The Biological Significance of Pyruvate Sensing and Uptake in Salmonella enterica Serovar Typhimurium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Oligonucleotides
2.2. Growth Conditions
2.3. Luciferase Reporter Assay for the Analysis of btsT and cstA Expression
2.4. External Pyruvate Determination
2.5. Pyruvate Uptake Measurement
2.6. Motility Assay
2.7. Chemotaxis Test
2.8. Stress Assay
2.9. Persister Formation
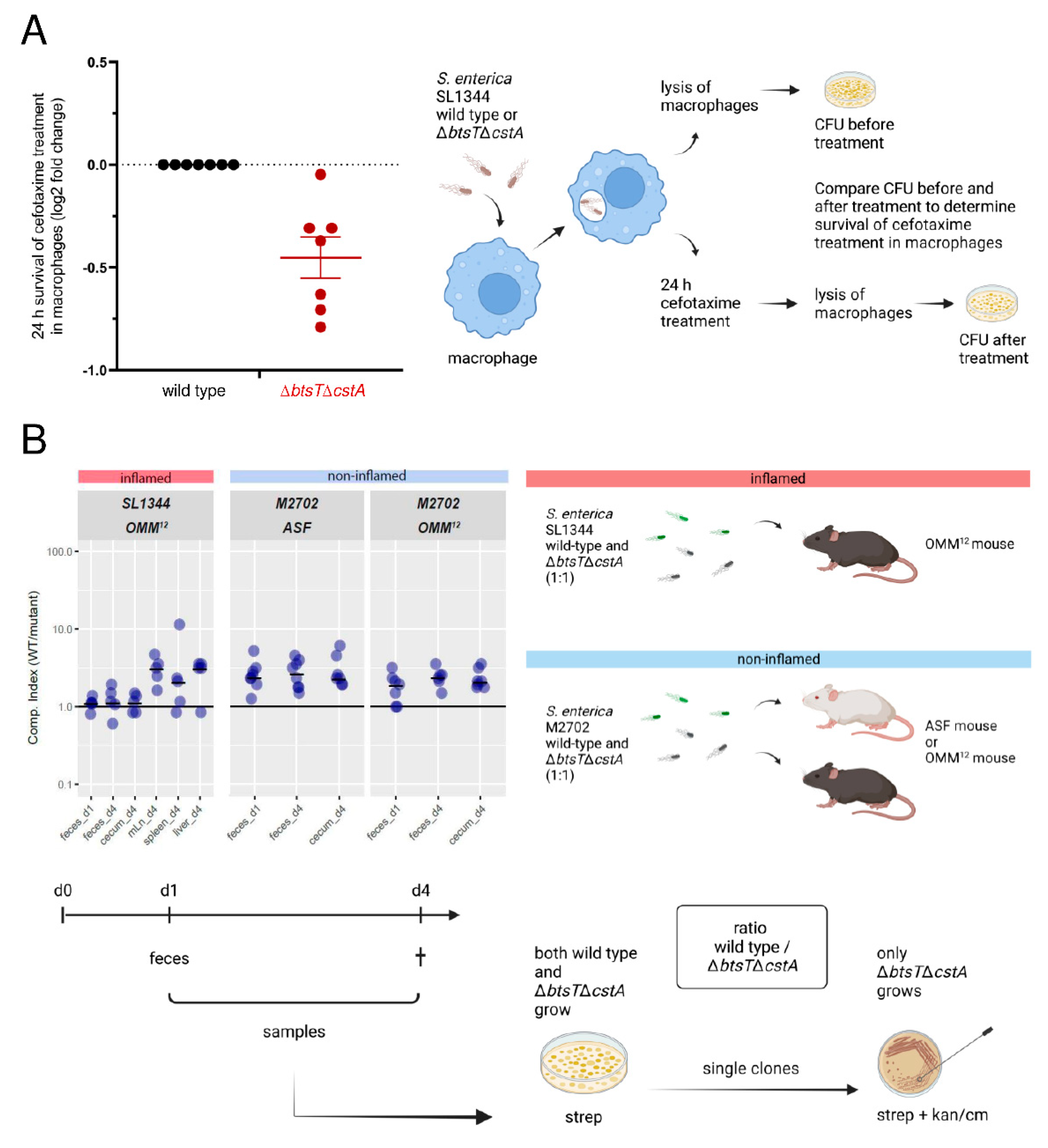
2.10. Intramacrophage Antibiotic Survival Assays
2.11. Infection of Gnotobiotic Mice
3. Results and Discussion
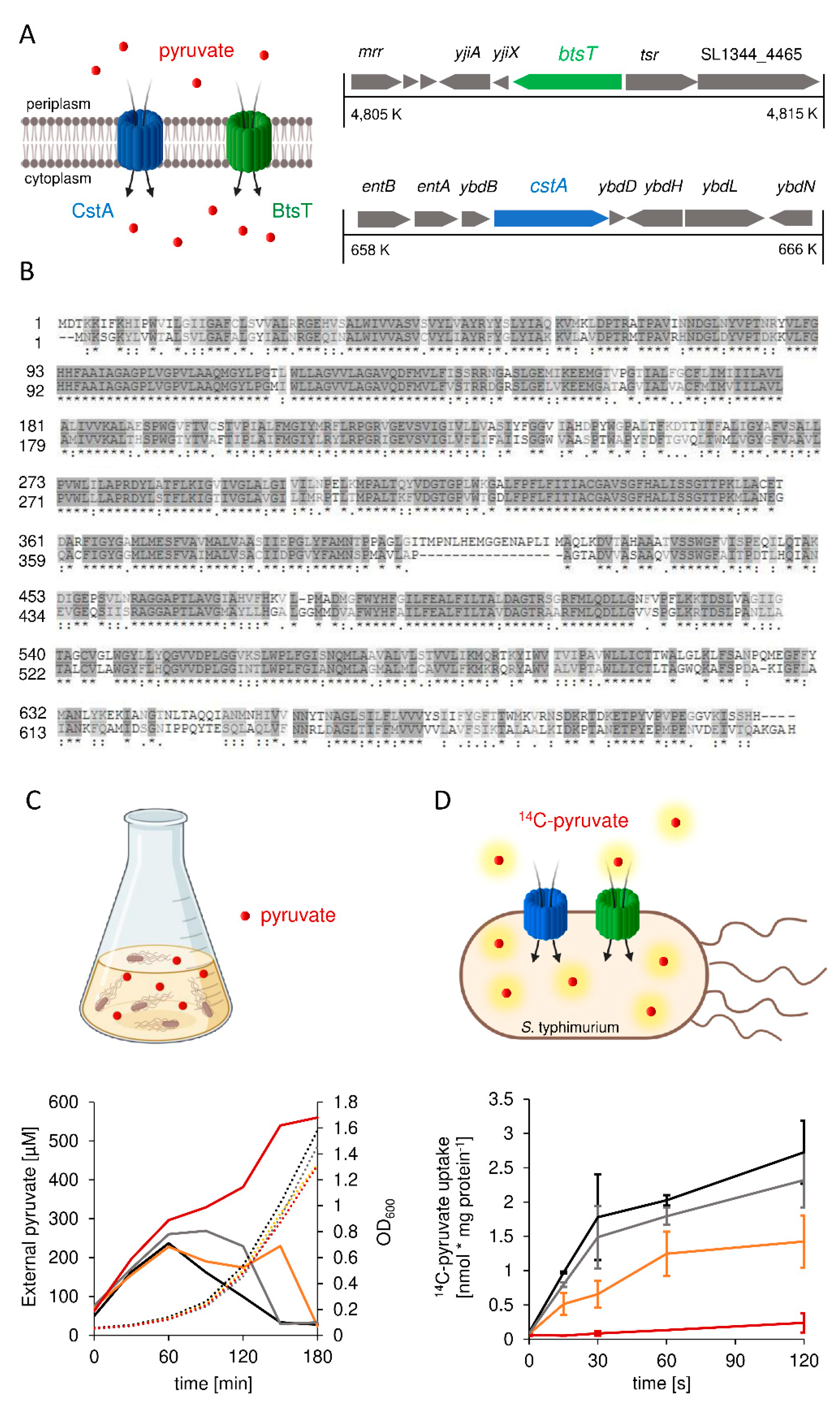
3.1. S. Typhimurium Possesses Two Pyruvate Transporters, BtsT and CstA
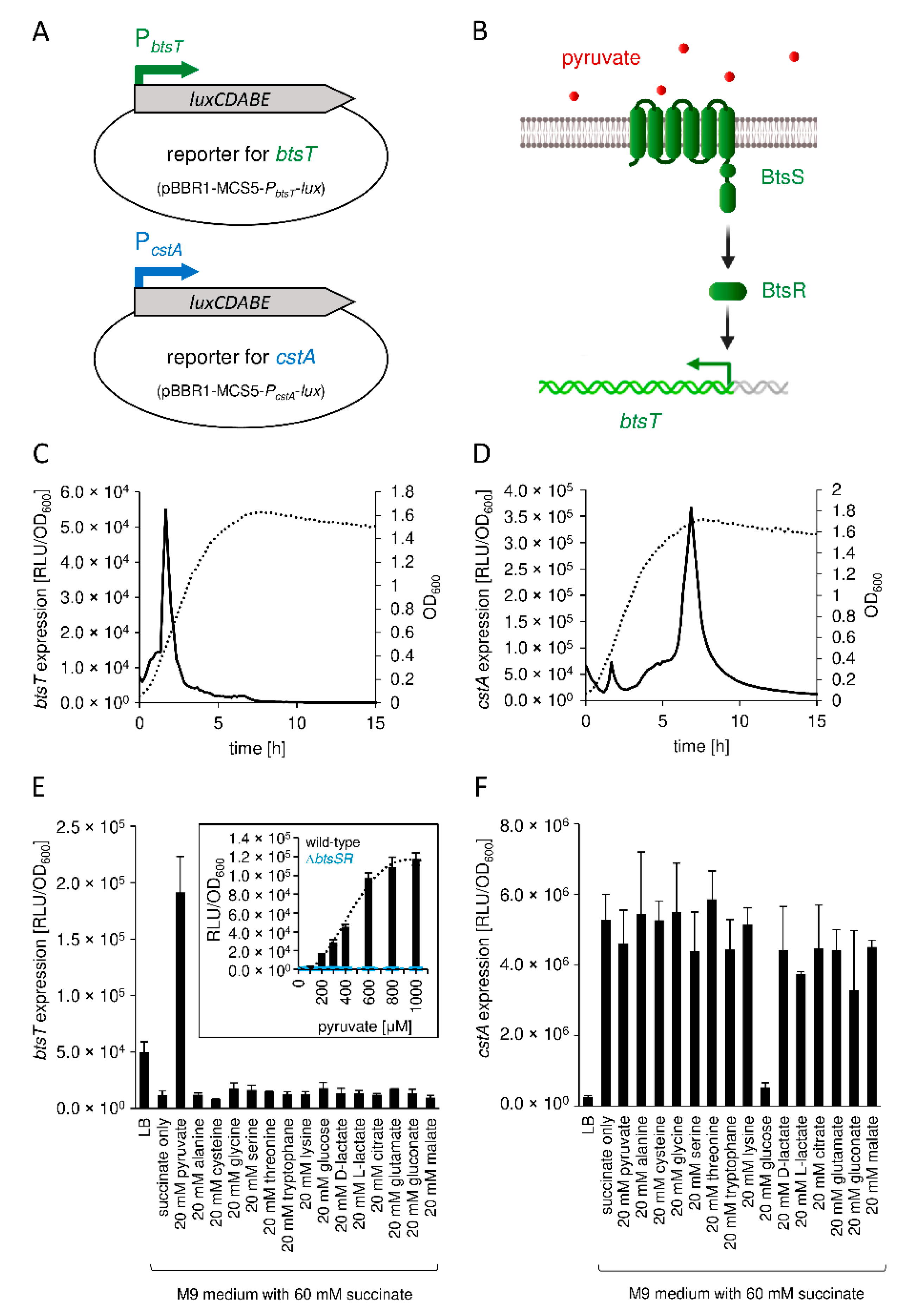
3.2. Expression of btsT Is Activated by the Histidine Kinase Response Regulator System BtsS/BtsR in the Presence of Pyruvate, whereas Expression of cstA Is Dependent on the Growth Phase
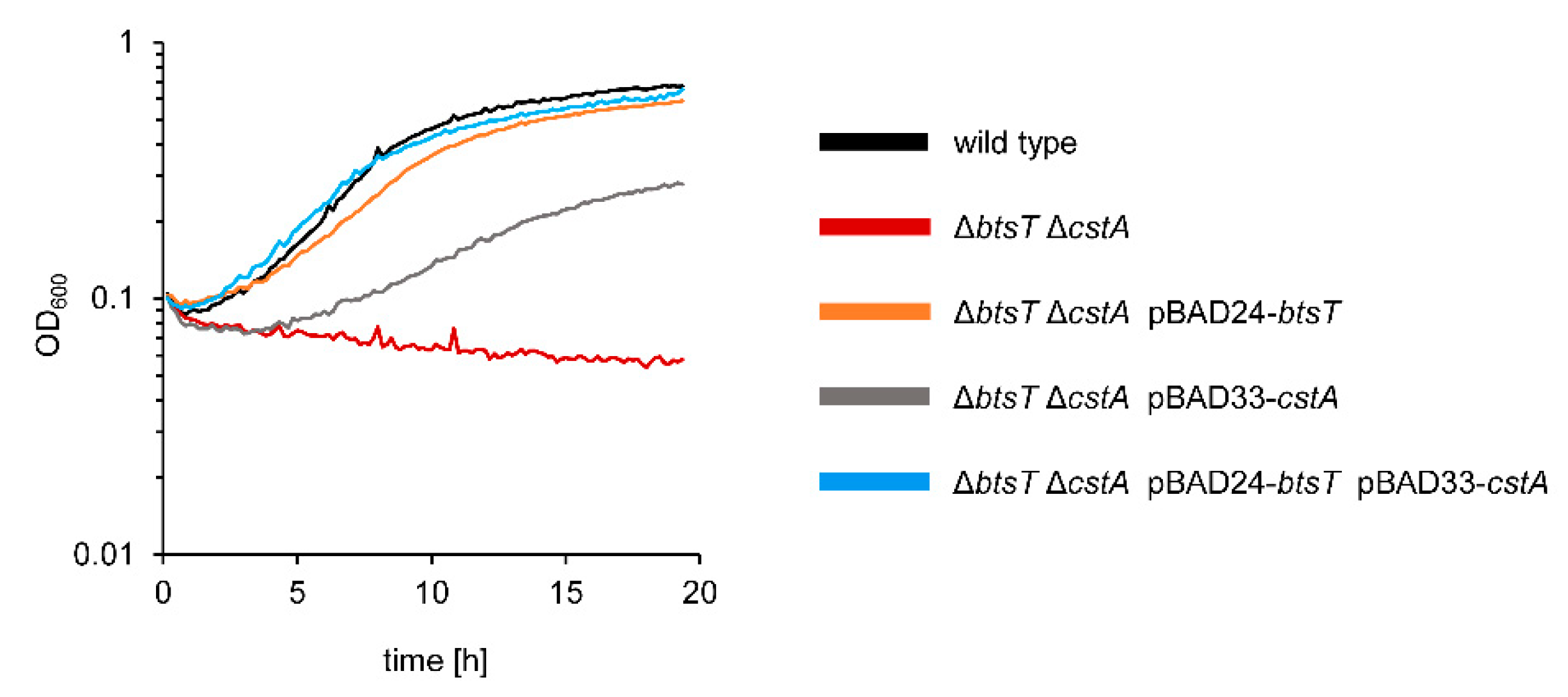
3.3. Pyruvate Uptake by BtsT and CstA Is Required for Growth on Pyruvate and Chemotaxis to Pyruvate
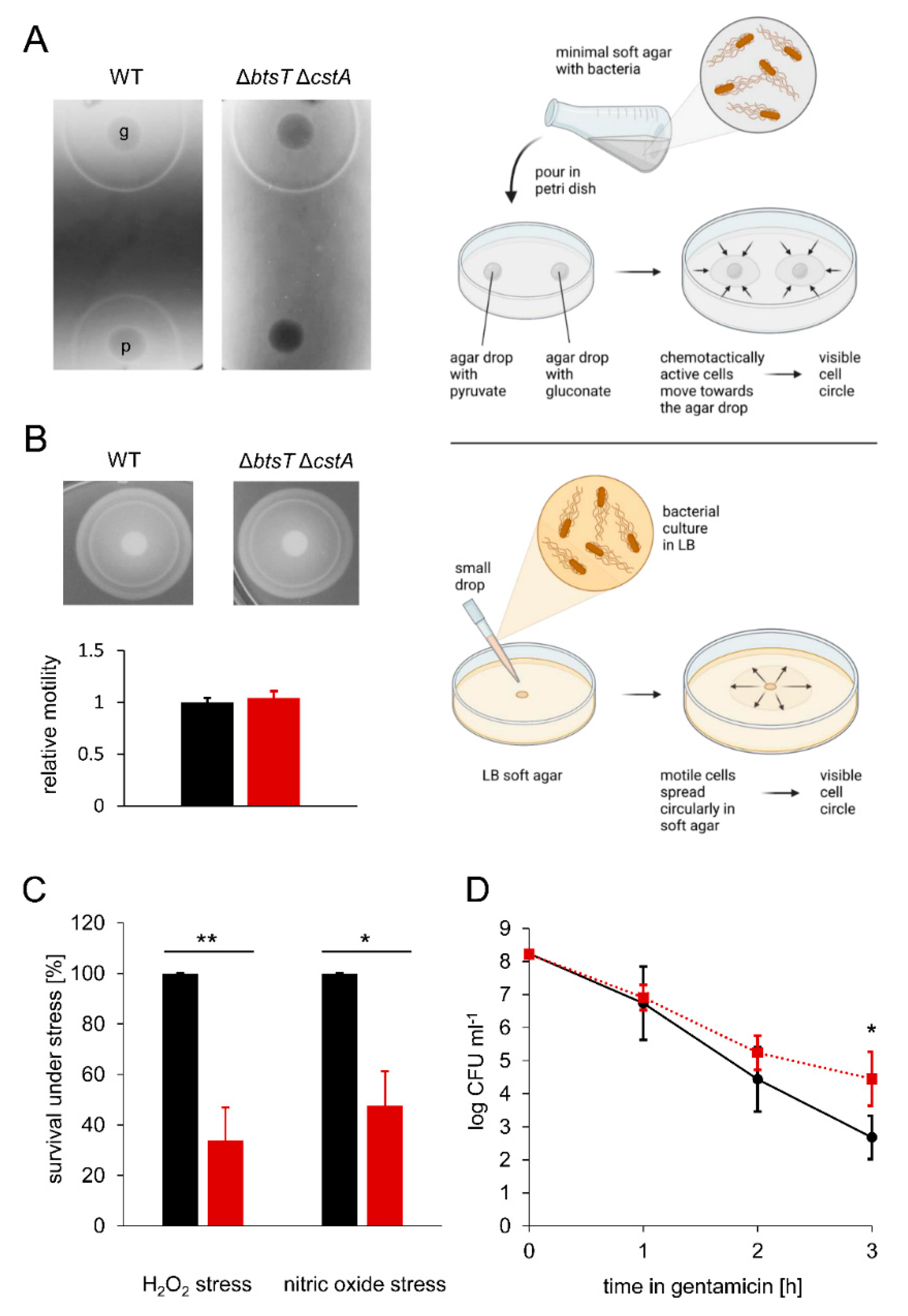
3.4. Pyruvate Uptake Is Important to Survive Oxidative Stress, Nitrosative Stress, and Antibiotic Treatment
3.5. Pyruvate Uptake Is Important to Recover from Intra-Macrophage Antibiotic Treatment
3.6. Mutants Lacking Pyruvate Transporters Show a Slight Disadvantage in Colonization and Systemic Infection of Gnotobiotic Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.T.; Bennett, G.N.; San, K.Y. The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab. Eng. 2001, 3, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Carter, J.; Ward, J.B.; Glaser, L. The effect of carbon and nitrogen sources on the level of metabolic intermediates in Escherichia coli. J. Biol. Chem. 1971, 246, 6511–6521. [Google Scholar] [CrossRef]
- Snoep, J.L.; de Graef, M.R.; de Mattos, M.J.T.; Neijssel, O.M. Pyruvate catabolism during transient state conditions in chemostat cultures of Enterococcus faecalis NCTC 775: Importance of internal pyruvate concentrations and NADH/NAD+ ratios. J. Gen. Microbiol. 1992, 138, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Behr, S.; Brameyer, S.; Witting, M.; Schmitt-Kopplin, P.; Jung, K. Comparative analysis of LytS/LytTR-type histidine kinase/response regulator systems in γ-proteobacteria. PLoS ONE 2017, 12, e0182993. [Google Scholar] [CrossRef]
- Chubukov, V.; Gerosa, L.; Kochanowski, K.; Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 2014, 12, 327–340. [Google Scholar] [CrossRef]
