The Use of Yeast in Biosensing
Abstract
1. Introduction
2. Yeast Biosensors in Protein–Protein Interactions
2.1. Yeast Two-Hybrid (Y2H) System
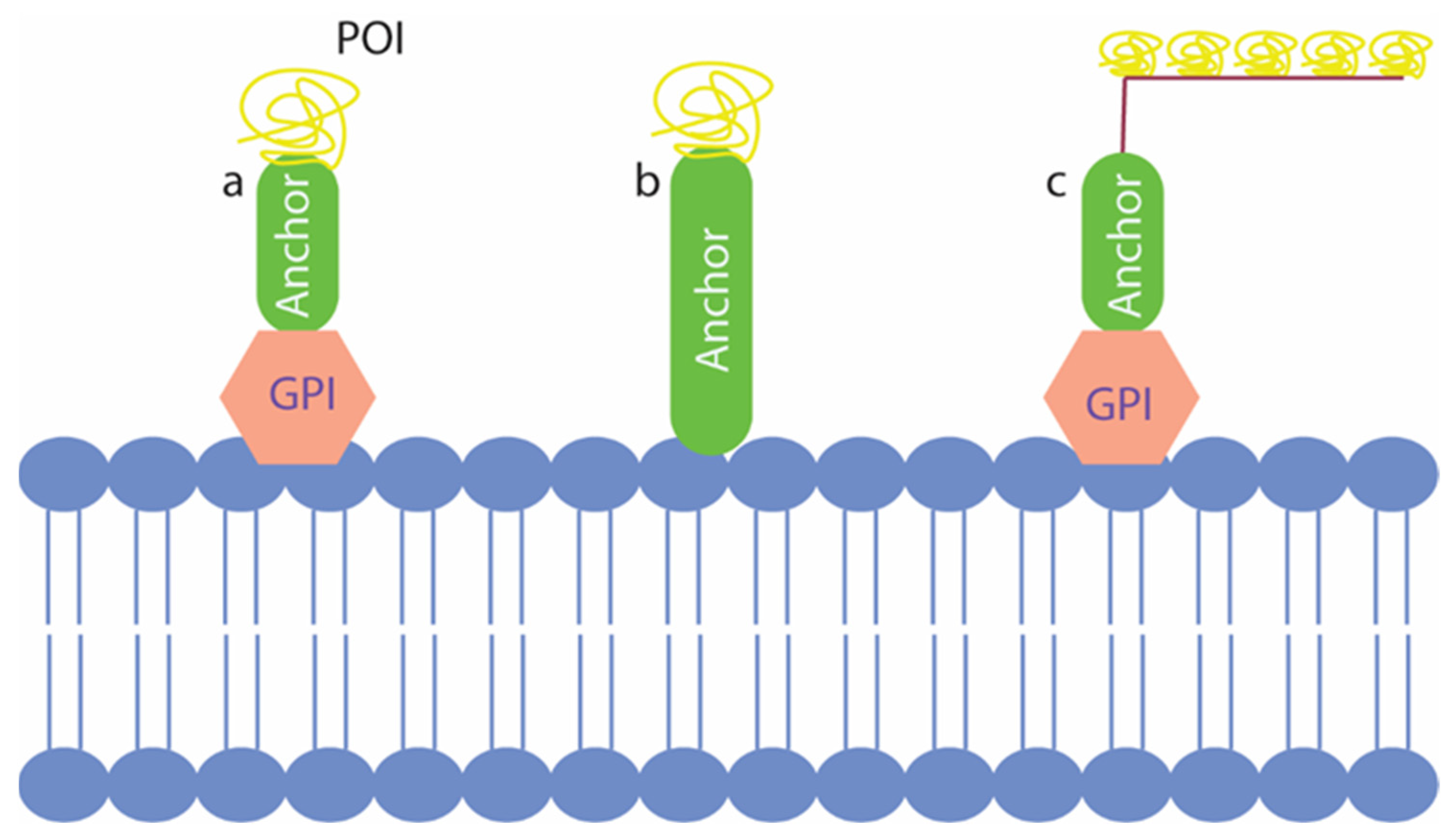
2.2. Yeast Surface Display
3. Yeast Reporters
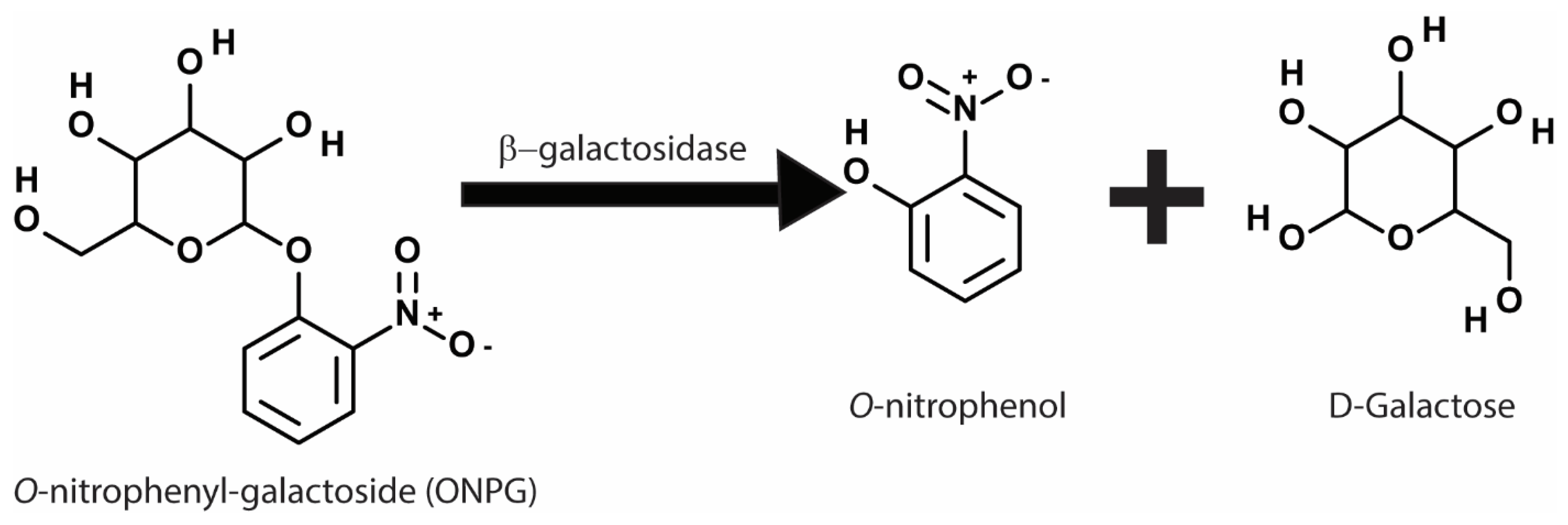
3.1. Biochemical Reporters
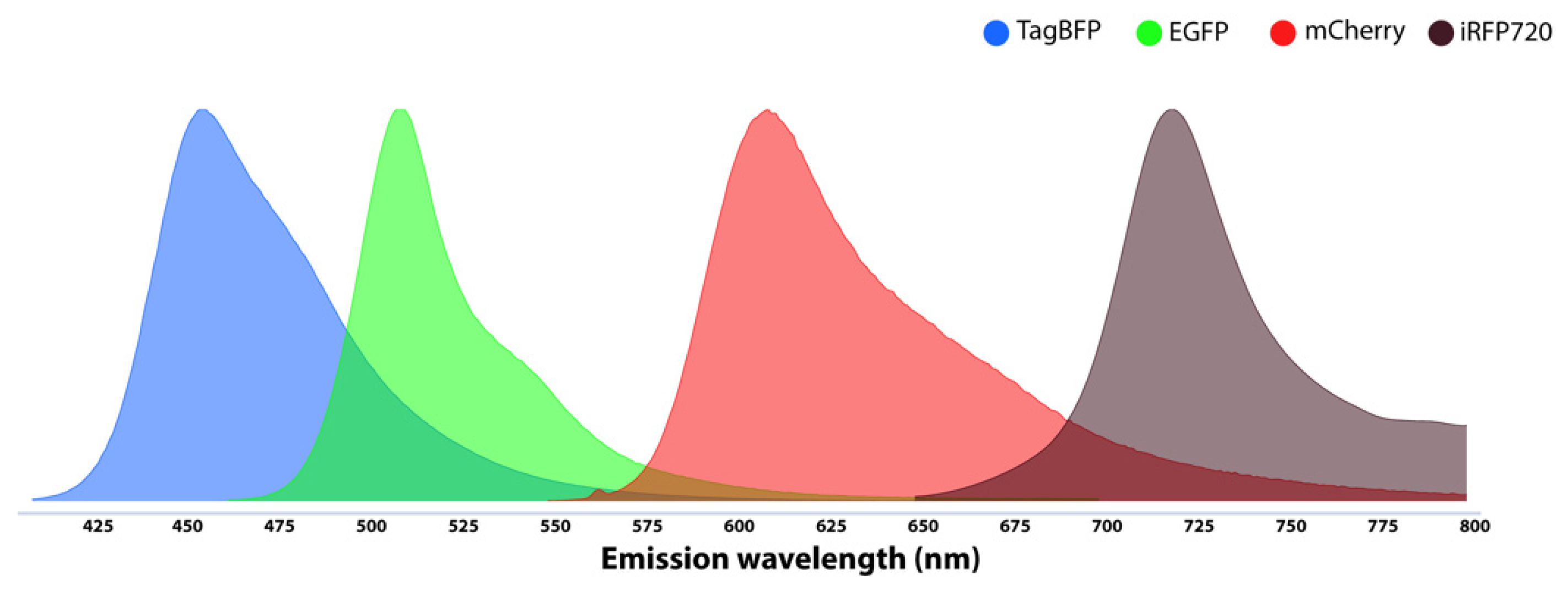
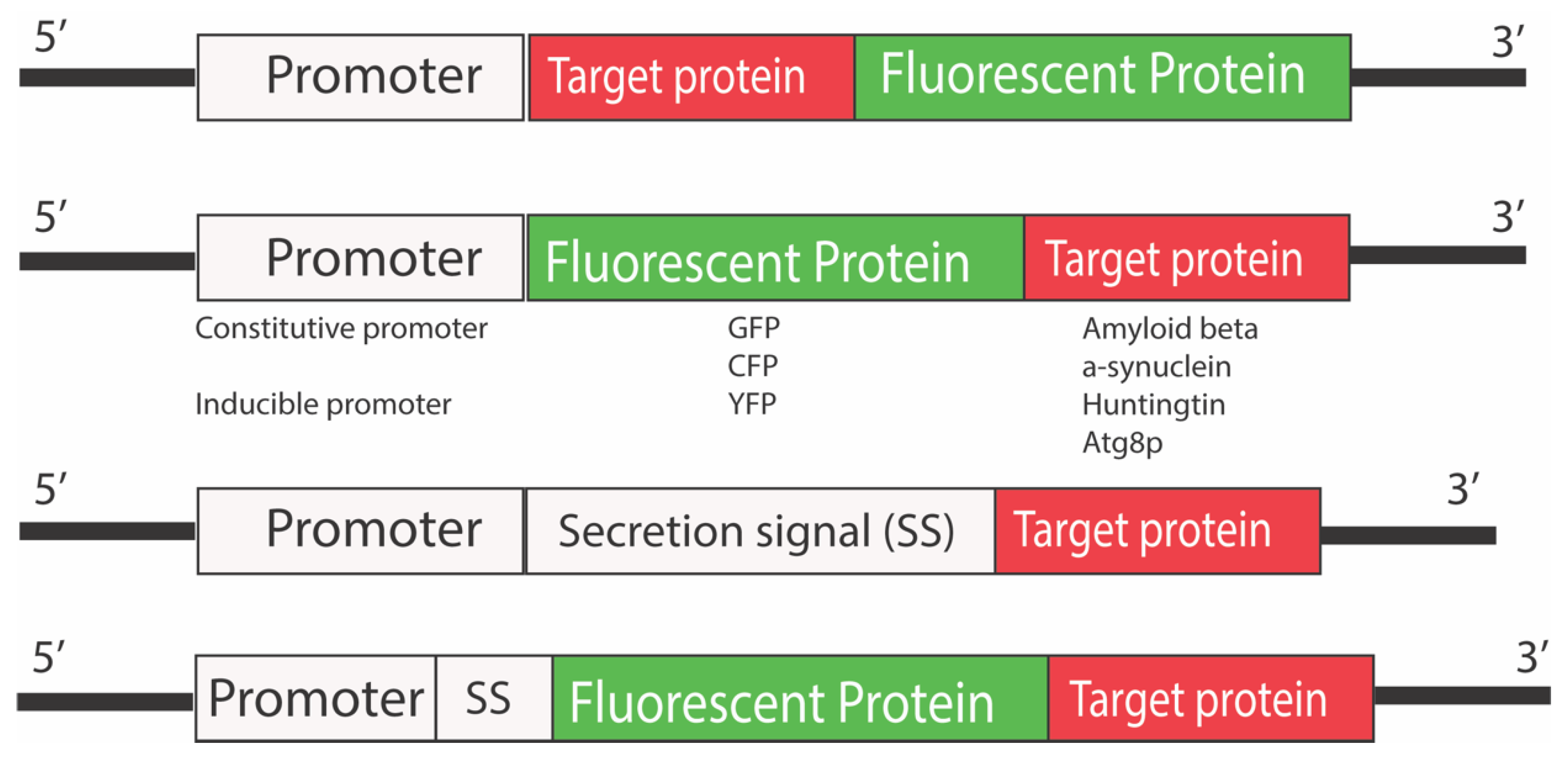
3.2. Fluorescent Reporters
3.2.1. Fluorescent Biosensors for Studying Neurodegenerative Diseases
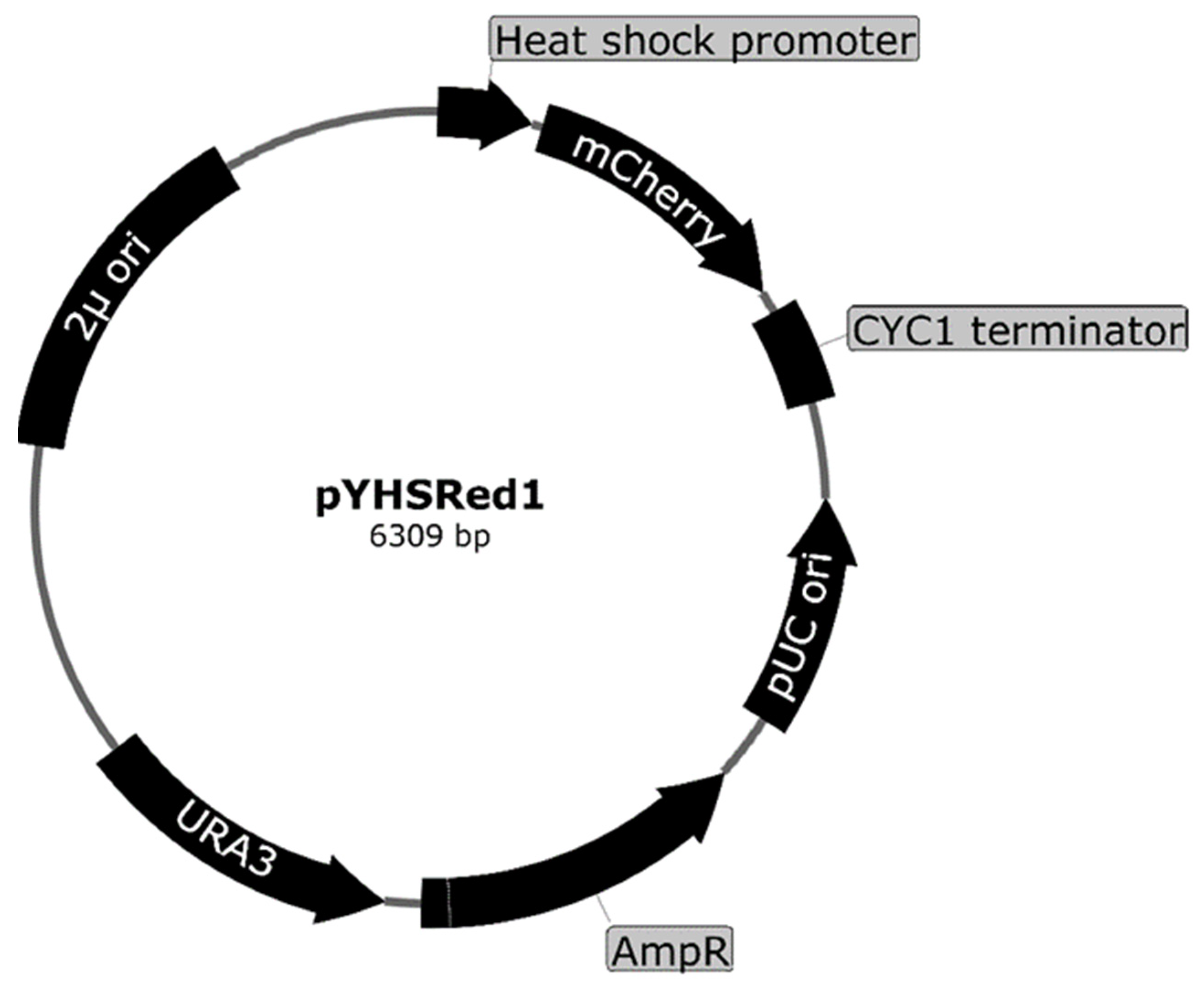
3.2.2. Fluorescence-Based Heat Shock Response Yeast Sensor
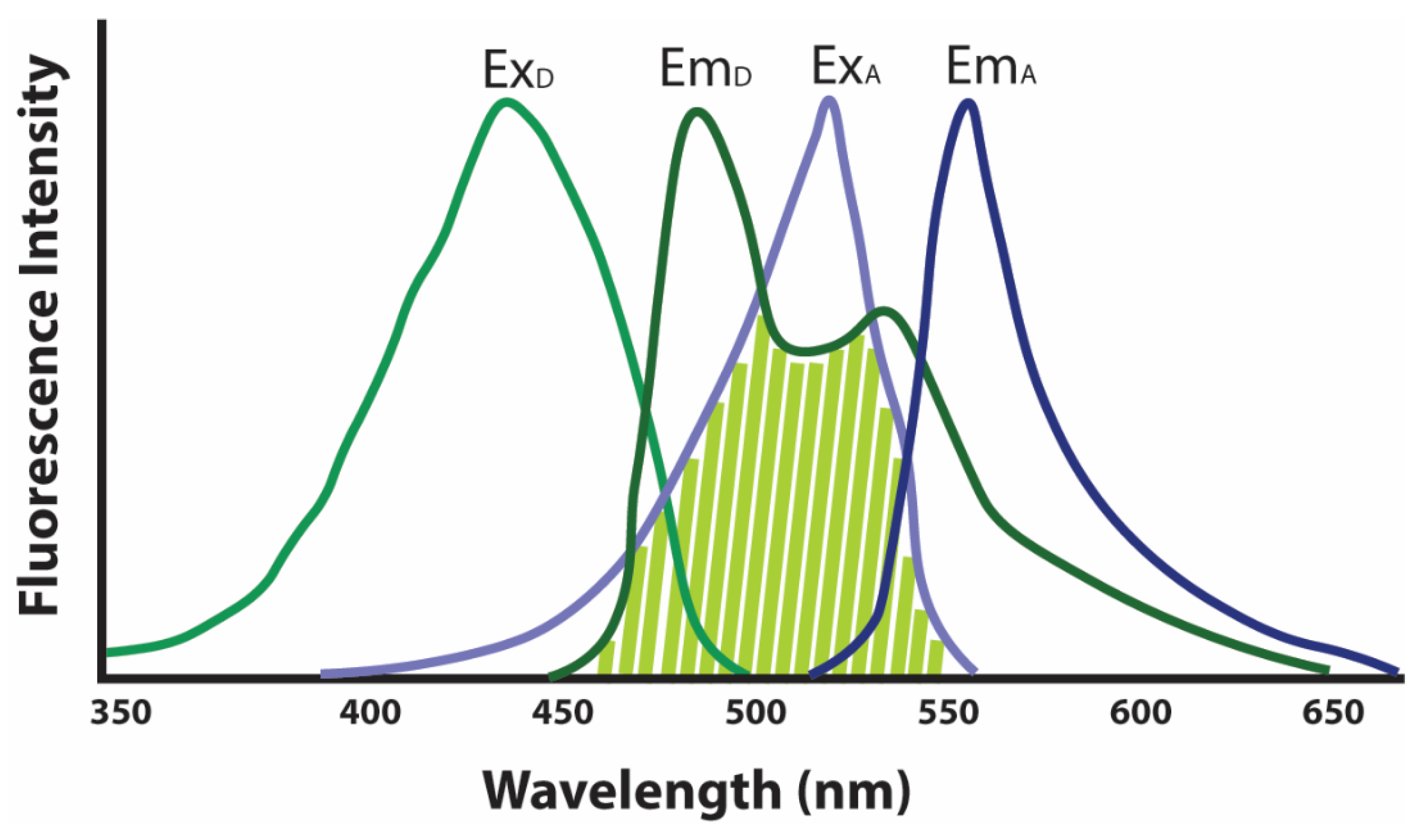
3.2.3. FRET Microscopy-Based Biosensors
3.2.4. Yeast G-Protein Coupled Receptor (GPCR)-Based Biosensors
4. Yeast Biosensors for Drug Discovery
5. Yeast Biosensors in the Environment and Biotech Industries
6. Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Macreadie, I.; Dhakal, S. The awesome power of yeast. Microbiol. Aust. 2022, 43, 19–21. [Google Scholar] [CrossRef]
- Dujon, B. The yeast genome project: What did we learn? Trends Genet. 1996, 12, 263–270. [Google Scholar] [CrossRef]
- Hazbun, T.R.; Malmström, L.; Anderson, S.; Graczyk, B.J.; Fox, B.; Riffle, M.; Sundin, B.A.; Aranda, J.D.; McDonald, W.H.; Chiu, C.-H.; et al. Assigning Function to Yeast Proteins by Integration of Technologies. Mol. Cell 2003, 12, 1353–1365. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Shields, D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef]
- Lum, P.Y.; Armour, C.D.; Stepaniants, S.B.; Cavet, G.; Wolf, M.K.; Butler, J.S.; Hinshaw, J.C.; Garnier, P.; Prestwich, G.D.; Leonardson, A.; et al. Discovering Modes of Action for Therapeutic Compounds Using a Genome-Wide Screen of Yeast Heterozygotes. Cell 2004, 116, 121–137. [Google Scholar] [CrossRef]
- Homma, T.; Iwahashi, H.; Komatsu, Y. Yeast gene expression during growth at low temperature. Cryobiology 2003, 46, 230–237. [Google Scholar] [CrossRef]
- Odani, M.; Komatsu, Y.; Oka, S.; Iwahashi, H. Screening of genes that respond to cryopreservation stress using yeast DNA microarray. Cryobiology 2003, 47, 155–164. [Google Scholar] [CrossRef]
- Cooper, S.; Shedden, K. Microarray analysis of gene expression during the cell cycle. Cell Chromosome 2003, 2, 1. [Google Scholar] [CrossRef]
- Billinton, N.; Barker, M.G.; Michel, C.E.; Knight, A.W.; Heyer, W.D.; Goddard, N.J.; Fielden, P.R.; Walmsley, R.M. Development of a green fluorescent protein reporter for a yeast genotoxicity biosensor. Biosens. Bioelectron. 1998, 13, 831–838. [Google Scholar] [CrossRef]
- Skruzny, M.; Pohl, E.; Abella, M. FRET Microscopy in Yeast. Biosensors 2019, 9, 122. [Google Scholar] [CrossRef]
- Ishii, J.; Tanaka, T.; Matsumura, S.; Tatematsu, K.; Kuroda, S.; Ogino, C.; Fukuda, H.; Kondo, A. Yeast-based fluorescence reporter assay of G protein-coupled receptor signalling for flow cytometric screening: FAR1-disruption recovers loss of episomal plasmid caused by signalling in yeast. J. Biochem. 2008, 143, 667–674. [Google Scholar] [CrossRef]
- Rao, V.S.; Srinivas, K.; Sujini, G.; Kumar, G. Protein-protein interaction detection: Methods and analysis. Int. J. Proteom. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Xing, S.; Wallmeroth, N.; Berendzen, K.W.; Grefen, C. Techniques for the Analysis of Protein-Protein Interactions in Vivo. Plant Physiol. 2016, 171, 727–758. [Google Scholar] [CrossRef]
- Ptashne, M. How eukaryotic transcriptional activators work. Nature 1988, 335, 683–689. [Google Scholar] [CrossRef]
