Revealing a New Family of D-2-Hydroxyglutarate Dehydrogenases in Escherichia coli and Pantoea ananatis Encoded by ydiJ
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Analytical Method
2.3. Bacterial Strains, Plasmids, and Growth Conditions
2.4. DNA Manipulation and Plasmid Construction
2.5. Screening for Genes Involved in D-2-HGA Utilization
2.6. DNA Analysis
2.7. Enzyme Overexpression and Purification
2.8. Measurement of Enzyme Activity
2.9. Characterization of the Recombinant D2HGDH (YdiJ)
2.10. Polyacrylamide Electrophoresis
2.11. Structure-Based Protein Sequence Alignment
2.12. Phylogenetic Analysis
3. Results and Discussion
3.1. Screening the Genomic P. ananatis Library for Genes That Conferred Fast Growth on D-2-HGA
3.2. Determination of Physiological Function of YdiJ In Vivo
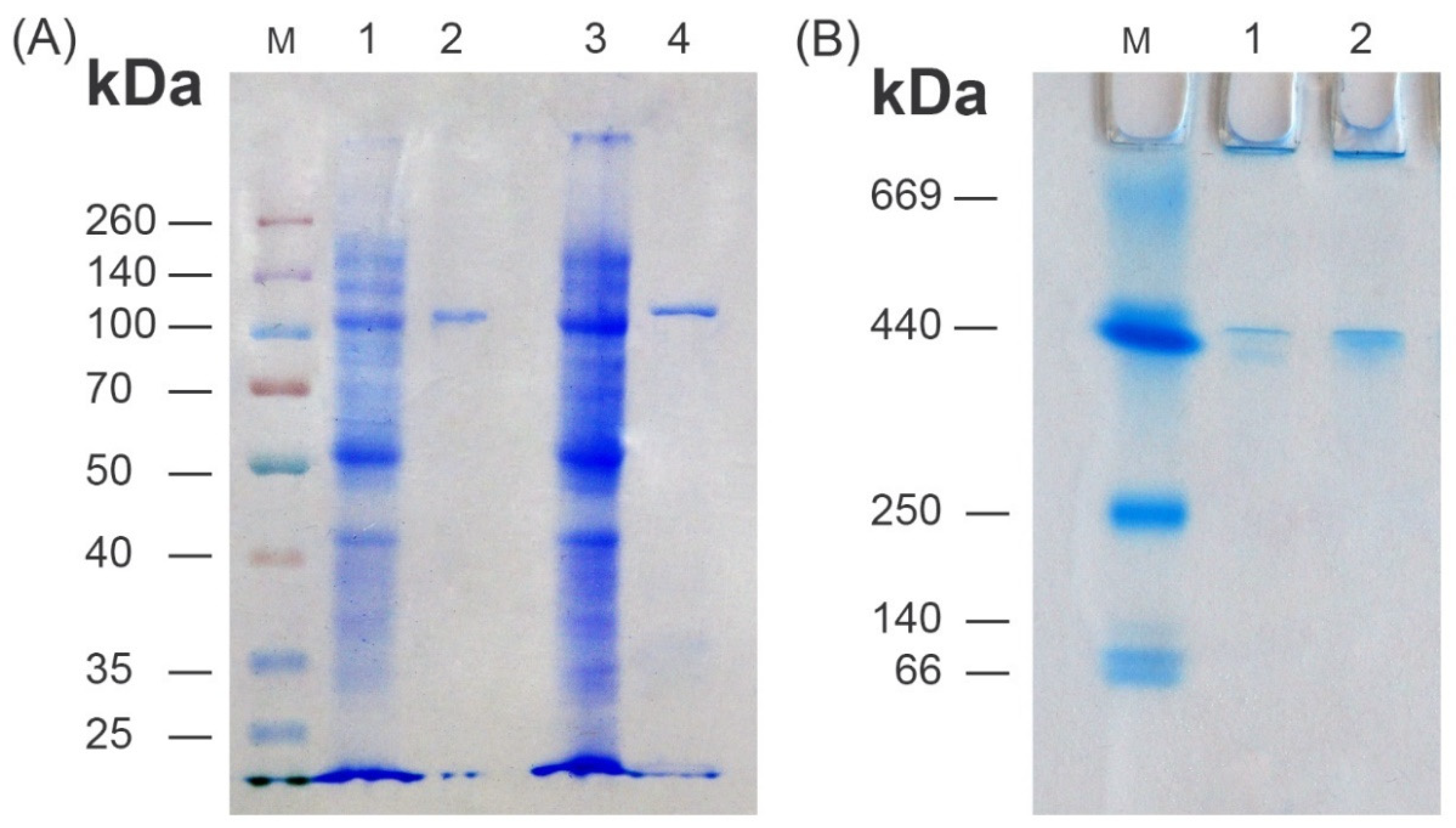
3.3. Recombinant Enzyme Purification and Characterization
3.4. Effects of pH and Temperature
3.5. Effects of Metal Ions on D2HGDH Activity
3.6. Substrate Specificity
3.7. Phylogenetic Analysis of YdiJ-Like Enzymes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, D.L.; Cox, M.M.; Lehninger, A. Lehninger: Principles of Biochemistry, 5th ed.; W. H. Freeman: New York, NY, USA, 2009. [Google Scholar]
- Grant, G.A. Contrasting catalytic and allosteric mechanisms for phosphoglycerate dehydrogenases. Arch. Biochem. Biophys. 2012, 519, 175–185. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, G.; Shi, J.; Koffas, M.A.G.; Xu, Z. Microbial Production of l-Serine from Renewable Feedstocks. Trends Biotechnol. 2018, 36, 700–712. [Google Scholar] [CrossRef]
- Mundhada, H.; Schneider, K.; Christensen, H.B.; Nielsen, A.T. Engineering of high yield production of L-serine in Escherichia coli. Biotechnol. Bioeng. 2016, 113, 807–816. [Google Scholar] [CrossRef]
- Gu, P.; Yang, F.; Su, T.; Li, F.; Li, Y.; Qi, Q. Construction of an L-serine producing Escherichia coli via metabolic engineering. J. Ind. Microbiol. Biotechnol. 2014, 41, 1443–1450. [Google Scholar] [CrossRef]
- Aboulwafa, M.; Saier, M.H., Jr. Dependency of sugar transport and phosphorylation by the phosphoenolpyruvate-dependent phosphotransferase system on membranous phosphatidyl glycerol in Escherichia coli: Studies with a pgsA mutant lacking phosphatidyl glycerophosphate synthase. Res. Microbiol. 2002, 153, 667–677. [Google Scholar] [CrossRef]
- Reid, M.A.; Allen, A.E.; Liu, S.; Liberti, M.V.; Liu, P.; Liu, X.; Dai, Z.; Gao, X.; Wang, Q.; Liu, Y.; et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat. Commun. 2018, 9, 5442. [Google Scholar] [CrossRef]