- Paczia, N.; Nilgen, A.; Lehmann, T.; Gätgens, J.; Wiechert, W.; Noack, S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Factories 2012, 11, 122. [Google Scholar] [CrossRef]
- Yasid, N.A.; Rolfe, M.D.; Green, J.; Williamson, M.P. Homeostasis of metabolites in Escherichia coli on transition from anaerobic to aerobic conditions and the transient secretion of pyruvate. R. Soc. Open Sci. 2016, 3, 160187. [Google Scholar] [CrossRef]
- Constantopoulos, G.; Barranger, J.A. Nonenzymatic decarboxylation of pyruvate. Anal. Biochem. 1984, 139, 353–358. [Google Scholar] [CrossRef]
- Kładna, A.; Marchlewicz, M.; Piechowska, T.; Kruk, I.; Aboul-Enein, H.Y. Reactivity of pyruvic acid and its derivatives towards reactive oxygen species. Luminescence 2015, 30, 1153–1158. [Google Scholar] [CrossRef]
- Varma, S.D.; Hegde, K.; Henein, M. Oxidative damage to mouse lens in culture. Protective effect of pyruvate. Biochim. Biophys. Acta 2003, 1621, 246–252. [Google Scholar] [CrossRef]
- O’Donnell-Tormey, J.; Nathan, C.F.; Lanks, K.; DeBoer, C.J.; de la Harpe, J. Secretion of pyruvate. An antioxidant defense of mammalian cells. J. Exp. Med. 1987, 165, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Pan, H.; Yang, D.; Rao, L.; Zhao, L.; Wang, Y.; Liao, X. Induction, detection, formation, and resuscitation of viable but non-culturable state microorganisms. Compr. Rev. Food Sci. 2020, 19, 149–183. [Google Scholar] [CrossRef] [PubMed]
- Göing, S.; Gasperotti, A.F.; Yang, Q.; Defoirdt, T.; Jung, K. Insights into a Pyruvate Sensing and Uptake System in Vibrio campbellii and Its Importance for Virulence. J. Bacteriol. 2021, 203, e0029621. [Google Scholar] [CrossRef]
- Mizunoe, Y.; Wai, S.N.; Takade, A.; Yoshida, S. Restoration of culturability of starvation-stressed and low-temperature-stressed Escherichia coli O157 cells by using H2O2-degrading compounds. Arch. Microbiol. 1999, 172, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Vilhena, C.; Kaganovitch, E.; Grünberger, A.; Motz, M.; Forné, I.; Kohlheyer, D.; Jung, K. Importance of pyruvate sensing and transport for the resuscitation of viable but nonculturable Escherichia coli K-12. J. Bacteriol. 2019, 201, e00610-00618. [Google Scholar] [CrossRef]
- Liao, H.; Jiang, L.; Zhang, R. Induction of a viable but non-culturable state in Salmonella Typhimurium by thermosonication and factors affecting resuscitation. FEMS Microbiol. Lett. 2018, 365, fnx249. [Google Scholar] [CrossRef] [PubMed]