- Brent, R.; Ptashne, M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature 1984, 312, 612–615. [Google Scholar] [CrossRef]
- Brent, R.; Ptashne, M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 1985, 43, 729–736. [Google Scholar] [CrossRef]
- Brent, R.; Finley, R.L., Jr. Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet. 1997, 31, 663–704. [Google Scholar] [CrossRef][Green Version]
- Fields, S.; Song, O.-k. A novel genetic system to detect protein–protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Brückner, A.; Polge, C.; Lentze, N.; Auerbach, D.; Schlattner, U. Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 2009, 10, 2763–2788. [Google Scholar] [CrossRef]
- Vidalain, P.O.; Jacob, Y.; Hagemeijer, M.C.; Jones, L.M.; Neveu, G.; Roussarie, J.P.; Rottier, P.J.; Tangy, F.; de Haan, C.A. A field-proven yeast two-hybrid protocol used to identify coronavirus-host protein-protein interactions. In Methods in Molecular Biology; Maier, H., Bickerton, E., Britton, P., Eds.; Humana Press: Totowa, NJ, USA, 2015; Volume 1282, pp. 213–229. [Google Scholar]
- Striebinger, H.; Koegl, M.; Bailer, S.M. A high-throughput yeast two-hybrid protocol to determine virus-host protein interactions. In Methods in Molecular Biology; Bailer, S., Lieber, D., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 1064, pp. 1–15. [Google Scholar]
- Cao, Y.-Q.; Yuan, L.; Zhao, Q.; Yuan, J.-L.; Miao, C.; Chang, Y.-F.; Wen, X.-T.; Wu, R.; Huang, X.-B.; Wen, Y.-P.; et al. Hsp40 Protein DNAJB6 Interacts with Viral NS3 and Inhibits the Replication of the Japanese Encephalitis Virus. Int. J. Mol. Sci. 2019, 20, 5719. [Google Scholar] [CrossRef]
- Gimferrer, I.; Ibáñez, A.; Farnós, M.; Sarrias, M.-R.; Fenutría, R.; Roselló, S.; Zimmermann, P.; David, G.; Vives, J.; Serra-Pagès, C.; et al. The Lymphocyte Receptor CD6 Interacts with Syntenin-1, a Scaffolding Protein Containing PDZ Domains. J. Immunol. 2005, 175, 1406. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.Q.; Wen, S.J.; Wang, H.Y.; Chen, L.Y. Screening the proteins that interact with calpain in a human heart cDNA library using a yeast two-hybrid system. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2002, 25, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Patrício, D.; Fardilha, M. The mammalian two-hybrid system as a powerful tool for high-throughput drug screening. Drug Discov. Today 2020, 25, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Stasi, M.; De Luca, M.; Bucci, C. Two-hybrid-based systems: Powerful tools for investigation of membrane traffic machineries. J. Biotechnol. 2015, 202, 105–117. [Google Scholar] [CrossRef]
- Vidal, M.; Endoh, H. Prospects for drug screening using the reverse two-hybrid system. Trends Biotechnol. 1999, 17, 374–381. [Google Scholar] [CrossRef]
- Vincent, O.; Gutierrez-Nogués, A.; Trejo-Herrero, A.; Navas, M.-A. A novel reverse two-hybrid method for the identification of missense mutations that disrupt protein–protein binding. Sci. Rep. 2020, 10, 21043. [Google Scholar] [CrossRef]
- Huang, J.; Schreiber, S.L. A yeast genetic system for selecting small molecule inhibitors of protein-protein interactions in nanodroplets. Proc. Natl. Acad. Sci. USA 1997, 94, 13396–13401. [Google Scholar] [CrossRef]
- Teymennet-Ramírez, K.V.; Martínez-Morales, F.; Trejo-Hernández, M.R. Yeast Surface Display System: Strategies for Improvement and Biotechnological Applications. Front. Bioeng. Biotechnol. 2021, 9, 794742. [Google Scholar] [CrossRef]
- Kondo, A.; Ueda, M. Yeast cell-surface display—Applications of molecular display. Appl. Microbiol. Biotechnol. 2004, 64, 28–40. [Google Scholar] [CrossRef]
