.; Leppla, S.H.; Vrentas, C.; Pomerantsev, A.P.; Liu, S. Anthrax Pathogenesis. Annu. Rev. Microbiol. 2015, 69, 185–208. [Google Scholar] [CrossRef]
- Helgason, E.; Okstad, O.A.; Caugant, D.A.; Johansen, H.A.; Fouet, A.; Mock, M.; Hegna, I.; Kolstø, A.B. Bacillus Anthracis, Bacillus Cereus, and Bacillus Thuringiensis--One Species on the Basis of Genetic Evidence. Appl. Environ. Microbiol. 2000, 66, 2627–2630. [Google Scholar] [CrossRef]
- Rasko, D.A.; Altherr, M.R.; Han, C.S.; Ravel, J. Genomics of the Bacillus cereus Group of Organisms. FEMS Microbiol. Rev. 2005, 29, 303–329. [Google Scholar] [CrossRef]
- Patiño-Navarrete, R.; Sanchis, V. Evolutionary Processes and Environmental Factors Underlying the Genetic Diversity and Lifestyles of Bacillus cereus Group Bacteria. Res. Microbiol. 2017, 168, 309–318. [Google Scholar] [CrossRef]
- Rasigade, J.-P.; Hollandt, F.; Wirth, T. Genes under Positive Selection in the Core Genome of Pathogenic Bacillus cereus Group Members. Infect. Genet. Evol. 2018, 65, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.; Sorokin, A.; Anderson, I.; Galleron, N.; Candelon, B.; Kapatral, V.; Bhattacharyya, A.; Reznik, G.; Mikhailova, N.; Lapidus, A.; et al. Genome Sequence of Bacillus cereus and Comparative Analysis with Bacillus anthracis. Nature 2003, 423, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Read, T.D.; Peterson, S.N.; Tourasse, N.; Baillie, L.W.; Paulsen, I.T.; Nelson, K.E.; Tettelin, H.; Fouts, D.E.; Eisen, J.A.; Gill, S.R.; et al. The Genome Sequence of Bacillus anthracis Ames and Comparison to Closely Related Bacteria. Nature 2003, 423, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L.; Jorgensen, J.Z.; Dolan, S.; Kolchinsky, R.; Rainey, F.A.; Lo, S.C. The Arthromitus Stage of Bacillus cereus: Intestinal Symbionts of Animals. Proc. Natl. Acad. Sci. USA 1998, 95, 1236–1241. [Google Scholar] [CrossRef]
- Wood, V.; Lock, A.; Harris, M.A.; Rutherford, K.; Bähler, J.; Oliver, S.G. Hidden in Plain Sight: What Remains to Be Discovered in the Eukaryotic Proteome? Open Biol. 2019, 9, 180241. [Google Scholar] [CrossRef]
- Kim, Y.; Koh, I.; Young Lim, M.; Chung, W.-H.; Rho, M. Pan-Genome Analysis of Bacillus for Microbiome Profiling. Sci. Rep. 2017, 7, 10984. [Google Scholar] [CrossRef]
- Link, L.; Sawyer, J.; Venkateswaran, K.; Nicholson, W. Extreme Spore UV Resistance of Bacillus pumilus Isolates Obtained from an Ultraclean Spacecraft Assembly Facility. Microb. Ecol. 2004, 47, 159–163. [Google Scholar] [CrossRef]
- Satomi, M.; La Duc, M.T.; Venkateswaran, K. Bacillus safensis Sp. Nov., Isolated from Spacecraft and Assembly-Facility Surfaces. Int. J. Syst. Evol. Microbiol. 2006, 56, 1735–1740. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Mekhedov, S.L.; Puigbo, P.; Smirnov, S.; Wolf, Y.I.; Rigden, D.J. Genomic Determinants of Sporulation in Bacilli and Clostridia: Towards the Minimal Set of Sporulation-Specific Genes. Environ. Microbiol. 2012, 14, 2870–2890. [Google Scholar] [CrossRef]
- Eijlander, R.T.; de Jong, A.; Krawczyk, A.O.; Holsappel, S.; Kuipers, O.P. SporeWeb: An Interactive Journey through the Complete Sporulation Cycle of Bacillus subtilis. Nucleic Acids Res. 2014, 42, D685–D691. [Google Scholar] [CrossRef][Green Version]
- Michna, R.H.; Zhu, B.; Mäder, U.; Stülke, J. SubtiWiki 2.0—An Integrated Database for the Model Organism Bacillus subtilis. Nucleic Acids Res. 2016, 44, D654–D662. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sella, S.R.B.R.; Vandenberghe, L.P.S.; Soccol, C.R. Life Cycle and Spore Resistance of Spore-Forming Bacillus atrophaeus. Microbiol. Res. 2014, 169, 931–939. [Google Scholar] [CrossRef] [PubMed]
- McKenney, P.T.; Driks, A.; Eichenberger, P. The Bacillus subtilis Endospore: Assembly and Functions of the Multilayered Coat. Nat. Rev. Microbiol. 2013, 11, 33–44. [Google Scholar] [CrossRef]
- Higgins, D.; Dworkin, J. Recent Progress in Bacillus subtilis Sporulation. FEMS Microbiol. Rev. 2012, 36, 131–148. [Google Scholar] [CrossRef]
- Zhu, B.; Stülke, J. SubtiWiki in 2018: From Genes and Proteins to Functional Network Annotation of the Model Organism Bacillus subtilis. Nucleic Acids Res. 2018, 46, D743–D748. [Google Scholar] [CrossRef]
- Meeske, A.J.; Rodrigues, C.D.A.; Brady, J.; Lim, H.C.; Bernhardt, T.G.; Rudner, D.Z. High-Throughput Genetic Screens Identify a Large and Diverse Collection of New Sporulation Genes in Bacillus subtilis. PLoS Biol. 2016, 14, e1002341. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).