- Bücker, R.; Heroven, A.K.; Becker, J.; Dersch, P.; Wittmann, C. The pyruvate-tricarboxylic acid cycle node: A focal point of virulence control in the enteric pathogen Yersinia pseudotuberculosis. J. Biol. Chem. 2014, 289, 30114–30132. [Google Scholar] [CrossRef]
- Schär, J.; Stoll, R.; Schauer, K.; Loeffler, D.I.; Eylert, E.; Joseph, B.; Eisenreich, W.; Fuchs, T.M.; Goebel, W. Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J. Bacteriol. 2010, 192, 1774–1784. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Pang, R.; Wu, Q.; Zhang, J.; Lei, T.; Li, Y.; Wang, J.; Ding, Y.; Chen, M.; Bai, J. Cold tolerance regulated by the pyruvate metabolism in Vibrio parahaemolyticus. Front. Microbiol. 2019, 10, 178. [Google Scholar] [CrossRef]
- Abernathy, J.; Corkill, C.; Hinojosa, C.; Li, X.; Zhou, H. Deletions in the pyruvate pathway of Salmonella Typhimurium alter SPI1-mediated gene expression and infectivity. J. Anim. Sci. Biotechnol. 2013, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- van Doorn, C.L.R.; Schouten, G.K.; van Veen, S.; Walburg, K.V.; Esselink, J.J.; Heemskerk, M.T.; Vrieling, F.; Ottenhoff, T.H.M. Pyruvate Dehydrogenase Kinase Inhibitor Dichloroacetate Improves Host Control of Salmonella enterica Serovar Typhimurium Infection in Human Macrophages. Front. Immunol. 2021, 12, 739938. [Google Scholar] [CrossRef] [PubMed]
- Goodwine, J.; Gil, J.; Doiron, A.; Valdes, J.; Solis, M.; Higa, A.; Davis, S.; Sauer, K. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci. Rep. 2019, 9, 3763. [Google Scholar] [CrossRef]
- Petrova, O.E.; Schurr, J.R.; Schurr, M.J.; Sauer, K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 2012, 86, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, Y.D.N.; Durand, B.A.R.; Hamiot, A.; Martin-Verstraete, I.; Oberkampf, M.; Monot, M.; Dupuy, B. Metabolic adaption to extracellular pyruvate triggers biofilm formation in Clostridioides difficile. ISME J. 2021, 15, 3623–3635. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.J.; Medina, C.B.; Barron, B.J.; Karvelyte, L.; Aaes, T.L.; Lambertz, I.; Perry, J.S.A.; Mehrotra, P.; Gonçalves, A.; Lemeire, K.; et al. Microbes exploit death-induced nutrient release by gut epithelial cells. Nature 2021, 596, 262–267. [Google Scholar] [CrossRef]
- Gasperotti, A.; Göing, S.; Fajardo-Ruiz, E.; Forné, I.; Jung, K. Function and regulation of the pyruvate transporter CstA in Escherichia coli. Int. J. Mol. Sci. 2020, 21, 9068. [Google Scholar] [CrossRef]