- Yang, N.; Yu, Z.; Jia, D.; Xie, Z.; Zhang, K.; Xia, Z.; Lei, L.; Qiao, M. The contribution of Pir protein family to yeast cell surface display. Appl. Microbiol. Biotechnol. 2014, 98, 2897–2905. [Google Scholar] [CrossRef]
- Kajiwara, K.; Aoki, W.; Ueda, M. Evaluation of the yeast surface display system for screening of functional nanobodies. AMB Express 2020, 10, 51. [Google Scholar] [CrossRef]
- Gai, S.A.; Wittrup, K.D. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 2007, 17, 467–473. [Google Scholar] [CrossRef]
- Tabañag, I.D.F.; Chu, I.M.; Wei, Y.-H.; Tsai, S.-L. The Role of Yeast-Surface-Display Techniques in Creating Biocatalysts for Consolidated BioProcessing. Catalysts 2018, 8, 94. [Google Scholar] [CrossRef]
- Lozančić, M.; Hossain, S.A.; Mrša, V.; Teparić, R. Surface Display—An Alternative to Classic Enzyme Immobilization. Catalysts 2019, 9, 728. [Google Scholar] [CrossRef]
- Zhao, J.-Z.; Xu, L.-M.; Liu, M.; Cao, Y.-S.; LaPatra, S.E.; Yin, J.-S.; Liu, H.-B.; Lu, T.-Y. Preliminary study of an oral vaccine against infectious hematopoietic necrosis virus using improved yeast surface display technology. Mol. Immunol. 2017, 85, 196–204. [Google Scholar] [CrossRef]
- Fan, S.; Liang, B.; Xiao, X.; Bai, L.; Tang, X.; Lojou, E.; Cosnier, S.; Liu, A. Controllable Display of Sequential Enzymes on Yeast Surface with Enhanced Biocatalytic Activity toward Efficient Enzymatic Biofuel Cells. J. Am. Chem. Soc. 2020, 142, 3222–3230. [Google Scholar] [CrossRef]
- Bidlingmaier, S.; Liu, B. Utilizing Yeast Surface Human Proteome Display Libraries to Identify Small Molecule-Protein Interactions. In Yeast Surface Display: Methods, Protocols, and Applications; Liu, B., Ed.; Springer: New York, NY, USA, 2015; pp. 203–214. [Google Scholar]
- Li, J.; Wuethrich, A.; Edwardraja, S.; Lobb, R.J.; Puttick, S.; Rose, S.; Howard, C.B.; Trau, M. Amplification-Free SARS-CoV-2 Detection Using Nanoyeast-scFv and Ultrasensitive Plasmonic Nanobox-Integrated Nanomixing Microassay. Anal. Chem. 2021, 93, 10251–10260. [Google Scholar] [CrossRef]
- Francino-Urdaniz, I.M.; Steiner, P.J.; Kirby, M.B.; Zhao, F.; Haas, C.M.; Barman, S.; Rhodes, E.R.; Leonard, A.C.; Peng, L.; Sprenger, K.G.; et al. One-shot identification of SARS-CoV-2 S RBD escape mutants using yeast screening. Cell Rep. 2021, 36, 109627. [Google Scholar] [CrossRef]
- Uchański, T.; Zögg, T.; Yin, J.; Yuan, D.; Wohlkönig, A.; Fischer, B.; Rosenbaum, D.M.; Kobilka, B.K.; Pardon, E.; Steyaert, J. An improved yeast surface display platform for the screening of nanobody immune libraries. Sci. Rep. 2019, 9, 382. [Google Scholar] [CrossRef]
- Adeniran, A.; Sherer, M.; Tyo, K.E. Yeast-based biosensors: Design and applications. FEMS Yeast Res. 2015, 15, 1–15. [Google Scholar] [CrossRef]
- Martin-Yken, H. Yeast-based biosensors: Current applications and new developments. Biosensors 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Macreadie, I. Protein Homeostasis Networks and the Use of Yeast to Guide Interventions in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 8014. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Macreadie, I. Chapter 1—Developing systems in yeast to address Alzheimer’s disease. In Methods in Microbiology; Gurtler, V., Ed.; Academic Press: Cambridge, MA, USA, 2022; Volume 51, pp. 1–43. [Google Scholar]
- Laskowska, E.; Matuszewska, E.; Kuczynska-Wisnik, D. Small Heat Shock Proteins and Protein-Misfolding Diseases. Curr. Pharm. Biotechnol. 2010, 11, 146–157. [Google Scholar] [CrossRef]
