Diversity of Mixotrophic Neutrophilic Thiosulfate- and Iron-Oxidizing Bacteria from Deep-Sea Hydrothermal Vents
Abstract
:1. Introduction
2. Material and Methods
2.1. Sampling, Enrichment and Isolation
2.2. Determination of Iron-Oxidation Capacity
2.3. Determination of Manganese-Oxidation Capacity
2.4. Determination of Thiosulfate-Oxidizing Capacity
2.5. Determination of Carbon-Fixation Capacity
2.6. Measurements of Fe(II), Fe(III), Sulfate, and Nitrate
2.7. Fluorescence Microscopy and Scanning Electron Microscopy/Energy Dispersive X-ray Spectroscopy (SEM/EDS)
2.8. Phylogenetic Analysis of the Isolated Strains
2.9. Genomic Analysis of the Isolated Strains
2.10. Nucleotide Sequence Accession Numbers for Strains
3. Results
3.1. Phylogenetic Affiliations
3.2. Growth Test with Different Electron Donors and Acceptors
3.2.1. Iron-Oxidizing Capacity
3.2.2. Manganese-Oxidizing Capacity
3.2.3. Thiosulfate-Oxidizing Capacity
3.3. Carbon Dioxide–Fixation Capacity
3.4. Genome Characteristics of Isolated Strains
4. Discussion
4.1. Widespread of Mixotrophic Bacterial Strains in the Hydrothermal Vents
4.2. Diverse Metabolism of Mixotrophic Bacterial Strains in the Hydrothermal Vents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyd, P.W.; Ellwood, M.J. The biogeochemical cycle of iron in the ocean. Nat. Geosci. 2010, 3, 675. [Google Scholar] [CrossRef]
- Jannasch, H.W. The Chemosynthetic support of life and the microbial diversity at Deep-Sea hydrothermal vents. Proc. R. Soc. Lond. Ser. B 1985, 225, 277. [Google Scholar]
- Edwards, K.J.; Bach, W.; McCollom, T.M.; Rogers, D.R. Neutrophilic iron-oxidizing bacteria in the ocean: Their habitats, diversity, and roles in mineral deposition, rock alteration, and biomass production in the deep-sea. Geomicrobiol. J. 2004, 21, 393–404. [Google Scholar] [CrossRef]
- Edwards, K.J.; Rogers, D.R.; Wirsen, C.O.; McCollom, T. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α-and γ-Proteobacteria from the deep sea. Appl. Environ. Microbiol. 2003, 69, 2906–2913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakharova, Y.R.; Parfenova, V.V.; Granina, L.Z.; Kravchenko, O.S.; Zemskaya, T.I. Distribution of iron- and manganese-oxidizing bacteria in the bottom sediments of Lake Baikal. Inland Water Biol. 2010, 3, 313–321. [Google Scholar] [CrossRef]
- Rowe, A.R.; Chellamuthu, P.; Lam, B.; Okamoto, A.; Nealson, K.H. Marine sediments microbes capable of electrode oxidation as a surrogate for lithotrophic insoluble substrate metabolism. Front. Microbiol. 2015, 5, 784. [Google Scholar] [CrossRef] [Green Version]
- Sudek, L.A.; Templeton, A.S.; Tebo, B.M.; Staudigel, H. Microbial ecology of Fe (hydr) oxide mats and basaltic rock from Vailulu’u Seamount, American Samoa. Geomicrobiol. J. 2009, 26, 581–596. [Google Scholar] [CrossRef]
- Emerson, D.; Floyd, M.M. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol. 2005, 397, 112–123. [Google Scholar]