- Pizer, L.I.; Potochny, M.L. Nutritional and regulatory aspects of serine metabolism in Escherichia coli. J. Bacteriol. 1964, 88, 611–619. [Google Scholar] [CrossRef]
- Grant, G.A. D-3-Phosphoglycerate dehydrogenase. Front. Mol. Biosci. 2018, 5, 110. [Google Scholar] [CrossRef]
- Schuller, D.J.; Fetter, C.H.; Banaszak, L.J.; Grant, G.A. Enhanced expression of the Escherichia coli serA gene in a plasmid vector: Purification, crystallization, and preliminary X-ray data of D-3-phosphoglycerate dehydrogenase. J. Biol. Chem. 1989, 264, 2645–2648. [Google Scholar] [CrossRef]
- Schuller, D.J.; Grant, G.A.; Banaszak, L.J. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat. Struct. Biol. 1995, 2, 69–76. [Google Scholar] [CrossRef]
- Merrill, D.K.; McAlexander, J.C.; Guynn, R.W. Equilibrium constants under physiological conditions for the reactions of D-3-phosphoglycerate dehydrogenase and L-phosphoserine aminotransferase. Arch. Biochem. Biophys. 1981, 212, 717–729. [Google Scholar] [CrossRef]
- Guynn, R.W.; Thames, H. Equilibrium constants under physiological conditions for the reactions of L-phosphoserine phosphatase and pyrophosphate: L-serine phosphotransferase. Arch. Biochem. Biophys. 1982, 215, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, M.; Gao, C.; Zhang, Y.Y.; Ge, Y.; Guo, S.; Guo, X.; Zhou, Z.; Liu, Q.; Zhang, Y.Y.; et al. Coupling between D-3-phosphoglycerate dehydrogenase and D-2-hydroxyglutarate dehydrogenase drives bacterial L-serine synthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E7574–E7582. [Google Scholar] [CrossRef]
- Zhao, G.; Winkler, M.E. A novel alpha-ketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J. Bacteriol. 1996, 178, 232–239. [Google Scholar] [CrossRef]
- Fan, J.; Teng, X.; Liu, L.; Mattaini, K.R.; Looper, R.E.; Vander Heiden, M.G.; Rabinowitz, J.D. Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate. ACS Chem. Biol. 2015, 10, 510–516. [Google Scholar] [CrossRef]
- Murtas, G.; Marcone, G.L.; Peracchi, A.; Zangelmi, E.; Pollegioni, L. Biochemical and Biophysical Characterization of Recombinant Human 3-Phosphoglycerate Dehydrogenase. Int. J. Mol. Sci. 2021, 22, 4231. [Google Scholar] [CrossRef]
- Becker-Kettern, J.; Paczia, N.; Conrotte, J.F.; Kay, D.P.; Guignard, C.; Jung, P.P.; Linster, C.L. Saccharomyces cerevisiae Forms D-2-Hydroxyglutarate and Couples Its Degradation to D-Lactate Formation via a Cytosolic Transhydrogenase. J. Biol. Chem. 2016, 291, 6036–6058. [Google Scholar] [CrossRef]