- Dhakal, S. ‘The awesome power of yeast’ in Alzheimer’s disease research. Microbiol. Aust. 2021, 42, 130–133. [Google Scholar] [CrossRef]
- Dhakal, S.; Macreadie, I. Tyramine and Amyloid Beta 42: A Toxic Synergy. Biomedicines 2020, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Rupp, S. LacZ assays in yeast. In Methods in Enzymology; Guthrie, C., Fink, G.R., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 350, pp. 112–131. [Google Scholar]
- Serebriiskii, I.G.; Golemis, E.A. Uses of lacZ to Study Gene Function: Evaluation of β-Galactosidase Assays Employed in the Yeast Two-Hybrid System. Anal. Biochem. 2000, 285, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Caine, J.; Sankovich, S.; Antony, H.; Waddington, L.; Macreadie, P.; Varghese, J.; Macreadie, I. Alzheimer’s Aβ fused to green fluorescent protein induces growth stress and a heat shock response. FEMS Yeast Res. 2007, 7, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Orij, R.; Brul, S.; Smits, G.J. Intracellular pH is a tightly controlled signal in yeast. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2011, 1810, 933–944. [Google Scholar] [CrossRef]
- Rosado, C.J.; Mijaljica, D.; Hatzinisiriou, I.; Prescott, M.; Devenish, R.J. Rosella: A fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 2008, 4, 205–213. [Google Scholar] [CrossRef]
- Luu, Y.N.; Macreadie, I. Development of Convenient System for Detecting Yeast Cell Stress, Including That of Amyloid Beta. Int. J. Mol. Sci. 2018, 19, 2136. [Google Scholar] [CrossRef]
- Pronk, J.T. Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 2002, 68, 2095–2100. [Google Scholar] [CrossRef]
- Williamson, D.H.; Fennell, D.J. [62] Visualization of yeast mitochondrial dna with the fluorescent stain “DAPI”. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1979; Volume 56, pp. 728–733. [Google Scholar]
- Darzynkiewicz, Z. Critical aspects in analysis of cellular DNA content. In Current Protocols Cytometry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; Chapter 7; pp. 7.2.1–7.2.8. [Google Scholar]
- Ferro, A.; Mestre, T.; Carneiro, P.; Sahumbaiev, I.; Seruca, R.; Sanches, J.M. Blue intensity matters for cell cycle profiling in fluorescence DAPI-stained images. Lab. Investig. 2017, 97, 615–625. [Google Scholar] [CrossRef]
- Christopher, J.A.; Stadler, C.; Martin, C.E.; Morgenstern, M.; Pan, Y.; Betsinger, C.N.; Rattray, D.G.; Mahdessian, D.; Gingras, A.-C.; Warscheid, B.; et al. Subcellular proteomics. Nat. Rev. Methods Primers 2021, 1, 32. [Google Scholar] [CrossRef]
- Marshall, J.; Molloy, R.; Moss, G.W.; Howe, J.R.; Hughes, T.E. The jellyfish green fluorescent protein: A new tool for studying ion channel expression and function. Neuron 1995, 14, 211–215. [Google Scholar] [CrossRef]
- Lambert, T.J. FPbase: A community-editable fluorescent protein database. Nat. Methods 2019, 16, 277–278. [Google Scholar] [CrossRef]
- Misra, A.; Chauhan, S.; Tanwar, P.; Singh, S.; Sharma, S.; Rai, S. Real-Time Compensation in Flow Cytometry: A Real Need of Time. Indian J. Hematol. Blood Transfus. 2018, 34, 585–588. [Google Scholar] [CrossRef]
- Mcdonald, J.B.; Dhakal, S.; Macreadie, I.G. Yeast contributions to Alzheimer’s Disease. J. Hum. Clin. Genet. 2020, 2. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Grizel, A.V.; Rubel, A.A.; Zelinsky, A.A.; Chandramowlishwaran, P.; Chernova, T.A. Application of yeast to studying amyloid and prion diseases. In Advances in Genetics; Kumar, D., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 105, pp. 293–380. [Google Scholar]