- Emerson, D.; Rentz, J.A.; Lilburn, G.T.; Davis, R.E.; Aldrich, H.; Chan, C.; Moyer, C.L. A novel lineage of Proteobacteria involved in formation of marine fe-oxidizing microbial mat communities. PLoS ONE 2007, 2, e667. [Google Scholar] [CrossRef] [Green Version]
- Hallbe, L.; Stahl, F.; Pedersen, K. Phylogeny and phenotypic characterization of the stalk-forming and iron-oxidizing bacterium Gallionella ferruginea. J. Gen. Microbiol. 1993, 139, 1531–1539. [Google Scholar] [CrossRef]
- Hanert, H.H. The Genus Gallionella. In The Prokaryotes: Volume 7: Proteobacteria: Delta, Epsilon Subclass; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 990–995. [Google Scholar]
- Woods, D.; Sokol, P. The Prokaryotes, Vol. 5. Proteobacteria: Alpha and Beta Subclasses; Springer: New York, NY, USA, 2006. [Google Scholar]
- Krumbein, W.; Altmann, H. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgoländer Wiss. Meeresunters. 1973, 25, 347–356. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Lyu, J.; Shao, Z. Sulfur metabolism of Hydrogenovibrio thermophilus strain S5 and its adaptations to deep-sea hydrothermal vent environment. Front. Microbiol. 2017, 8, 2513. [Google Scholar] [CrossRef] [PubMed]
- Orcutt, B.N.; Sylvan, J.B.; Rogers, D.R.; Delaney, J.; Lee, R.W.; Girguis, P.R. Carbon fixation by basalt-hosted microbial communities. Front. Microbiol. 2015, 6, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stookey, L.L. Ferrozine—A new spectrophotometric reagent for Iron. Anal. Chem. 1970, 42, 779–781. [Google Scholar] [CrossRef] [Green Version]
- Josephson, B. A volumetric method for the determination of sulphur and sulphate ion. Analyst 1939, 64, 181–185. [Google Scholar] [CrossRef]
- Small, H.; Stevens, T.S.; Bauman, W.C. Novel ion exchange chromatographic method using conductimetric detection. Anal. Chem. 1975, 47, 1801–1809. [Google Scholar] [CrossRef]
- Cox, R.D. Determination of nitrate and nitrite at the parts per billion level by chemiluminescence. Anal. Chem. 1980, 52, 332–335. [Google Scholar] [CrossRef]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef] [Green Version]
- Hiraishi, A. Direct automated sequencing of 16s rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 1992, 15, 210–213. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, 354–357. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In Methods in Molecular Biology; Humana: New York, NY, USA, 2019; Volume 1962, pp. 1–14. [Google Scholar]
- Rodriguez, R.L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar]
- Makita, H. Iron-oxidizing bacteria in marine environments: Recent progresses and future directions. World J. Microbiol. Biotechnol. 2018, 34, 110. [Google Scholar] [CrossRef]
