Comparison of Virulence between Two Main Clones (ST11 and ST307) of Klebsiella pneumoniae Isolates from South Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. K. pneumoniae Isolates and Genotyping
2.2. Antibiotic Susceptibility Testing and String Test
2.3. Serum Resistance Assay
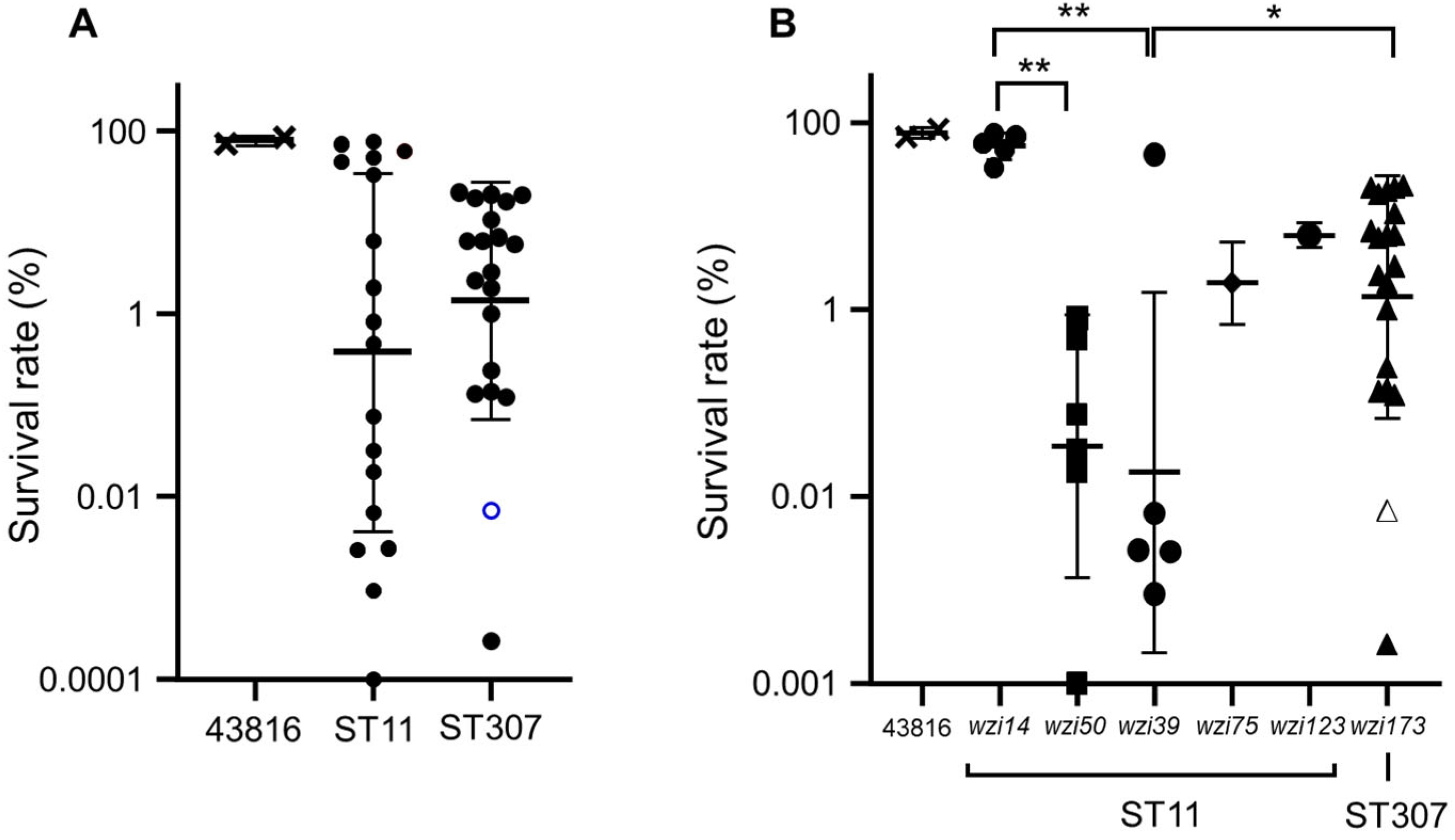
2.4. Macrophage Infection Assay
2.5. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updates 2016, 29, 30–46. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, G.R.; Jeong, J.; Kim, S.; Shin, J.H. Prevalence and characteristics of carbapenemase-producing Enterobacteriaceae in three tertiary-care Korean university hospitals between 2017–2018. Jpn J. Infect. Dis. 2020, 73, 431–436. [Google Scholar] [CrossRef]
- Rhee, J.Y.; Park, Y.K.; Shin, J.Y.; Choi, J.Y.; Lee, M.Y.; Peck, K.R.; Song, J.H.; Ko, K.S. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob. Agents Chemother. 2010, 54, 2278–2279. [Google Scholar] [CrossRef]
- D’Souza, R.; Pinto, N.A.; Hwang, I.; Younjee, H.; Cho, Y.; Kim, H.; Yong, D.; Choi, J.; Lee, K.; Chong, Y. Molecular epidemiology and resistome analysis of multidrug-resistant ST11 Klebsiella pneumoniae strain containing multiple copies of extended-spectrum β-lactamase genes using whole-genome sequencing. New Microbiol. 2017, 40, 38–44. [Google Scholar]
- Kim, J.O.; Song, S.A.; Yoon, E.J.; Shin, J.H.; Lee, H.; Jeong, S.H.; Lee, K. Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagn. Microbiol. Infect. Dis 2017, 87, 343–348. [Google Scholar] [CrossRef]
- Yoon, E.J.; Kim, J.O.; Kim, D.; Lee, H.; Yang, J.W.; Lee, K.J.; Jeong, S.H. Klebsiella pneumoniae carbapenemase producers in South Korea between 2013 and 2015. Front. Microbiol. 2018, 9, 56. [Google Scholar] [CrossRef]
- Ko, K.S. The contribution of capsule polysaccharide genes to virulence of Klebsiella pneumoniae. Virulence 2017, 8, 485–486. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef]
- Kim, J.H.; Cho, Y.Y.; Choi, J.Y.; Wi, Y.M.; Ko, K.S. Two distinct genotypes of KPC-2-producing Klebsiella pneumoniae isolates from South Korea. Antibiotics 2021, 10, 911. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Roderova, M.; Kolar, M. Primer evaluation for PCR and its application for detection of carbaepenmase Enterobacteriaceae. Jundishapur. J. Microbiol. 2016, 9, e29314. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brisse, S.; Passet, V.; Haugaard, A.B.; Babosan, A.; Kassis-Chikhani, N.; Struve, C.; Decré, D. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 2013, 51, 4073–4078. [Google Scholar] [CrossRef] [PubMed]
- Compain, F.; Babosan, A.; Brisse, S.; Genel, N.; Audo, J.; Ailloud, F.; Kassis-Chikhani, N.; Arlet, G.; Decré, D. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J. Clin. Microbiol. 2014, 52, 4377–4380. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.R.; Molton, J.S.; Wyres, K.L.; Gorrie, C.; Wong, J.; Hoh, C.H.; Teo, J.; Kalimunddin, S.; Lye, D.C.; Archuleta, S.; et al. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci. Rep. 2016, 6, 29316. [Google Scholar] [CrossRef]