- Bayer, T.; Wirths, O. Intracellular accumulation of amyloid-beta—A predictor for synaptic dysfunction and neuron loss in Alzheimer’s disease. Front. Aging Neurosci. 2010, 2, 8. [Google Scholar] [CrossRef]
- Dhakal, S.; Kushairi, N.; Phan, C.W.; Adhikari, B.; Sabaratnam, V.; Macreadie, I. Dietary Polyphenols: A Multifactorial Strategy to Target Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 5090. [Google Scholar] [CrossRef]
- Dhakal, S.; Ramsland, P.A.; Adhikari, B.; Macreadie, I. Trans-Chalcone Plus Baicalein Synergistically Reduce Intracellular Amyloid Beta (Aβ42) and protect from Aβ42 Induced Oxidative Damage in Yeast Models of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 9456. [Google Scholar] [CrossRef]
- Dhakal, S.; Subhan, M.; Fraser, J.M.; Gardiner, K.; Macreadie, I. Simvastatin Efficiently Reduces Levels of Alzheimer’s Amyloid Beta in Yeast. Int. J. Mol. Sci. 2019, 20, 3531. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, P.R.; Verdile, G.; Barr, R.K.; Gupta, V.; Steele, J.W.; Lachenmayer, M.L.; Yue, Z.; Ehrlich, M.E.; Petsko, G.; Ju, S.; et al. Latrepirdine (dimebon) enhances autophagy and reduces intracellular GFP-Aβ42 levels in yeast. J. Alzheimer’s Dis. 2012, 32, 949–967. [Google Scholar] [CrossRef]
- Antony, H.; Macreadie, I. Alzheimer’s Disease, the Importance of AB and the Hopes for Chemo Preventatives. In Medicinal Chemistry Research Progress; Nova Science: New York, NY, USA, 2009; pp. 7–65. [Google Scholar]
- Dokladny, K.; Myers, O.B.; Moseley, P.L. Heat shock response and autophagy—cooperation and control. Autophagy 2015, 11, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Mizukami, Y.; Sakurai, H. Identification of a Novel Class of Target Genes and a Novel Type of Binding Sequence of Heat Shock Transcription Factor in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 11911–11919. [Google Scholar] [CrossRef]
- Botman, D.; de Groot, D.H.; Schmidt, P.; Goedhart, J.; Teusink, B. In vivo characterisation of fluorescent proteins in budding yeast. Sci. Rep. 2019, 9, 2234. [Google Scholar] [CrossRef] [PubMed]
- Margineanu, A.; Chan, J.J.; Kelly, D.J.; Warren, S.C.; Flatters, D.; Kumar, S.; Katan, M.; Dunsby, C.W.; French, P.M.W. Screening for protein-protein interactions using Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM). Sci. Rep. 2016, 6, 28186. [Google Scholar] [CrossRef] [PubMed]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef] [PubMed]
- Bajar, B.T.; Wang, E.S.; Lam, A.J.; Kim, B.B.; Jacobs, C.L.; Howe, E.S.; Davidson, M.W.; Lin, M.Z.; Chu, J. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 2016, 6, 20889. [Google Scholar] [CrossRef]
- Damelin, M.; Silver, P.A. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell 2000, 5, 133–140. [Google Scholar] [CrossRef]
- McDonald, N.A.; Lind, A.L.; Smith, S.E.; Li, R.; Gould, K.L. Nanoscale architecture of the Schizosaccharomyces pombe contractile ring. eLife 2017, 6, e288652017. [Google Scholar] [CrossRef]
- Skruzny, M.; Pohl, E.; Gnoth, S.; Malengo, G.; Sourjik, V. The protein architecture of the endocytic coat analyzed by FRET microscopy. Mol. Syst. Biol. 2020, 16, e9009. [Google Scholar] [CrossRef]
- Viswanath, S.; Bonomi, M.; Kim, S.J.; Klenchin, V.A.; Taylor, K.C.; Yabut, K.C.; Umbreit, N.T.; Van Epps, H.A.; Meehl, J.; Jones, M.H.; et al. The molecular architecture of the yeast spindle pole body core determined by Bayesian integrative modeling. Mol. Biol. Cell 2017, 28, 3298–3314. [Google Scholar] [CrossRef]