- Smith, A.; Popa, R.; Fisk, M.; Nielsen, M.; Wheat, C.G.; Jannasch, H.W.; Fisher, A.T.; Becker, K.; Sievert, S.M.; Flores, G. In situ enrichment of ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem. Geophys. Geosyst. 2011, 12, Q06007. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, J.; Bach, W.; Edwards, K.J.; Orcutt, B.N.; Wang, F. Nitrogen stimulates the growth of subsurface basalt-associated microorganisms at the western flank of the Mid-Atlantic Ridge. Front. Microbiol. 2016, 7, 633. [Google Scholar] [CrossRef] [Green Version]
- Bonis, B.M.; Gralnick, J.A. Marinobacter subterrani, a genetically tractable neutrophilic Fe (II)-oxidizing strain isolated from the Soudan Iron Mine. Front. Microbiol. 2015, 6, 719. [Google Scholar] [CrossRef]
- Choi, B.-R.; Pham, V.H.; Park, S.-J.; Kim, S.-J.; Roh, D.-H.; Rhee, S.-K. Characterization of facultative sulfur-oxidizing Marinobacter sp. BR13 isolated from marine sediment of Yellow Sea, Korea. J. Korean Soc. Appl. Biol. Chem. 2009, 52, 309–314. [Google Scholar] [CrossRef]
- Templeton, A.S.; Staudigel, H.; Tebo, B.M. Diverse Mn (II)-oxidizing bacteria isolated from submarine basalts at Loihi Seamount. Geomicrobiol. J. 2005, 22, 127–139. [Google Scholar] [CrossRef]
- Homann, V.V.; Sandy, M.; Tincu, J.A.; Templeton, A.S.; Tebo, B.M.; Butler, A. Loihichelins A-F, a suite of amphiphilic siderophores produced by the marine bacterium Halomonas LOB-5. J. Nat. Prod. 2009, 72, 884–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaye, J.Z.; Sylvan, J.B.; Edwards, K.J.; Baross, J.A. Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol. Ecol. 2011, 75, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Xiao, E.; Krumins, V.; Dong, Y.; Xiao, T.; Ning, Z.; Chen, H.; Xiao, Q. Characterization of the microbial community composition and the distribution of Fe-metabolizing bacteria in a creek contaminated by acid mine drainage. Appl. Microbiol. Biotechnol. 2016, 100, 8523–8535. [Google Scholar] [CrossRef]
- Podgorsek, L.; Petri, R.; Imhoff, J.F. Cultured and genetic diversity, and activities of sulfur-oxidizing bacteria in low-temperature hydrothermal fluids of the North Fiji Basin. Marine Ecol. Prog. Ser. 2004, 266, 65–76. [Google Scholar] [CrossRef]
- Xu, X.; Yang, G.; Wang, F.; Li, M. Ferro-oxidase producing conditions of Pseudomonas sp. J. Microbiol. 2009, 29, 16–19. [Google Scholar]
- Hedrich, S.; Schlömann, M.; Johnson, D.B. The iron-oxidizing proteobacteria. Microbiology 2011, 157, 1551–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blöthe, M.; Roden, E.E. Composition and activity of an autotrophic Fe (II)-oxidizing, nitrate-reducing enrichment culture. Appl. Environ. Microbiol. 2009, 75, 6937–6940. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jiang, L.; Li, S.; Zhong, T.; Lai, Q.; Shao, Z. Diversity of culturable sulfur-oxidizing bacteria in deep-sea hydrothermal vent environments of the South Atlantic. Wei Sheng Wu Xue Bao Acta Microbiol. Sin. 2016, 56, 88–100. [Google Scholar]