- Hwang, S.; Choe, D.; Yoo, M.; Cho, S.; Kim, S.C.; Cho, S.; Cho, B.K. Peptide transporter CstA imports pyruvate in Escherichia coli K-12. J. Bacteriol. 2018, 200, e00771-00717. [Google Scholar] [CrossRef] [PubMed]
- Kristoficova, I.; Vilhena, C.; Behr, S.; Jung, K. BtsT, a novel and specific pyruvate/H+ symporter in Escherichia coli. J. Bacteriol. 2018, 200, e00599-00517. [Google Scholar] [CrossRef]
- Behr, S.; Fried, L.; Jung, K. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J. Bacteriol. 2014, 196, 2023–2029. [Google Scholar] [CrossRef]
- Behr, S.; Kristoficova, I.; Witting, M.; Breland, E.J.; Eberly, A.R.; Sachs, C.; Schmitt-Kopplin, P.; Hadjifrangiskou, M.; Jung, K. Identification of a high-affinity pyruvate receptor in Escherichia coli. Sci. Rep. 2017, 7, 1388. [Google Scholar] [CrossRef] [Green Version]
- Fried, L.; Behr, S.; Jung, K. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J. Bacteriol. 2013, 195, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.E.; Matin, A. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 1991, 218, 129–140. [Google Scholar] [CrossRef]
- Hosie, A.H.; Allaway, D.; Poole, P.S. A monocarboxylate permease of Rhizobium leguminosarum is the first member of a new subfamily of transporters. J. Bacteriol. 2002, 184, 5436–5448. [Google Scholar] [CrossRef]
- Jolkver, E.; Emer, D.; Ballan, S.; Krämer, R.; Eikmanns, B.J.; Marin, K. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 2009, 191, 940–948. [Google Scholar] [CrossRef]
- Charbonnier, T.; Le Coq, D.; McGovern, S.; Calabre, M.; Delumeau, O.; Aymerich, S.; Jules, M. Molecular and Physiological Logics of the Pyruvate-Induced Response of a Novel Transporter in Bacillus subtilis. mBio 2017, 8, e00976-17. [Google Scholar] [CrossRef]
- Ahn, S.J.; Deep, K.; Turner, M.E.; Ishkov, I.; Waters, A.; Hagen, S.J.; Rice, K.C. Characterization of LrgAB as a stationary phase-specific pyruvate uptake system in Streptococcus mutans. BMC Microbiol. 2019, 19, 223. [Google Scholar] [CrossRef]
- Popa, G.L.; Papa, M.I. Salmonella spp. Infection—A continuous threat worldwide. Germs 2021, 11, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, M.R.; Schmitz, G.E.; Downs, D.M. YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium. J. Bacteriol. 2008, 190, 3057–3062. [Google Scholar] [CrossRef]