- Miller, A.; Chen, J.; Takasuka, T.E.; Jacobi, J.L.; Kaufman, P.D.; Irudayaraj, J.M.; Kirchmaier, A.L. Proliferating cell nuclear antigen (PCNA) is required for cell cycle-regulated silent chromatin on replicated and nonreplicated genes. J. Biol. Chem. 2010, 285, 35142–35154. [Google Scholar] [CrossRef]
- Bhaumik, S.R.; Raha, T.; Aiello, D.P.; Green, M.R. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004, 18, 333–343. [Google Scholar] [CrossRef]
- Dye, B.T.; Schell, K.; Miller, D.J.; Ahlquist, P. Detecting protein-protein interaction in live yeast by flow cytometry. Cytometry. Part A J. Int. Soc. Anal. Cytol. 2005, 63, 77–86. [Google Scholar] [CrossRef]
- Ernst, S.; Batisse, C.; Zarrabi, N.; Böttcher, B.; Börsch, M. Regulatory assembly of the vacuolar proton pump VoV1-ATPase in yeast cells by FLIM-FRET. Proc. SPIE 2010, 7569, 113–120. [Google Scholar]
- Luo, J.; Sun, P.; Siwko, S.; Liu, M.; Xiao, J. The role of GPCRs in bone diseases and dysfunctions. Bone Res. 2019, 7, 19. [Google Scholar] [CrossRef]
- Takeda, S.; Kadowaki, S.; Haga, T.; Takaesu, H.; Mitaku, S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002, 520, 97–101. [Google Scholar] [CrossRef]
- Pierce, K.L.; Premont, R.T.; Lefkowitz, R.J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 639–650. [Google Scholar] [CrossRef]
- Rosenbaum, D.M.; Rasmussen, S.G.F.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, Y.; Lu, R.; Xin, F.; Liu, G. Synthetic biology applications of the yeast mating signal pathway. Trends Biotechnol. 2022, 40, 620–631. [Google Scholar] [CrossRef]
- Versele, M.; Lemaire, K.; Thevelein, J.M. Sex and sugar in yeast: Two distinct GPCR systems. EMBO Rep. 2001, 2, 574–579. [Google Scholar] [CrossRef]
- Liu, R.; Wong, W.; Ijzerman, A.P. Human G protein-coupled receptor studies in Saccharomyces cerevisiae. Biochem. Pharmacol. 2016, 114, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Vavitsas, K. Turning G protein-coupled receptors into tunable biosensors. Synth. Biol. 2019, 4, ysz011. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Bhattacharyya, S.; Peralta-Yahya, P. GPCR-Based Chemical Biosensors for Medium-Chain Fatty Acids. ACS Synth. Biol. 2015, 4, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Conklin, B.R.; Blin, N.; Yun, J.; Wess, J. Identification of a receptor/G-protein contact site critical for signaling specificity and G-protein activation. Proc. Natl. Acad. Sci. USA 1995, 92, 11642–11646. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, K.; Leelahakorn, N.; Almeida, A.; Zhao, Y.; Hansen, L.R.; Nikolajsen, I.E.; Andersen, J.B.; Givskov, M.; Staerk, D.; Bak, S.; et al. A GPCR-based yeast biosensor for biomedical, biotechnological, and point-of-use cannabinoid determination. Nat. Commun. 2022, 13, 3664. [Google Scholar] [CrossRef] [PubMed]
- Bean, B.D.M.; Mulvihill, C.J.; Garge, R.K.; Boutz, D.R.; Rousseau, O.; Floyd, B.M.; Cheney, W.; Gardner, E.C.; Ellington, A.D.; Marcotte, E.M.; et al. Functional expression of opioid receptors and other human GPCRs in yeast engineered to produce human sterols. Nat. Commun. 2022, 13, 2882. [Google Scholar] [CrossRef]
- Lengger, B.; Jensen, M.K. Engineering G protein-coupled receptor signalling in yeast for biotechnological and medical purposes. FEMS Yeast Res. 2020, 20, foz087. [Google Scholar] [CrossRef]
- Lengger, B.; Hoch-Schneider, E.E.; Jensen, C.N.; Jakočiūnas, T.; Petersen, A.A.; Frimurer, T.M.; Jensen, E.D.; Jensen, M.K. Serotonin G Protein-Coupled Receptor-Based Biosensing Modalities in Yeast. ACS Sens. 2022, 7, 1323–1335. [Google Scholar] [CrossRef]