- Johnson, D.B.; Bacelar-Nicolau, P.; Okibe, N.; Thomas, A.; Hallberg, K.B. Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: Heterotrophic, iron-oxidizing, extremely acidophilic actinobacteria. Int. J. Syst. Evol. Microbiol. 2009, 59, 1082–1089. [Google Scholar] [CrossRef]
- Rajasabapathy, R.; Mohandass, C.; Dastager, S.G.; Liu, Q.; Li, W.-J.; Colaço, A. Citreicella manganoxidans sp. nov., a novel manganese oxidizing bacterium isolated from a shallow water hydrothermal vent in Espalamaca (Azores). Antonie Leeuwenhoek 2015, 108, 1433–1439. [Google Scholar] [CrossRef]
- Kepkay, P.; Nealson, K. Growth of a manganese oxidizing Pseudomonas sp. in continuous culture. Arch. Microbiol. 1987, 148, 63–67. [Google Scholar] [CrossRef]
- Okazaki, M.; Sugita, T.; Shimizu, M.; Ohode, Y.; Iwamoto, K.; de Vrind-de Jong, E.W.; de Vrind, J.P.; Corstjens, P.L. Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl. Environ. Microbiol. 1997, 63, 4793–4799. [Google Scholar] [CrossRef] [Green Version]
- Brouwers, G.-J.; de Vrind, J.P.; Corstjens, P.L.; Cornelis, P.; Baysse, C.; de Vrind-de Jong, E.W. cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 1999, 65, 1762–1768. [Google Scholar] [CrossRef] [Green Version]
- Murray, K.J.; Mozafarzadeh, M.L.; Tebo, B.M. Cr (III) oxidation and Cr toxicity in cultures of the manganese (II)-oxidizing Pseudomonas putida strain GB-1. Geomicrobiol. J. 2005, 22, 151–159. [Google Scholar] [CrossRef]
- Toner, B.; Fakra, S.; Villalobos, M.; Warwick, T.; Sposito, G. Spatially resolved characterization of biogenic manganese oxide production within a bacterial biofilm. Appl. Environ. Microbiol. 2005, 71, 1300–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denkmann, K.; Grein, F.; Zigann, R.; Siemen, A.; Bergmann, J.; van Helmont, S.; Nicolai, A.; Pereira, I.A.; Dahl, C. Thiosulfate dehydrogenase: A widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 2012, 14, 2673–2688. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Long, Y.; Yan, H.; Cai, H.; Li, Y.; Wang, X. Gene cloning, identification, and characterization of the multicopper oxidase CumA from Pseudomonas sp. 593. Biotechnol. Appl. Biochem. 2017, 64, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.; Guiral, M.; Malarte, G.; Ledgham, F.; Leroy, G.; Brugna, M.; Giudici-Orticoni, M.-T. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 2008, 283, 25803–25811. [Google Scholar] [CrossRef]
- Barco, R.A.; Emerson, D.; Sylvan, J.B.; Orcutt, B.N.; Jacobson Meyers, M.E.; Ramírez, G.A.; Zhong, J.D.; Edwards, K.J. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl. Environ. Microbiol. 2015, 81, 5927–5937. [Google Scholar] [CrossRef] [Green Version]
- McAllister, S.M.; Polson, S.W.; Butterfield, D.A.; Glazer, B.T.; Sylvan, J.B.; Chan, C.S. Validating the Cyc2 neutrophilic Fe oxidation pathway using meta-omics of Zetaproteobacteria iron mats at marine hydrothermal vents. bioRxiv 2019. bioRxiv:722066. [Google Scholar]
