Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review
Abstract
1. Introduction
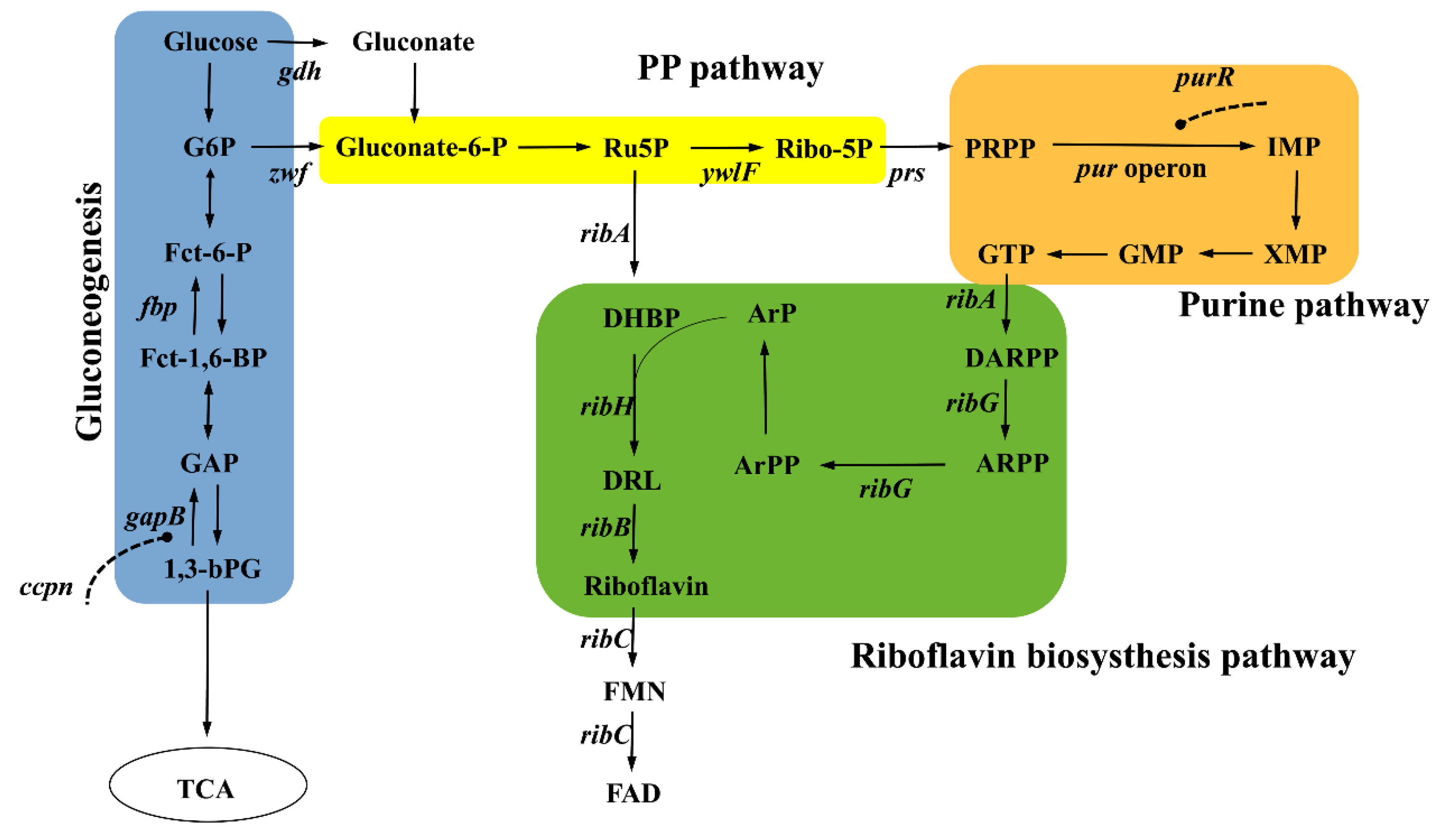
2. Riboflavin Biosynthesis Pathway in B. subtilis
2.1. Upstream Synthetic Pathways—Precursors Ru5P and GTP Supply
2.2. Downstream Synthetic Pathways—Direct Riboflavin Biosynthesis
3. Regulation on Riboflavin Synthesis
3.1. Upstream Pathway—Regulation of GTP Synthesis Module
3.2. Downstream Pathways—Regulation of the Riboflavin Synthesis Module
4. Metabolic Engineering Strategies to Increase Riboflavin Production
4.1. Engineering Upstream Synthetic Pathway
4.1.1. Enhancing Pentose Phosphate Pathway for Ribulose-5-Phosphate Supply
4.1.2. Enhancing De Novo Purine Synthesis Pathway
4.2. Engineering the Downstream Synthetic Pathway—Riboflavin Synthesis Module
4.3. Enhancement of Energy Supply to Increase Riboflavin Production
| Target Gene | Method | Strain Background | VB2 Improvement a | VB2 Titers or Yields b | Reference |
|---|---|---|---|---|---|
| ribA | VegI promoter | B. subtilis RB50::[pRF69]n[pRF93]m Ade+ | 1.25 | 17.5 g/L | [69] |
| rib operon | Multiple copies, VegI promoter | B. subtilis RB9 | 280 | 14 g/L (0.02–0.05 g/L) | [5] |
| zwf | Pxyl promoter | B. subtilis RH33 | 1.25 | 0.05 g/g Glc (0.04 g/g Glc) | [52] |
| zwf | Site-directed mutagenesis | B. subtilis RH33 | 1.11 | 0.052 mmol/g CDW/h (0.047 mmol/g CDW/h) | [56] |
| zwf | Double site-directed mutagenesis with gnd | B. subtilis RH33 | 1.17 | 0.055 mmol/g CDW/h (0.047 mmol/g CDW/h) | [56] |
| gdh | P43 promoter | B. subtilis RH33::[pRB63]n | 1.60 | 0.047 g/g CDW (0.03 g/g CDW) | [53] |
| gapB, fbp | P43 promoter | B. subtilis RH33 | 1.27 | 13.36 g/L (10.5 g/L) | [58] |
| ccpn | Deletion | B. subtilis RB50::pRF69 | 1.63 | 0.062 g/g Glc (0.038 g/g glc) | [59] |
| prs, ywlF | P43 promoter | B. subtilis RH33 | 1.25 | 15 g/L (12 g/L) | [61] |
| purR, purF | purR deletion; purF overexpression | B. subtilis 168 | 3 | 826 mg/L (275 mg/L) | [24] |
| purFMNHD | P43 promoter | B. subtilis RH33 | 1.24 | 0.031 g/g Glc (0.025 g/g Glc) | [24] |
| cyd | Deletion | B. subtilis RH50::[pRB69]n | 1.38 | 12.3 g/L (8.9 g/L) | [75] |
| cyd | Deletion | B. subtilis PK | 1.4 | 0.07 mmol/g CDW/h (0.05 mmol/g CDW/h) | [76] |
| pta, alsS | pta deletion alsS overexpression | B. subtilis RH33::[pRB63]n | 1.5 | 0.045 g/g CDW (0.03 g/g CDW) | [78] |
5. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abbas, C.A.; Sibirny, A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 2011, 75, 321–360. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, S.K.; Park, E.Y.; Revuelta, J.L.; Becker, J.; Wittmann, C. Biotechnology of riboflavin. Appl. Microbiol. Biotechnol. 2016, 100, 2107–2119. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Wang, H.; Yuan, F.; Xu, Z.; Yang, K.; Li, Z.; Chen, Y.; Fan, K. Improvement of stress tolerance and riboflavin production of Bacillus subtilis by introduction of heat shock proteins from thermophilic bacillus strains. Appl. Microbiol. Biotechnol. 2019, 103, 4455–4465. [Google Scholar] [CrossRef] [PubMed]
- Averianova, L.A.; Balabanova, L.A.; Son, O.M.; Podvolotskaya, A.B.; Tekutyeva, L.A. Production of vitamin B2 (riboflavin) by microorganisms: An overview. Front. Bioeng. Biotechnol. 2020, 8, 570828. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.B.; Sloma, A.; Hermann, T.; Theriault, K.; Zachgo, E.; Erdenberger, T.; Hannett, N.; Chatterjee, N.P.; Williams, V., II; Rufo, G.A., Jr.; et al. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol. Biotechnol. 1999, 22, 8–18. [Google Scholar] [CrossRef]
- Lee, K.H.; Park, Y.H.; Han, J.K.; Park, J.H.; Lee, K.H.; Choi, H. Microorganism for Producing Riboflavin and Method for Producing Riboflavin Using the Same: US Patent. US7078222B2, 18 July 2006. [Google Scholar]
- Sauer, U.; Cameron, D.C.; Bailey, J.E. Metabolic capacity of Bacillus subtilis for the production of purine nucleosides, riboflavin, and folic acid. Biotechnol. Bioeng. 1998, 59, 227–238. [Google Scholar] [CrossRef]
- Shen, T.; Wang, J.Y. Biochemistry; Higher Education Press: Beijing, China, 1993. [Google Scholar]
- Zamboni, N.; Fischer, E.; Laudert, D.; Laudert, D.; Aymerich, S.; Hohmann, H.; Sauer, U. The Bacillus subtilis ygjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase pathway. J. Bacteriol. 2004, 186, 4528–4534. [Google Scholar] [CrossRef]
- Shi, T.; Wang, Y.; Wang, Z.; Wang, G.; Liu, D.; Fu, J.; Chen, T.; Zhao, X. Deregulation of purine pathway in Bacillus subtilis and its use in riboflavin biosynthesis. Microb. Cell Fact. 2014, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Ebbole, D.J.; Zalkin, H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 1987, 262, 8274–8287. [Google Scholar] [CrossRef]
- Kappock, T.J.; Ealick, S.E.; Stubbe, J.A. Modular evolution of the purine biosynthetic pathway. Curr. Opin. Chem. Biol. 2000, 4, 567–572. [Google Scholar] [CrossRef]