- Hughes, J.; Rees, S.; Kalindjian, S.; Philpott, K. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Lentze, N.; Auerbach, D. The yeast two-hybrid system and its role in drug discovery. Expert Opin. Ther. Targets 2008, 12, 505–515. [Google Scholar] [CrossRef]
- McMahon, C.; Baier, A.S.; Pascolutti, R.; Wegrecki, M.; Zheng, S.; Ong, J.X.; Erlandson, S.C.; Hilger, D.; Rasmussen, S.G.F.; Ring, A.M.; et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 2018, 25, 289–296. [Google Scholar] [CrossRef]
- Castelli, L.A.; Nguyen, N.P.; Macreadie, I.G. Sulfa drug screening in yeast: Fifteen sulfa drugs compete with p-aminobenzoate in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2001, 199, 181–184. [Google Scholar] [CrossRef]
- Porzoor, A.; Macreadie, I. Yeast as a Model for Studies on Aβ Aggregation Toxicity in Alzheimer’s Disease, Autophagic Responses, and Drug Screening. In Systems Biology of Alzheimer’s Disease; Castrillo, J.I., Oliver, S.G., Eds.; Springer: New York, NY, USA, 2016; pp. 217–226. [Google Scholar]
- McDonald, J.B.; Dhakal, S.; Macreadie, I. A Toxic Synergy between Aluminium and Amyloid Beta in Yeast. Int. J. Mol. Sci. 2021, 22, 1835. [Google Scholar] [CrossRef]
- Yang, H.-C.; Simon, V.; Swayne, T.C.; Pon, L. Chapter 19 Visualization of mitochondrial movement in yeast. In Methods in Cell Biology; Academic Press: Cambridge, MA, USA, 2001; Volume 65, pp. 333–351. [Google Scholar]
- Davey, H.M.; Hexley, P. Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environ. Microbiol. 2011, 13, 163–171. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Dhakal, S.; Macreadie, I. Potential contributions of trace amines in Alzheimer’s disease and therapeutic prospects. Neural Regen. Res. 2021, 16, 1394–1396. [Google Scholar]
- Trosok, S.P.; Driscoll, B.T.; Luong, J.H. Mediated microbial biosensor using a novel yeast strain for wastewater BOD measurement. Appl. Microbiol. Biotechnol. 2001, 56, 550–554. [Google Scholar] [CrossRef]
- Hikuma, M.; Suzuki, H.; Yasuda, Y.; Karube, I.; Suzuki, S. Amperometric estimation of BOD by using living immobilized yeasts. Eur. J. Appl. Microbiol. Biotechnol. 1979, 8, 289–297. [Google Scholar] [CrossRef]
- Jarque, S.; Bittner, M.; Blaha, L.; Hilscherova, K. Yeast Biosensors for Detection of Environmental Pollutants: Current State and Limitations. Trends Biotechnol. 2016, 34, 408–419. [Google Scholar] [CrossRef]
- Jouanneau, S.; Recoules, L.; Durand, M.J.; Boukabache, A.; Picot, V.; Primault, Y.; Lakel, A.; Sengelin, M.; Barillon, B.; Thouand, G. Methods for assessing biochemical oxygen demand (BOD): A review. Water Res. 2014, 49, 62–82. [Google Scholar] [CrossRef] [PubMed]
- Tag, K.; Riedel, K.; Bauer, H.-J.; Hanke, G.; Baronian, K.H.R.; Kunze, G. Amperometric detection of Cu2+ by yeast biosensors using flow injection analysis (FIA). Sens. Actuators B Chem. 2007, 122, 403–409. [Google Scholar] [CrossRef]
- Matsuura, H.; Yamamoto, Y.; Muraoka, M.; Akaishi, K.; Hori, Y.; Uemura, K.; Tsuji, N.; Harada, K.; Hirata, K.; Bamba, T.; et al. Development of surface-engineered yeast cells displaying phytochelatin synthase and their application to cadmium biosensors by the combined use of pyrene-excimer fluorescence. Biotechnol. Prog. 2013, 29, 1197–1202. [Google Scholar] [CrossRef]
- Park, J.N.; Sohn, M.J.; Oh, D.B.; Kwon, O.; Rhee, S.K.; Hur, C.G.; Lee, S.Y.; Gellissen, G.; Kang, H.A. Identification of the cadmium-inducible Hansenula polymorpha SEO1 gene promoter by transcriptome analysis and its application to whole-cell heavy-metal detection systems. Appl. Environ. Microbiol. 2007, 73, 5990–6000. [Google Scholar] [CrossRef]