- Okamura, E.; Hirai, M.Y. Novel regulatory mechanism of serine biosynthesis associated with 3-phosphoglycerate dehydrogenase in Arabidopsis thaliana. Sci. Rep. 2017, 7, 3533. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, M.; Cao, M.; Zhang, W.; Kang, Z.; Xu, P.; Ma, C.; Gao, C. d-2-Hydroxyglutarate dehydrogenase plays a dual role in l-serine biosynthesis and d-malate utilization in the bacterium Pseudomonas stutzeri. J. Biol. Chem. 2018, 293, 15513–15523. [Google Scholar] [CrossRef]
- Xiao, D.; Zhang, W.; Guo, X.; Liu, Y.; Hu, C.; Guo, S.; Kang, Z.; Xu, X.; Ma, C.; Gao, C.; et al. A D-2-hydroxyglutarate biosensor based on specific transcriptional regulator DhdR. Nat. Commun. 2021, 12, 7108. [Google Scholar] [CrossRef]
- Gusyatiner, M.M.; Ziyatdinov, M.K. 2-Hydroxyglutarate production is necessary for the reaction catalyzed by 3-phosphoglycerate dehydrogenase in Escherichia coli. Ref. J. Chem. 2015, 5, 21–29. [Google Scholar] [CrossRef]
- Xu, X.L.; Grant, G.A. Determinants of substrate specificity in D-3-phosphoglycerate dehydrogenase. Conversion of the M. tuberculosis enzyme from one that does not use α-ketoglutarate as a substrate to one that does. Arch. Biochem. Biophys. 2019, 671, 218–224. [Google Scholar] [CrossRef]
- Grant, G.A. Elucidation of a Self-Sustaining Cycle in Escherichia coli l-Serine Biosynthesis That Results in the Conservation of the Coenzyme, NAD+. Biochemistry 2018, 57, 1798–1806. [Google Scholar] [CrossRef]
- Quaye, J.A.; Gadda, G. Kinetic and Bioinformatic Characterization of d-2-Hydroxyglutarate Dehydrogenase from Pseudomonas aeruginosa PAO1. Biochemistry 2020, 59, 4833–4844. [Google Scholar] [CrossRef]
- Wu, B.; Li, Z.; Kang, Z.; Ma, C.; Song, H.; Lu, F.; Zhu, Z. An Enzymatic Biosensor for the Detection of D-2-Hydroxyglutaric Acid in Serum and Urine. Biosensors 2022, 12, 66. [Google Scholar] [CrossRef]
- Engqvist, M.; Drincovich, M.F.; Flügge, U.I.; Maurino, V.G. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and β-oxidation pathways. J. Biol. Chem. 2009, 284, 25026–25037. [Google Scholar] [CrossRef]
- Yang, J.; Zhu, H.; Zhang, T.; Ding, J. Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations. Cell Discov. 2021, 7, 3. [Google Scholar] [CrossRef]
- Toplak, M.; Brunner, J.; Schmidt, J.; Macheroux, P. Biochemical characterization of human D-2-hydroxyglutarate dehydrogenase and two disease related variants reveals the molecular cause of D-2-hydroxyglutaric aciduria. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140255. [Google Scholar] [CrossRef]