- Khaertynov, K.; Anokhin, V.A.; Rizvanov, A.A.; Davidyuk, Y.N.; Semyeonova, D.R.; Lubin, S.A.; Skvortsova, N.N. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front. Med. 2018, 5, 225. [Google Scholar] [CrossRef]
- Performance Standards for Antimicrobial Susceptibility Testing; 31st Informational Supplement. Document M100-S31; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2021.
- Shon, A.S.; Bajwa, R.P.S.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef]
- Lee, H.; Baek, J.Y.; Kim, S.Y.; Jo, H.; Kang, K.; Ko, J.H.; Cho, S.Y.; Chung, D.R.; Peck, K.R.; Song, J.H.; et al. Comparison of virulence between matt and mucoid colonies of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 isolated from a single patient. J. Microbiol. 2018, 56, 665–672. [Google Scholar] [CrossRef]
- Cejas, D.; Elena, A.; Guevara Nuñez, D.; Sevillano Platero, P.; De Paulis, A.; Magariños, F.; Alfonso, C.; Berger, M.A.; Fernández-Canigia, L.; Gutkind, G.; et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019, 18, 238–242. [Google Scholar] [CrossRef]
- Rojas, R.; Macesic, N.; Tolari, G.; Guzman, A.; Uhlemann, A.C. Multidrug-resistant Klebsiella pneumoniae ST307 in traveler returning from Puerto Rico to Dominican Republic. Emerg. Infect. Dis. 2019, 25, 1583–1585. [Google Scholar] [CrossRef]
- Magi, G.; Tontarelli, F.; Caucci, S.; Sante, L.D.; Brenciani, A.; Morroni, G.; Giovanetti, E.; Menzo, S.; Mingoia, M. High prevalence of carbapenem-resistant Klebsiella pneumoniae ST307 recovered from fecal samples in an Italian hospital. Future Microbiol. 2021, 16, 703–711. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Chen, G.; Lin, M.; Chen, Y.; He, R.; Galvão, K.N.; El-Gawad El-Sayed Ahmed, M.A.; Roberts, A.P.; Wu, Y.; et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg. Microbes Infect. 2021, 10, 700–709. [Google Scholar] [CrossRef]
- Lee, M.; Choi, T.J. Antimicrobial resistance caused by KPC-2 encoded by promiscuous plasmids of the Klebsiella pneumoniae ST307 strain. Ann. Lab. Med. 2021, 41, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Kerdsin, A.; Akeda, Y.; Hamada, S. Detection of plasmid-mediated colistin-resistant and carbapenem-resistant genes by multiplex PCR. MethodsX 2018, 5, 532–536. [Google Scholar] [CrossRef]
- Weill, F.X.; Lailler, R.; Praud KKérouanton, A.; Fabre, L.; Brisabois, A.; Grimont, P.A.D.; Cloeckaert, A. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 2004, 42, 5767–5773. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Genotype | Carbapenem | Isolate Number | Source | MBL and ESBL | wzi Allele | Virulence Genes |
|---|---|---|---|---|---|---|
| ST11 | Carbapenem-resistant | SCH2104-31 | Bile | KPC-2 | 14 | rmpA2, ybtS, iutA |
| SCH2104-32 | Voided urine | KPC-2, CTX-M-15 | 14 | rmpA2, ybtS, iutA | ||