- Richter, I.; Fidler, A.E. Detection of marine microalgal biotoxins using bioassays based on functional expression of tunicate xenobiotic receptors in yeast. Toxicon 2015, 95, 13–22. [Google Scholar] [CrossRef]
- Schappert, K.T.; Khachatourians, G.G. A yeast bioassay for T-2 toxin. J. Microbiol. Methods 1984, 3, 43–46. [Google Scholar] [CrossRef]
- Engler, K.H.; Coker, R.; Evans, I.H. A novel colorimetric yeast bioassay for detecting trichothecene mycotoxins. J. Microbiol. Methods 1999, 35, 207–218. [Google Scholar] [CrossRef]
- Mitterbauer, R.; Weindorfer, H.; Safaie, N.; Krska, R.; Lemmens, M.; Ruckenbauer, P.; Kuchler, K.; Adam, G. A Sensitive and Inexpensive Yeast Bioassay for the Mycotoxin Zearalenone and Other Compounds with Estrogenic Activity. Appl. Environ. Microbiol. 2003, 69, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Varela, J.N.; Yadav, V.G. A Pichia biosensor for high-throughput analyses of compounds that can influence mosquito behavior. Microbiol. Open 2021, 10, e11392021. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Zhai, H.; Hou, J. Biosensors design in yeast and applications in metabolic engineering. FEMS Yeast Res. 2019, 19, foz082. [Google Scholar] [CrossRef]
- Ehrenworth, A.M.; Claiborne, T.; Peralta-Yahya, P. Medium-Throughput Screen of Microbially Produced Serotonin via a G-Protein-Coupled Receptor-Based Sensor. Biochemistry 2017, 56, 5471–5475. [Google Scholar] [CrossRef]
- Chou, H.H.; Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef]
- Laurent, J.M.; Young, J.H.; Kachroo, A.H.; Marcotte, E.M. Efforts to make and apply humanized yeast. Brief. Funct. Genom. 2016, 15, 155–163. [Google Scholar] [CrossRef]


| Fluorescent Protein | Organism | Molecular Weight (kDa) | Excitation Maxima (nm) | Emission Maxima (nm) | Brightness | pKa |
|---|---|---|---|---|---|---|
| mTagBFP2 | Entacmaea quadricolor | 26.7 | 399 | 454 | 32.38 | 2.7 |
| CFP | Aequorea victoria | 26.9 | 456 | 480 | NA | NA |
| Cerulean | Aequorea victoria | 26.8 | 433 | 475 | 26.66 | 4.7 |
| mTurquoise2 | Aequorea victoria | 26.9 | 434 | 474 | 27.90 | 3.1 |
| mKeima | Montipora sp. 20 | 25.1 | 440 | 620 | 3.46 | 6.5 |
| EGFP | Aequorea victoria | 26.9 | 488 | 507 | 33.54 | 6.0 |
| EYFP | Aequorea victoria | 27.0 | 513 | 527 | 44.89 | 6.9 |
| Venus | Aequorea victoria | 26.8 | 515 | 528 | 52.55 | 6.0 |
| mKO1 | Verrillofungia concinna | 24.5 | 548 | 549 | 30.96 | 5.0 |
| tDimer2 | Discosoma spp. | 52.7 | 552 | 579 | 81.60 | 4.8 |
| tdTomato | Discosoma spp. | 54.2 | 554 | 581 | 95.22 | 4.7 |
| DsRed/RFP | Discosoma spp. | 25.9 | 558 | 583 | 49.30 | NA |
| mRuby2 | Entacmaea quadricolor | 26.5 | 559 | 600 | 42.94 | 4.4 |
| mStrawberry | Discosoma spp. | 26.6 | 574 | 596 | 26.10 | 4.5 |
| mCherry | Discosoma spp. | 26.7 | 587 | 610 | 15.84 | 4.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhakal, S.; Macreadie, I. The Use of Yeast in Biosensing. Microorganisms 2022, 10, 1772. https://doi.org/10.3390/microorganisms10091772
Dhakal S, Macreadie I. The Use of Yeast in Biosensing. Microorganisms. 2022; 10(9):1772. https://doi.org/10.3390/microorganisms10091772
Chicago/Turabian StyleDhakal, Sudip, and Ian Macreadie. 2022. "The Use of Yeast in Biosensing" Microorganisms 10, no. 9: 1772. https://doi.org/10.3390/microorganisms10091772
APA StyleDhakal, S., & Macreadie, I. (2022). The Use of Yeast in Biosensing. Microorganisms, 10(9), 1772. https://doi.org/10.3390/microorganisms10091772