- Bacher, A.; Eberhardt, S.; Fischer, M.; Kis, K.; Richter, G. Biosynthesis of vitamin b2 (riboflavin). Annu. Rev. Nutr. 2000, 20, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Hümbelin, M.; Griesser, V.; Keller, T.; Schurter, W.; Haiker, M.; Hohmann, H.-P.; Ritz, H.; Richter, G.; Bacher, A.; van Loon, A.P.G.M. GTP cyclohydrolase II and 3,4-dihydroxy-2-butanone 4-phosphate synthase are rate-limiting enzymes in riboflavin synthesis of an industrial Bacillus subtilis strain used for riboflavin production. J. Ind. Microbiol. Biotechnol. 1999, 22, 1–7. [Google Scholar] [CrossRef]
- Richter, G.; Fischer, M.; Krieger, C.; Eberhardt, S.; Lüttgen, H.; Gerstenschläger, I.; Bacher, A. Biosynthesis of riboflavin: Characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 1997, 179, 2022–2028. [Google Scholar] [CrossRef] [PubMed]
- Burrows, R.B.; Brown, G.M. Presence of Escherichia coli of a deaminase and a reductase involved in biosynthesis of riboflavin. J. Bacteriol. 1978, 136, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Hollander, I.; Brown, G.M. Biosynthesis of riboflavin: Reductase and deaminase of Ashbya gossypii. Mol. Cell. Biol. Res. Commun. 1979, 89, 759–763. [Google Scholar] [CrossRef]
- Haase, I.; Gräwert, T.; Illarionov, B.; Bacher, A.; Fischer, M. Recent Advances in Riboflavin Biosynthesis. Flavins and Flavoproteins; Springer: New York, NY, USA, 2014; pp. 15–40. [Google Scholar]
- Bacher, A.; Mailänder, B. Biosynthesis of riboflavin in Bacillus subtilis: Function and genetic control of the riboflavin synthase complex. J. Bacteriol. 1978, 134, 476–482. [Google Scholar] [CrossRef]
- Kis, K.; Volk, R.; Bacher, A. Biosynthesis of riboflavin. Studies on the reaction mechanism of 6,7-dimethyl-8-ribityllumazine synthase. Biochemistry 1995, 34, 2883–2892. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.; Neuberger, G.; Fujii, I.; Bown, D.H.; Keller, P.J.; Floss, H.G.; Bacher, A. Biosynthesis of riboflavin. Enzymatic formation of 6,7-dimethyl-8-ribityllumazine from pentose phosphates. Mol. Cell Biol. Res. Commun. 1986, 139, 3661–3669. [Google Scholar] [CrossRef]
- Yakimov, A.P.; Seregina, T.A.; Kholodnyak, A.A.; Kreneva, R.A.; Mironov, A.S.; Perumov, D.A.; Timkovskii, A.L. Possible function of the ribT gene of Bacillus subtilis: Theoretical prediction, cloning, and expression. Acta Naturae 2014, 6, 106–109. [Google Scholar] [CrossRef]