- Garai, P.; Lahiri, A.; Ghosh, D.; Chatterjee, J.; Chakravortty, D. Peptide utilizing carbon starvation gene yjiY is required for flagella mediated infection caused by Salmonella. Microbiology 2016, 162, 100–116. [Google Scholar] [CrossRef] [PubMed]
- Tenor, J.L.; McCormick, B.A.; Ausubel, F.M.; Aballay, A. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 2004, 14, 1018–1024. [Google Scholar] [CrossRef] [Green Version]
- Wong, V.K.; Pickard, D.J.; Barquist, L.; Sivaraman, K.; Page, A.J.; Hart, P.J.; Arends, M.J.; Holt, K.E.; Kane, L.; Mottram, L.F.; et al. Characterization of the yehUT two-component regulatory system of Salmonella enterica Serovar Typhi and Typhimurium. PLoS ONE 2013, 8, e84567. [Google Scholar] [CrossRef]
- Holt, K.E.; Parkhill, J.; Mazzoni, C.J.; Roumagnac, P.; Weill, F.X.; Goodhead, I.; Rance, R.; Baker, S.; Maskell, D.J.; Wain, J.; et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 2008, 40, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Karlinsey, J.E. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 2007, 421, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Brameyer, S.; Hoyer, E.; Bibinger, S.; Burdack, K.; Lassak, J.; Jung, K. Molecular design of a signaling system influences noise in protein abundance under acid stress in different γ-Proteobacteria. J. Bacteriol. 2020, 202, e00121-20. [Google Scholar] [CrossRef]
- Lassak, J.; Henche, A.L.; Binnenkade, L.; Thormann, K.M. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2010, 76, 3263–3274. [Google Scholar] [CrossRef]
- Hoiseth, S.K.; Stocker, B.A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 1981, 291, 238–239. [Google Scholar] [CrossRef]
- Maier, L.; Vyas, R.; Cordova, C.D.; Lindsay, H.; Schmidt, T.S.; Brugiroux, S.; Periaswamy, B.; Bauer, R.; Sturm, A.; Schreiber, F.; et al. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 2013, 14, 641–651. [Google Scholar] [CrossRef]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Gödeke, J.; Heun, M.; Bubendorfer, S.; Paul, K.; Thormann, K.M. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Appl. Environ. Microbiol. 2011, 77, 5342–5351. [Google Scholar] [CrossRef] [Green Version]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