- Bathe, S.; Norris, P.R. Ferrous iron-and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 2007, 73, 2491–2497. [Google Scholar] [CrossRef] [Green Version]
- Croal, L.R.; Jiao, Y.; Newman, D.K. The fox operon from Rhodobacter strain SW2 promotes phototrophic Fe (II) oxidation in Rhodobacter capsulatus SB1003. J. Bacteriol. 2007, 189, 1774–1782. [Google Scholar] [CrossRef] [Green Version]
- Ilbert, M.; Bonnefoy, V. Insight into the evolution of the iron oxidation pathways. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1827, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, Z.; Belchik, S.M.; Edwards, M.J.; Liu, C.; Kennedy, D.W.; Merkley, E.D.; Lipton, M.S.; Butt, J.N.; Richardson, D.J. Identification and characterization of MtoA: A decaheme c-type cytochrome of the neutrophilic Fe (II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 2012, 3, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, Y.; Newman, D.K. The pio operon is essential for phototrophic Fe (II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 2007, 189, 1765–1773. [Google Scholar] [CrossRef] [Green Version]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranz, R.G.; Richard-Fogal, C.; Taylor, J.-S.; Frawley, E.R. Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 2009, 73, 510–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutherland, M.C.; Rankin, J.A.; Kranz, R.G. Heme trafficking and modifications during system I cytochrome c biogenesis: Insights from heme redox potentials of Ccm proteins. Biochemistry 2016, 55, 3150–3156. [Google Scholar] [CrossRef] [Green Version]
- Enoch, H.G.; Lester, R.L. The role of a novel cytochrome b-containing nitrate reductase and quinone in the in vitro reconstruction of formate-nitrate reductase activity of E. coli. Biochem. Biophys. Res. Commun. 1974, 61, 1234–1241. [Google Scholar] [CrossRef]
- Meier, D.V.; Bach, W.; Girguis, P.R.; Gruber-Vodicka, H.R.; Reeves, E.P.; Richter, M.; Vidoudez, C.; Amann, R.; Meyerdierks, A. Heterotrophic Proteobacteria in the vicinity of diffuse hydrothermal venting. Environ. Microbiol. 2016, 18, 4348–4368. [Google Scholar] [CrossRef]
- Kaye, J.Z.; Marquez, M.C.; Ventosa, A.; Baross, J.A. Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: Halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evolut. Microbiol. 2004, 54, 499–511. [Google Scholar] [CrossRef] [Green Version]
- Raguénès, G.; Christen, R.; Guezennec, J.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Evolut. Microbiol. 1997, 47, 989–995. [Google Scholar]
- Takai, K.; Sugai, A.; Itoh, T.; Horikoshi, K. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evolut. Microbiol. 2000, 50, 489–500. [Google Scholar] [CrossRef]