| SCH2106-16 | Rectal swab | KPC-2 | 50 | ybtS | ||
| SCH2107-22 | Catheterized urine | KPC-2, CTX-M-15 | 50 | ybtS | ||
| SCH2108-30 | Sputum | KPC-2 | 14 | rmpA2, ybtS, iutA | ||
| SCH2104-17 | Catheterized urine | KPC-2 | 14 | ybtS | ||
| SCH2108-43 | Sputum | KPC-2 | 50 | |||
| SCH2108-44 | Bronchial washing | KPC-2 | 75 | ybtS | ||
| Carbapenem-susceptible | SCH2107-06 | Bile | CTX-M-9 | 50 | ybtS, clbB | |
| SCH2108-31 | ND a | CTX-M-15 | 14 | ybtS | ||
| B0706-169 | ND | CTX-M-15 | 50 | ybtS, clbB | ||
| B0708-216 | ND | CTX-M-15 | 39 | |||
| K01-Bact-08-03094 | ND | CTX-M-15 | 39 | |||
| K01-Bact-08-10058 | ND | CTX-M-15 | 50 | ybtS, clbB | ||
| K01-Bact-08-12164 | ND | CTX-M-15 | 39 | |||
| K01-Bact-08-12216 | ND | CTX-M-15 | 39 | |||
| K01-Bact-08-12226 | ND | CTX-M-15 | 39 | |||
| 726 | Urine | CTX-M-15 | 123 | ybtS, clbB | ||
| ST307 | Carbapenem-resistant | SCH2101-15 | Ascitic fluid | KPC-2, CTX-M-15 | 173 | ybtS |
| SCH2102-16 | Bile | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2102-30 | Rectal swab | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2104-07 | Voided urine | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2104-33 | Rectal swab | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2105-10 | Catheterized urine | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2105-20 | Rectal swab | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2106-08 | Rectal swab | KPC-2, CTX-M-15 | 173 b | ybtS | ||
| SCH2108-07 | Blood | KPC-2, CTX-M-15 | 173 | ybtS | ||
| SCH2109-15 | Sputum | KPC-2, CTX-M-15 | 173 | ybtS | ||
| Carbapenem-susceptible | SCH2012-07 | Blood | CTX-M-15 | 173 | ybtS | |
| SCH2012-19 | Blood | CTX-M-15 | 173 | ybtS | ||
| SCH2107-01 | Blood | CTX-M-15 | 173 | ybtS | ||
| SCH2107-07 | Blood | CTX-M-15 | 173 | ybtS | ||
| SCH2107-08 | Blood | CTX-M-15 | 173 | |||
| SCH2107-19 | Blood | None | 173 | |||
| 925 | Peritoneal fluid | None | 173 | ybtS | ||
| SCH-CR31 | Peritoneal fluid | None | 173 | ybtS | ||
| 633 | Stool | None | 173 | |||
| SCH2106-18 | Blood | None | 173 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, Y.Y.; Kim, J.H.; Kim, H.; Lee, J.; Im, S.J.; Ko, K.S. Comparison of Virulence between Two Main Clones (ST11 and ST307) of Klebsiella pneumoniae Isolates from South Korea. Microorganisms 2022, 10, 1827. https://doi.org/10.3390/microorganisms10091827
Cho YY, Kim JH, Kim H, Lee J, Im SJ, Ko KS. Comparison of Virulence between Two Main Clones (ST11 and ST307) of Klebsiella pneumoniae Isolates from South Korea. Microorganisms. 2022; 10(9):1827. https://doi.org/10.3390/microorganisms10091827
Chicago/Turabian StyleCho, Yun Young, Jee Hong Kim, Hyunkeun Kim, Junghwa Lee, Se Jin Im, and Kwan Soo Ko. 2022. "Comparison of Virulence between Two Main Clones (ST11 and ST307) of Klebsiella pneumoniae Isolates from South Korea" Microorganisms 10, no. 9: 1827. https://doi.org/10.3390/microorganisms10091827