- Pop, A.; Struys, E.A.; Jansen, E.E.W.; Fernandez, M.R.; Kanhai, W.A.; van Dooren, S.J.M.; Ozturk, S.; van Oostendorp, J.; Lennertz, P.; Kranendijk, M.; et al. D-2-hydroxyglutaric aciduria Type I: Functional analysis of D2HGDH missense variants. Hum. Mutat. 2019, 40, 975–982. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Hara, Y.; Kadotani, N.; Izui, H.; Katashkina, J.I.; Kuvaeva, T.M.; Andreeva, I.G.; Golubeva, L.I.; Malko, D.B.; Makeev, V.J.; Mashko, S.V.; et al. The complete genome sequence of Pantoea ananatis AJ13355, an organism with great biotechnological potential. Appl. Microbiol. Biotechnol. 2012, 93, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Katashkina, J.I.; Hara, Y.; Golubeva, L.I.; Andreeva, I.G.; Kuvaeva, T.M.; Mashko, S.V. Use of the lambda Red-recombineering method for genetic engineering of Pantoea ananatis. BMC Mol. Biol. 2009, 10, 34. [Google Scholar] [CrossRef]
- Sycheva, E.V.; Serebryanyy, V.A.; Yampolskaya, T.A.; Preobrazhenskaya, E.S.; Stoynova, N.V. Mutant Acetolactate Synthase and Method for Producing Branched-Chain L-Amino Acids. European Patent 1942183, 28 October 2009. [Google Scholar]
- Achouri, Y.; Noël, G.; Vertommen, D.; Rider, M.H.; Veiga-Da-Cunha, M.; Van Schaftingen, E. Identification of a dehydrogenase acting on D-2-hydroxyglutarate. Biochem. J. 2004, 381 Pt 1, 35–42. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Takumi, K.; Nonaka, G. An L-Amino Acid-Producing Bacterium and a Method for Producing an L-Amino Acid. European Patent EP 2218729 B1, 9 July 2014. [Google Scholar]
- Al-Rabiee, R.; Zhang, Y.; Grant, G.A. The mechanism of velocity modulated allosteric regulation in D-3-phosphoglycerate dehydrogenase. Site-directed mutagenesis of effector binding site residues. J. Biol. Chem. 1996, 271, 23235–23238. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Quayle, J.R. Phenazine ethosulfate as a preferred electron acceptor to phenazine methosulfate in dye-linked enzyme assays. Anal. Biochem. 1979, 99, 112–117. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Métoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Estellon, J.; Ollagnier de Choudens, S.; Smadja, M.; Fontecave, M.; Vandenbrouck, Y. An integrative computational model for large-scale identification of metalloproteins in microbial genomes: A focus on iron-sulfur cluster proteins. Metallomics 2014, 6, 1913–1930. [Google Scholar] [CrossRef]
- Ewing, T.A.; Fraaije, M.W.; Mattevi, A.; van Berkel, W.J.H. The VAO/PCMH flavoprotein family. Arch. Biochem. Biophys. 2017, 632, 104–117. [Google Scholar] [CrossRef]
- Ewing, T.A.; Gygli, G.; Fraaije, M.W.; van Berkel, W.J.H. Vanillyl Alcohol Oxidase. Enzymes 2020, 47, 87–116. [Google Scholar] [CrossRef]