- Vetriani, C.; Chew, Y.S.; Miller, S.M.; Yagi, J.; Coombs, J.; Lutz, R.A.; Barkay, T. Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 2005, 71, 220–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaye, J.Z.; Baross, J.A. High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol. Ecol. 2000, 32, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Nakawake, M.; Kita, J.; Yamanaka, T.; Utsumi, M.; Okamura, K.; Ishibashi, J.-i.; Ohkuma, M.; Yamagishi, A. Characteristics of microbial communities in crustal fluids in a deep-sea hydrothermal field of the Suiyo Seamount. Front. Microbiol. 2013, 4, 85. [Google Scholar] [CrossRef] [Green Version]
- Chiu, B.K.; Kato, S.; McAllister, S.M.; Field, E.K.; Chan, C.S. Novel pelagic iron-oxidizing Zetaproteobacteria from the Chesapeake Bay oxic–anoxic transition zone. Front. Microbiol. 2017, 8, 1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAllister, S.M.; Davis, R.E.; McBeth, J.M.; Tebo, B.M.; Emerson, D.; Moyer, C.L. Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl. Environ. Microbiol. 2011, 77, 5445–5457. [Google Scholar] [CrossRef] [Green Version]
- Emerson, D.; Moyer, C.L. Microbiology of seamounts: Common patterns observed in community structure. Oceanography 2010, 23, 148–163. [Google Scholar] [CrossRef]
- Emerson, D.; Moyer, C.L. Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl. Eenviron. Microbiol. 2002, 68, 3085–3093. [Google Scholar] [CrossRef]
- Mori, J.F.; Scott, J.J.; Hager, K.W.; Moyer, C.L.; Küsel, K.; Emerson, D. Physiological and ecological implications of an iron-or hydrogen-oxidizing member of the Zetaproteobacteria, Ghiorsea bivora, gen. nov., sp. nov. ISME J. 2017, 11, 2624–2636. [Google Scholar] [CrossRef] [Green Version]
- Litchman, E. Resource competition and the ecological success of phytoplankton. In Evolution of Primary Producers in the Sea; Paul, G., Falkowski, A., Knoll, H., Eds.; Academic Press: New York, NY, USA, 2007; pp. 351–375. [Google Scholar]
- Segerer, A.H.; Burggraf, S.; Fiala, G.; Huber, G.; Huber, R.; Pley, U.; Stetter, K.O. Life in hot springs and hydrothermal vents. Orig. Life Evol. Biosph. 1993, 23, 77–90. [Google Scholar] [CrossRef] [Green Version]
- Dick, G.J. The microbiomes of deep-sea hydrothermal vents: Distributed globally, shaped locally. Nat. Rev. Microbiol. 2019, 17, 271–283. [Google Scholar] [CrossRef]
- Morris, R.M.; Spietz, R.L. The Physiology and Biogeochemistry of SUP05. Annu. Rev. Mar. Sci. 2022, 14, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y. Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 2003, 72, 641–653. [Google Scholar] [CrossRef]
- Du, R.; Gao, D.; Wang, Y.; Liu, L.; Cheng, J.; Liu, J.; Zhang, X.H.; Yu, M. Heterotrophic sulfur oxidation of Halomonas titanicae SOB56 and its habitat adaptation to the hydrothermal environment. Front. Microbiol. 2022, 13, 888833. [Google Scholar] [CrossRef] [PubMed]