- Harwood, C.R.; Cutting, S.M. Molecular Biological Methods for Bacillus; Harwood, C.R.C., Simon, M., Eds.; Wiley: Chichester, UK, 1990. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Darias, J.A.; García-Fontana, C.; Lugo, A.C.; Rico-Jiménez, M.; Krell, T. Qualitative and quantitative assays for flagellum-mediated chemotaxis. Methods Mol. Biol. 2014, 1149, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Kraxenberger, T.; Fried, L.; Behr, S.; Jung, K. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J. Bacteriol. 2012, 194, 4272–4284. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Grosse, K.; Sourjik, V. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, 12159–12164. [Google Scholar] [CrossRef] [PubMed]
- Somavanshi, R.; Ghosh, B.; Sourjik, V. Sugar influx sensing by the phosphotransferase system of Escherichia coli. PLoS Biol. 2016, 14, e2000074. [Google Scholar] [CrossRef]
- Richardson, A.R.; Payne, E.C.; Younger, N.; Karlinsey, J.E.; Thomas, V.C.; Becker, L.A.; Navarre, W.W.; Castor, M.E.; Libby, S.J.; Fang, F.C. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell Host Microbe 2011, 10, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Kröger, C.; Colgan, A.; Srikumar, S.; Händler, K.; Sivasankaran, S.K.; Hammarlöf, D.L.; Canals, R.; Grissom, J.E.; Conway, T.; Hokamp, K.; et al. An Infection-Relevant Transcriptomic Compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 2013, 14, 683–695. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Vilhena, C.; Kaganovitch, E.; Shin, J.Y.; Grünberger, A.; Behr, S.; Kristoficova, I.; Brameyer, S.; Kohlheyer, D.; Jung, K. A single-cell view of the BtsSR/YpdAB pyruvate sensing network in Escherichia coli and its biological relevance. J. Bacteriol. 2018, 200, e00536-00517. [Google Scholar] [CrossRef]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Stapels, D.A.C.; Hill, P.W.S.; Westermann, A.J.; Fisher, R.A.; Thurston, T.L.; Saliba, A.E.; Blommestein, I.; Vogel, J.; Helaine, S. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 2018, 362, 1156–1160. [Google Scholar] [CrossRef]
- Löber, S.; Jäckel, D.; Kaiser, N.; Hensel, M. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 2006, 296, 435–447. [Google Scholar] [CrossRef]
- Brugiroux, S.; Beutler, M.; Pfann, C.; Garzetti, D.; Ruscheweyh, H.J.; Ring, D.; Diehl, M.; Herp, S.; Lötscher, Y.; Hussain, S.; et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2016, 2, 16215. [Google Scholar] [CrossRef]
- Eberl, C.; Weiss, A.S.; Jochum, L.M.; Durai Raj, A.C.; Ring, D.; Hussain, S.; Herp, S.; Meng, C.; Kleigrewe, K.; Gigl, M.; et al. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe 2021, 29, 1680–1692.e1687. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Clavel, T.; Lagkouvardos, I.; Blaut, M.; Stecher, B. The mouse gut microbiome revisited: From complex diversity to model ecosystems. Int. J. Med. Microbiol. 2016, 306, 316–327. [Google Scholar] [CrossRef]