- Du, X.; Hu, H. The Roles of 2-Hydroxyglutarate. Front. Cell Dev. Biol. 2021, 9, 651317. [Google Scholar] [CrossRef] [PubMed]
- Frimmersdorf, E.; Horatzek, S.; Pelnikevich, A.; Wiehlmann, L.; Schomburg, D. How Pseudomonas aeruginosa adapts to various environments: A metabolomic approach. Environ. Microbiol. 2010, 12, 1734–1747. [Google Scholar] [CrossRef] [PubMed]
- Winder, C.L.; Dunn, W.B.; Schuler, S.; Broadhurst, D.; Jarvis, R.; Stephens, G.M.; Goodacre, R. Global metabolic profiling of Escherichia coli cultures: An evaluation of methods for quenching and extraction of intracellular metabolites. Anal. Chem. 2008, 80, 2939–2948. [Google Scholar] [CrossRef] [PubMed]
- Samsonov, V.V.; Kiryukhin, M.Y.; Ziyatdinov, M.K.; Samsonova, S.A.; Rostova, Y.G. Method for Producing L-Amino Acids Using a Bacterium Belonging to the Family Enterobacteriaceae Having Overexpressed ydiJ Gene. Patent WO-2020171227-A1, 27 August 2020. [Google Scholar]








| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| Escherichia coli | ||
| MG1655 | Wild type | VKPM a |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK–, mK+) phoA supE44 λ–thi-1 gyrA96 relA1 | Novagen |
| BL21(DE3) | F–ompT hsdSB (rB–, mB–) gal dcm (DE3) | Novagen |
| MG1655 Pnlp8-ydiJEc | MG1655 with enhanced expression of ydiJ | This study |
| MG1655 ΔydiJ::Km | MG1655 with inactivated ydiJ | This study |
| Pantoea ananatis | ||
| SC 17 (AJ13355) | Wild type | [32] |
| SC17(0) | Mutant resistant to lambda Red recombinase selected from SC17 | [33] |
| SC17(0) ΔydiJ::Km | SC17(0) with inactivated ydiJ | This study |
| SC17(0) Pnlp8-ydiJPa | SC17(0) with enhanced expression of ydiJ | This study |
| Plasmids | ||
| pMIV-5JS | Vector, SC101 ori, Cmr empty vector | [34] |
| pMIV-Pnlp8serA348(Ec) | serA348 (encodes feedback-resistant enzyme [35]) from E. coli cloned in pMIV-5JS; the gene is flanked by P nlp8 (constitutive derivative of nlpD promoter) and TrrnB (rrnB terminator) | This study |
| pSTV29 | Cloning vector; Cmr | TaKaRa Bio |
| pET28b(+) | Expression vector, Kmr | Novagen |
| pET-ydiJ_Pa1 | pET28b(+) with cloned ydiJ from P. ananatis with 6His on N-end | This study |
| pET-ydiJ_Pa2 | pET28b(+) with cloned ydiJ from P. ananatis with 6His on C-end | This study |
| pET-ydiJ_Ec1 | pET28b(+) with cloned ydiJ from E. coli with 6His on N-end | This study |
| pET-ydiJ_Ec2 | pET28b(+) with cloned ydiJ from E. coli with 6His on C-end | This study |
| pMW-λattL-KmR-λattR | Source of antibiotic resistance marker Km | [33] |