- Straub, K.L.; Benz, M.; Schink, B.; Widdel, F. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl. Environ. Microbiol. 1996, 62, 1458–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.; Shao, S.; Huang, T.; Ma, F.; Yang, S.; Zhou, Z.; Zheng, S. Anaerobic nitrate-dependent iron (II) oxidation by a novel autotrophic bacterium, Pseudomonas sp. SZF15. J. Environ. Chem. Eng. 2015, 3, 2187–2193. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Teske, A.; Robertson, L.A.; Kuenen, J.G. Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria, belonging to the Pseudomonas stutzeri group. FEMS Microbiol. Ecol. 1999, 30, 113–123. [Google Scholar] [CrossRef]
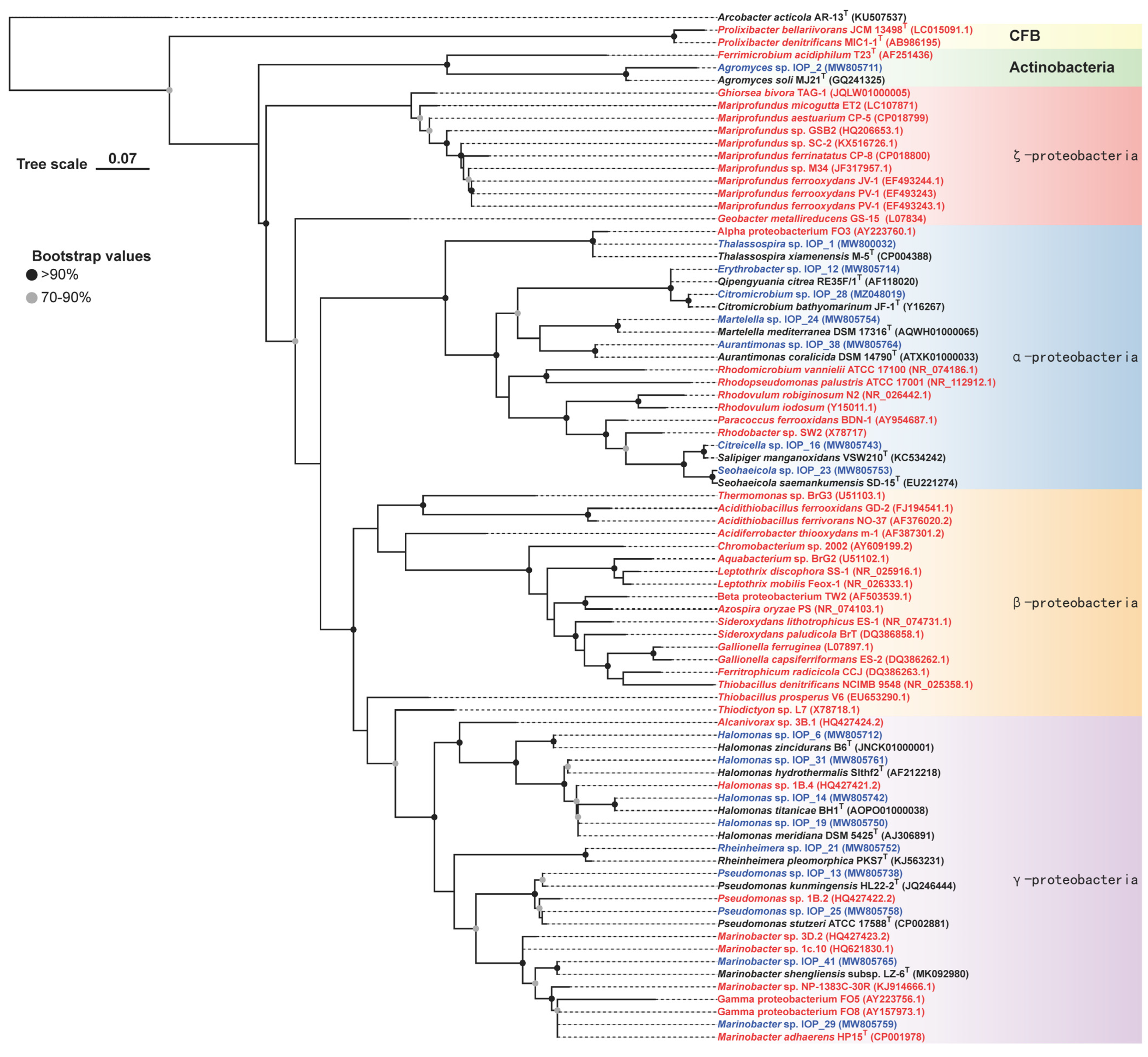


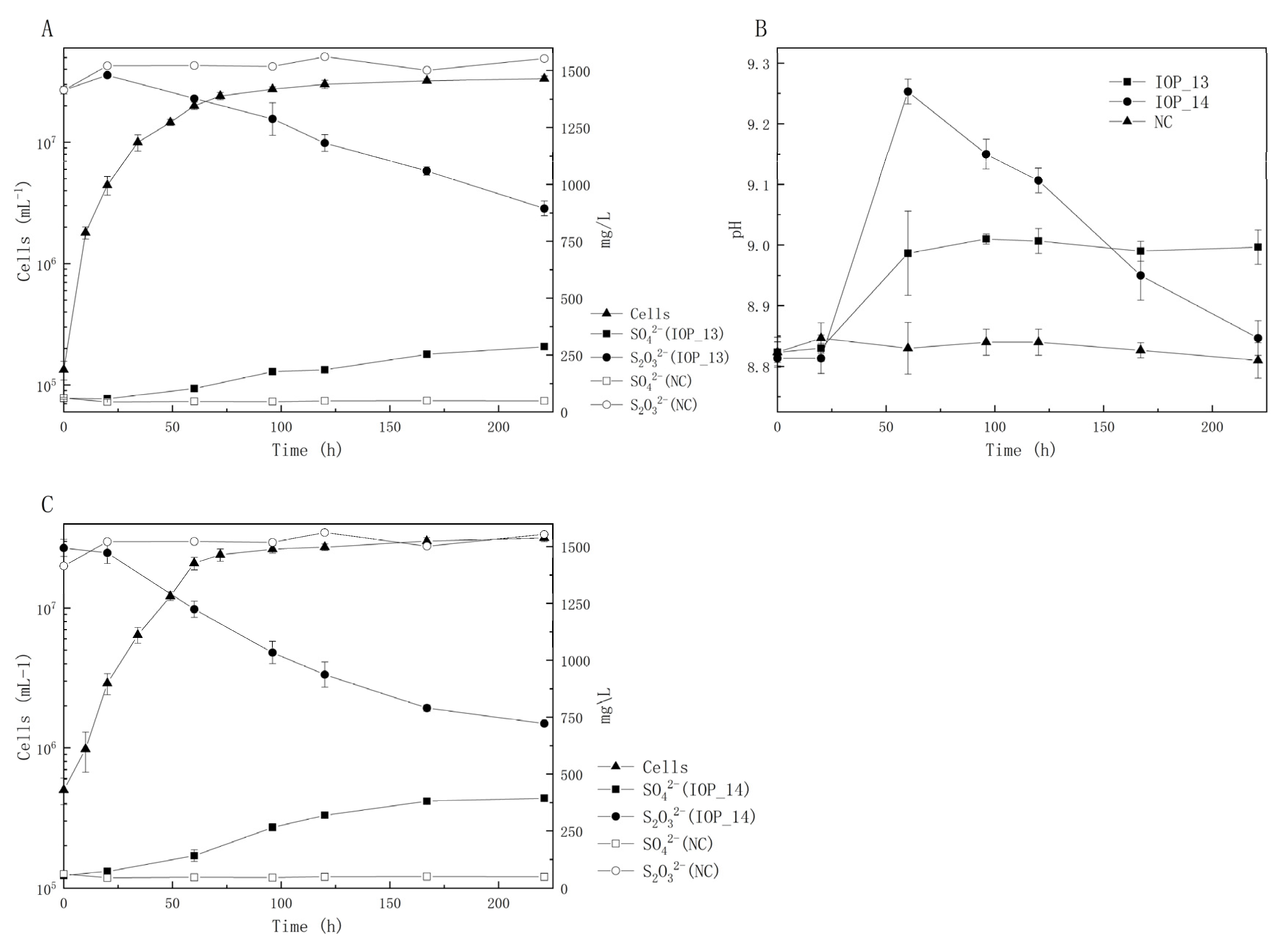
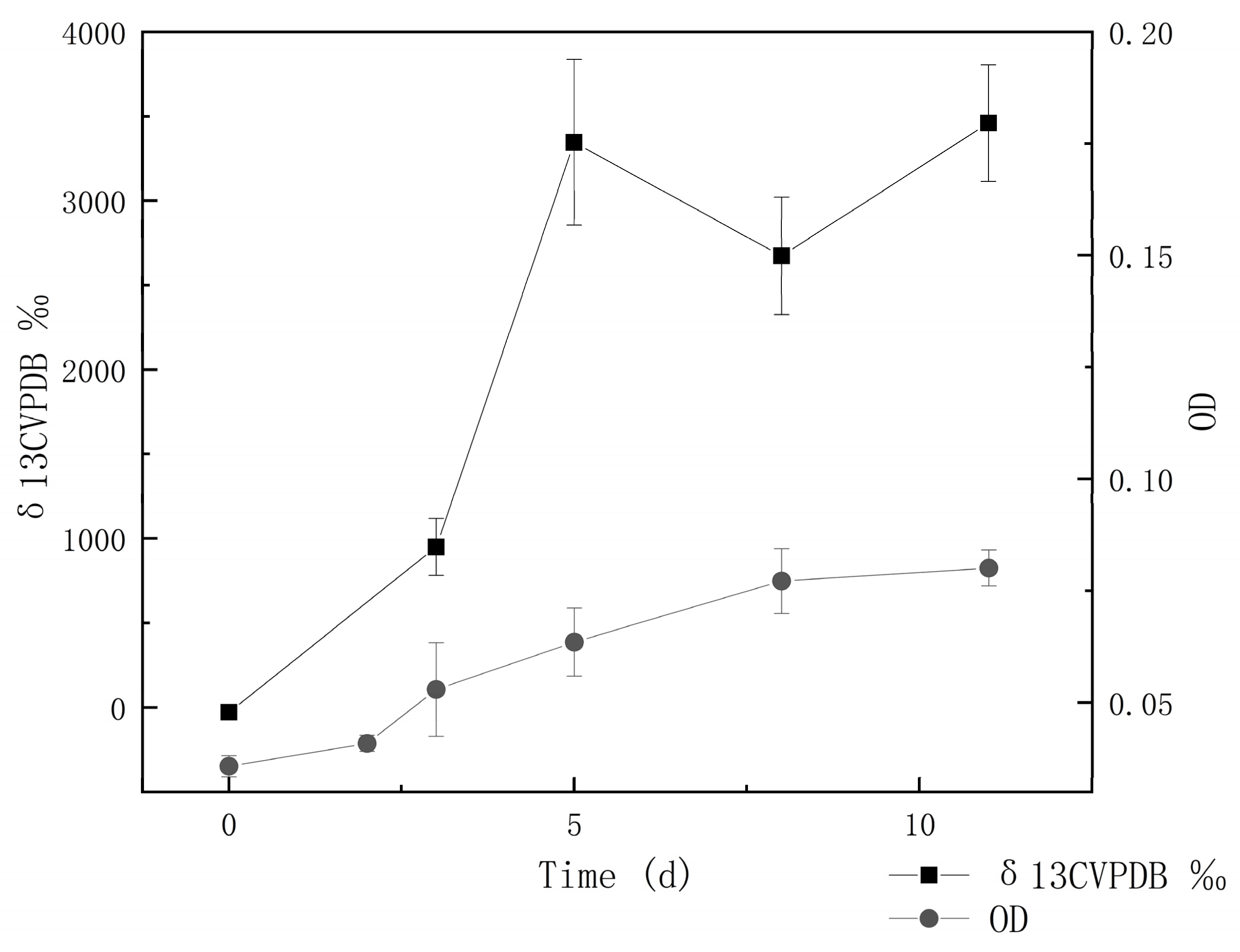

| MCCC Deposition No. | Strains | Source | Closest Species | 16S rDNA Identity (%) | 16S rDNA GenBank Accession | Genome GenBank Accession |
|---|---|---|---|---|---|---|
| M28193 | IOP_1 | mussels | Thalassospira xiamenensis M-5(T) | 98.79 | MW800032 | JAINWB000000000 |
| MCCC 1A14002 | IOP_2 | mussels | Agromyces soli MJ21(T) | 94.19 | MW805711 | JAINWC000000000 |
| MCCC 1A14012 | IOP_6 | sulfides | Halomonas zincidurans B6 (T) | 98.64 | MW805712 | JAIRBO000000000 |
| MCCC 1A13998 | IOP_12 | hydrothermal sediments | Qipengyuania citrea RE35F/1(T) | 98.78 | MW805714 | JAINWE000000000 |
| M28194 | IOP_13 | hydrothermal sediments | Pseudomonas kunmingensis HL22-2(T) | 98.61 | MW805738 | JAINWF000000000 |
| M28195 | IOP_14 | plume | Halomonas titanicae BH1(T) | 98.48 | MW805742 | JAINWD000000000 |
| MCCC 1A13999 | IOP_16 | plume | Salipiger manganoxidans VSW210(T) | 99.48 | MW805743 | JAINWH000000000 |
| MCCC 1A14001 | IOP_19 | sulfides | Halomonas meridiana DSM 5425 (T) | 98.82 | MW805750 | JAINWP000000000 |
| MCCC 1A14003 | IOP_21 | sulfides | Rheinheimera pleomorphica PKS7 (T) | 99.22 | MW805752 | JAINWG000000000 |
| MCCC 1A14004 | IOP_23 | sulfides | Seohaeicola saemankumensis SD-15 (T) | 99.04 | MW805753 | JAINWI000000000 |
| MCCC 1A14005 | IOP_24 | sulfides | Martelella mediterranea DSM 17316 (T) | 98.42 | MW805754 | JAINWJ000000000 |
| M28196 | IOP_25 | sulfides | Pseudomonas stutzeri ATCC 17588(T) | 98.55 | MW805758 | JAINWK000000000 |
| MCCC 1A14006 | IOP_28 | sulfides | Citromicrobium bathyomarinum JF-1(T) | 99.93 | MZ048019 | JAINWL000000000 |
| MCCC 1A14007 | IOP_29 | sulfides | Marinobacter adhaerens HP15(T) | 100.00 | MW805759 | JAINWM000000000 |
| MCCC 1A14008 | IOP_31 | sulfides | Halomonas hydrothermalis Slthf2(T) | 99.64 | MW805761 | JAIRBP000000000 |
| MCCC 1A14010 | IOP_38 | sulfides | Aurantimonas coralicida DSM 14790(T) | 99.93 | MW805764 | JAINWN000000000 |
| MCCC 1A14013 | IOP_41 | sulfides | Marinobacter shengliensis LZ-6(T) | 99.86 | MW805765 | JAINWO000000000 |