- Weiss, A.S.; Burrichter, A.G.; Durai Raj, A.C.; von Strempel, A.; Meng, C.; Kleigrewe, K.; Münch, P.C.; Rössler, L.; Huber, C.; Eisenreich, W.; et al. In vitro interaction network of a synthetic gut bacterial community. ISME J. 2021, 16, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Pontrelli, S.; Szabo, R.; Pollak, S.; Schwartzman, J.; Ledezma-Tejeida, D.; Cordero, O.X.; Sauer, U. Metabolic cross-feeding structures the assembly of polysaccharide degrading communities. Sci. Adv. 2022, 8, eabk3076. [Google Scholar] [CrossRef] [PubMed]
- Girinathan, B.P.; DiBenedetto, N.; Worley, J.N.; Peltier, J.; Arrieta-Ortiz, M.L.; Immanuel, S.R.C.; Lavin, R.; Delaney, M.L.; Cummins, C.K.; Hoffman, M.; et al. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 2021, 29, 1693–1708 e1697. [Google Scholar] [CrossRef]
- Ducret, A.; Quardokus, E.M.; Brun, Y.V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 2016, 1, 16077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Strain or Plasmid | Genotype or Description | Reference |
|---|---|---|
| S. Typhimurium strains | ||
| SL1344 | Wild type; strep R | [48] |
| LT2 | Wild type | DSMZ #17058 |
| SL1344 ΔbtsT | Mutant with in-frame deletion of btsT (SL1344_4463); strep R | this study |
| SL1344 ΔcstA | Mutant with in-frame replacement of cstA (SL1344_0588) by a kanamycin resistance cassette; strep R kan R | this study |
| SL1344 ΔbtsT ΔcstA | Mutant with in-frame deletion of btsT (SL1344_4463) and replacement of cstA (SL1344_0588) by a kanamycin resistance cassette; strep R kan R | this study |
| SL1344 ΔbtsSR | Mutant with in-frame deletion of btsS (SL1344_2137) and btsR (SL1344_2136); strep R | this study |
| M2702 | Non-virulent SL1344 strain, ΔinvG ΔssaV; strep R | [49] |
| M2702 ΔbtsT ΔcstA | Non-virulent mutant with in-frame deletion of cstA (SL1344_0588) and replacement of btsT (SL1344_4463) by a chloramphenicol resistance cassette; strep R cm R | this study |
| E. colistrains | ||
| DH5α λpir | Cloning strain; endA1 hsdR17 glnV44 thi-1 recA1 gyrA96 relA1 φ80′lacΔ(lacZ)M15 Δ(lacZYA-argF)U169 zdg-232::Tn10 uidA::pir+ | [50] |
| WM3064 | Conjugation strain; thrB1004 pro thi rpsL hsdS lacZ ΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir] | W. Metcalf, University of Illinois |