| pMW-λattR-KmR-λattL-Pnlp8 | Source of antibiotic resistance marker Km (for enhancing gene expression) | This study |
| pRSFRedTER | For red-dependent integration of PCR fragments in Pantoea ananatis | [33] |
| pKD46 | For red-dependent integration of PCR fragments in E. coli | [36] |
| Enzyme Family | Mw Calculated | Km (D-2-HGA) | Vmax (D-2-HGA) | kcat | kcat /Km | Reference |
|---|---|---|---|---|---|---|
| VAO/PCMH Flavoprotein [43,44] | Da | μM | μM/min·mg −1 | s−1 | s−1/mM−1 | |
| D2HGDH P. stutzeri | 51,086.14 | 170 ± 20 | 4.56 ± 0.60 | 7.9 ± 1.05 | 45.4 | [14,20] |
| D2HGDH A. thaliana | 51,286.52 | ~580 | NR | ~0.8 | 1.37 | [27] |
| Dld2 S. cervisiae | 59,282.45 | 28 ± 8 | NR | 0.18 ± 0.03 | 7 | [18] |
| Dld3 S. cervisiae | 55,241.00 | 130 ± 9 | NR | 6.6 ± 0.5 | 50 ± 2 | [18] |
| D2HGDH P. aeruginosa | 51,286.52 | 60 | NR | 11.2± 0.4 | 186 | [25] |
| D2HGDH A. denitrificans | 50,405.42 | 31.6 | 40.6 | 6.9 | 215 | [21] |
| D2HGDH R. solanacearum | 50,444.64 | 433 | NR | 4.86 | 11 | [26] |
| D2HGDH Homo sapiens | 56,416.06 | 38 ± 0.3 120 ± 10 | NR 2.29 ± 0.03 | 0.51 ± 0.01 1.98 ± 0.03 | 13 17 | [29] [28] |
| Fe4S4 FAD-linked oxidoreductase [42] | ||||||
| D2HGDH (YdiJ) E. coli | 113,247.68 | 83 ± 5 | 1.15 | 13.9 | 170 | This study |
| D2HGDH (YdiJ) P. ananatis | 113,453.43 | 208 ± 10 | 1.17 | 9.7 | 50 | This study |
| Metal Ions | Relative Activity (%) | |
|---|---|---|
| E. coli | P. ananatis | |
| None | 100.0 | 100.0 |
| Mg2+ | 95.0 | 93.0 |
| Zn2+ | 73.0 | 34.0 |
| Mn2+ | 0 | 0 |
| Co2+ | 0 | 0 |
| Ni2+ | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samsonov, V.V.; Kuznetsova, A.A.; Rostova, J.G.; Samsonova, S.A.; Ziyatdinov, M.K.; Kiriukhin, M.Y. Revealing a New Family of D-2-Hydroxyglutarate Dehydrogenases in Escherichia coli and Pantoea ananatis Encoded by ydiJ. Microorganisms 2022, 10, 1766. https://doi.org/10.3390/microorganisms10091766
Samsonov VV, Kuznetsova AA, Rostova JG, Samsonova SA, Ziyatdinov MK, Kiriukhin MY. Revealing a New Family of D-2-Hydroxyglutarate Dehydrogenases in Escherichia coli and Pantoea ananatis Encoded by ydiJ. Microorganisms. 2022; 10(9):1766. https://doi.org/10.3390/microorganisms10091766
Chicago/Turabian StyleSamsonov, Victor V., Anna A. Kuznetsova, Julia G. Rostova, Svetlana A. Samsonova, Mikhail K. Ziyatdinov, and Michael Y. Kiriukhin. 2022. "Revealing a New Family of D-2-Hydroxyglutarate Dehydrogenases in Escherichia coli and Pantoea ananatis Encoded by ydiJ" Microorganisms 10, no. 9: 1766. https://doi.org/10.3390/microorganisms10091766