| Strains | Most Similar Type Species | Iron Oxidiation | Manganese Oxidation | Sulfur Oxidation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Autotrophic-Microaerobic | Hetertrophic-Anaerobic | Heterotrophic | Autotrophic | Heterotrophic | ||||||
| Fe0 | FeS | FeCO3 | Pyrite | Basalt | FeCl2 | MnCl2 | S2O32− | S2O32− | ||
| Gammaproteobacteria | ||||||||||
| IOP_6 | Halomonas zincidurans B6 (T) | + | + | + | − | + | + | − | + | − |
| IOP_14 | Halomonas titanicae BH1(T) | + | + | + | − | − | + | − | + | + |
| IOP_19 | Halomonas meridiana DSM 5425 (T) | + | + | + | − | − | + | − | + | + |
| IOP_31 | Halomonas hydrothermalis Slthf2(T) | + | + | + | − | − | + | − | + | − |
| IOP_13 | Pseudomonas kunmingensis HL22-2(T) | + | + | + | + | − | + | + | + | − |
| IOP_25 | Pseudomonas stutzeri ATCC 17588(T) | + | + | + | + | − | + | + | + | − |
| IOP_29 | Marinobacter adhaerens HP15(T) | + | + | + | + | − | + | − | + | + |
| IOP_41 | Marinobacter shengliensis LZ-6(T) | + | + | + | − | − | + | − | + | − |
| IOP_21 | Rheinheimera pleomorphica PKS7 (T) | − | + | − | − | − | + | − | − | + |
| Alphaproteobacteria | ||||||||||
| IOP_1 | Thalassospira xiamenensis M-5(T) | + | + | + | − | − | − | − | + | + |
| IOP_12 | Qipengyuania citrea RE35F/1(T) | − | + | − | + | − | − | − | − | + |
| IOP_16 | Salipiger manganoxidans VSW210(T) | + | + | − | − | − | + | + | − | + |
| IOP_23 | Seohaeicola saemankumensis SD-15 (T) | + | + | − | + | − | + | − | − | + |
| IOP_24 | Martelella mediterranea DSM 17316 (T) | + | + | − | + | − | + | − | − | + |
| IOP_28 | Citromicrobium bathyomarinum JF-1(T) | − | + | − | − | − | + | − | + | − |
| IOP_38 | Aurantimonas coralicida DSM 14790(T) | + | + | + | + | − | − | − | + | + |
| Actinobacteria | ||||||||||
| IOP_2 | Agromyces soli MJ21(T) | − | + | − | + | + | − | − | + | − |
| NC1 | − | − | − | − | − | − | − | − | − | |
| NC2 | − | − | − | − | − | − | − | − | − | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Zeng, X.; Xu, F.; Shao, Z. Diversity of Mixotrophic Neutrophilic Thiosulfate- and Iron-Oxidizing Bacteria from Deep-Sea Hydrothermal Vents. Microorganisms 2023, 11, 100. https://doi.org/10.3390/microorganisms11010100
He Y, Zeng X, Xu F, Shao Z. Diversity of Mixotrophic Neutrophilic Thiosulfate- and Iron-Oxidizing Bacteria from Deep-Sea Hydrothermal Vents. Microorganisms. 2023; 11(1):100. https://doi.org/10.3390/microorganisms11010100
Chicago/Turabian StyleHe, Yang, Xiang Zeng, Fei Xu, and Zongze Shao. 2023. "Diversity of Mixotrophic Neutrophilic Thiosulfate- and Iron-Oxidizing Bacteria from Deep-Sea Hydrothermal Vents" Microorganisms 11, no. 1: 100. https://doi.org/10.3390/microorganisms11010100
APA StyleHe, Y., Zeng, X., Xu, F., & Shao, Z. (2023). Diversity of Mixotrophic Neutrophilic Thiosulfate- and Iron-Oxidizing Bacteria from Deep-Sea Hydrothermal Vents. Microorganisms, 11(1), 100. https://doi.org/10.3390/microorganisms11010100