| Plasmids | ||
| pNPTS138-R6KT | Plasmid backbone for in-frame deletions; mobRP4+; sacB, kan R | [47] |
| pNPTS138-R6KT-ΔcstA | Plasmid for in-frame deletion of cstA in SL1344; kan R | this study |
| pNPTS138-R6KT-ΔbtsT::cm R | Plasmid for in-frame replacement of btsT by a chloramphenicol resistance cassette in SL1344; kan R cm R | this study |
| pBBR1-MCS5-lux | Plasmid backbone to insert a promoter sequence upstream of luxCDABE for a luciferase-based reporter assay; gent R | [51] |
| pBBR1-MCS5-PbtsT-lux | Luciferase-based reporter plasmid with the promoter region of SL1344 btsT upstream of luxCDABE; gent R | this study |
| pBBR1-MCS5-PcstA-lux | Luciferase-based reporter plasmid with the promoter region of SL1344 cstA upstream of luxCDABE; gent R | this study |
| pBAD24 | Plasmid backbone for expression; amp R | [52] |
| pBAD24-btsT | Expression plasmid for SL1344 btsT; amp R | this study |
| pBAD33 | Plasmid backbone for expression; cm R | [52] |
| pBAD33-cstA | Expression plasmid for SL1344 cstA; cm R | this study |
| pKD46 | λ-red recombinase expressing plasmid; amp R | [45] |
| pKD4 | Template plasmid for kanamycin resistance cassette (FRT-aminoglycoside phosphotransferase-FRT); kan R | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paulini, S.; Fabiani, F.D.; Weiss, A.S.; Moldoveanu, A.L.; Helaine, S.; Stecher, B.; Jung, K. The Biological Significance of Pyruvate Sensing and Uptake in Salmonella enterica Serovar Typhimurium. Microorganisms 2022, 10, 1751. https://doi.org/10.3390/microorganisms10091751
Paulini S, Fabiani FD, Weiss AS, Moldoveanu AL, Helaine S, Stecher B, Jung K. The Biological Significance of Pyruvate Sensing and Uptake in Salmonella enterica Serovar Typhimurium. Microorganisms. 2022; 10(9):1751. https://doi.org/10.3390/microorganisms10091751
Chicago/Turabian StylePaulini, Stephanie, Florian D. Fabiani, Anna S. Weiss, Ana Laura Moldoveanu, Sophie Helaine, Bärbel Stecher, and Kirsten Jung. 2022. "The Biological Significance of Pyruvate Sensing and Uptake in Salmonella enterica Serovar Typhimurium" Microorganisms 10, no. 9: 1751. https://doi.org/10.3390/microorganisms10091751
APA StylePaulini, S., Fabiani, F. D., Weiss, A. S., Moldoveanu, A. L., Helaine, S., Stecher, B., & Jung, K. (2022). The Biological Significance of Pyruvate Sensing and Uptake in Salmonella enterica Serovar Typhimurium. Microorganisms, 10(9), 1751. https://doi.org/10.3390/microorganisms10091751