- You, J.; Pan, X.; Yang, C.; Du, Y.; Osire, T.; Yang, T.; Zhang, X.; Xu, M.; Xu, G.; Rao, Z. Microbial production of riboflavin: Biotechnological advances and perspectives. Metab. Eng. 2021, 68, 46–58. [Google Scholar] [CrossRef]
- Shi, S.; Shen, Z.; Chen, X.; Chen, T.; Zhao, X. Increased production of riboflavin by metabolic engineering of the purine pathway in Bacillus subtilis. Biochen. Eng. J. 2009, 46, 28–33. [Google Scholar] [CrossRef]
- Bera, A.K.; Zhu, J.; Zalkin, H.; Smith, J.L. Functional dissection of the Bacillus subtilis pug operator site. J. Bacteriol. 2003, 185, 4099–4109. [Google Scholar] [CrossRef]
- Shin, B.S.; Stein, A.; Zalkin, H. Interaction of Bacillus subtilis purine repressor with DNA. J. Bacteriol. 1997, 179, 7394–7402. [Google Scholar] [CrossRef] [PubMed]
- Mironov, V.N.; Kraev, A.S.; Chikindas, M.L.; Chernov, B.K.; Stepanov, A.I.; Skryabin, K.G. Functional organization of the riboflavin biosynthesis operon from Bacillus subtilis SHgw. Mol. Gen. Genet. 1994, 242, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Arnvig, K.; Hove-Jensen, B.; Switzer, R.L. Purification and properties of phosphoribosyl- diphosphate synthetase from Bacillus subtilis. Eur. J. Biochem. 2010, 192, 195–200. [Google Scholar] [CrossRef]
- Weng, M.; Nagy, P.L.; Zalkin, H. Identification of the Bacillus subtilis pug operon repressor. Proc. Natl. Acad. Sci. USA 1995, 92, 7455–7459. [Google Scholar] [CrossRef]
- Smith, J.L.; Zaluzec, E.J.; Wery, J.P.; Niu, L.; Switzer, R.L.; Zalkin, H.; Satow, Y. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science 1994, 264, 1427–1433. [Google Scholar] [CrossRef]
- Mandal, M.; Boese, B.; Barrick, J.E.; Winkler, W.C.; Breaker, R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 2003, 113, 577–586. [Google Scholar] [CrossRef]
- Christiansen, L.C.; Schou, S.; Nygaard, P.; Saxild, H.H. Xanthine metabolism in Bacillus subtilis: Characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 1997, 179, 2540–2550. [Google Scholar] [CrossRef]
- Denis, V.; Daignan-Fornier, B. Synthesis of glutamine, glycine and 10-formyl tetrahydrofolate is coregulated with purine biosynthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 1998, 259, 246–255. [Google Scholar] [CrossRef]
- Saxild, H.H.; Nygaard, P. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J. Bacteriol. 1987, 169, 2977–2983. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Shiio, I. Improved inosine production and derepression of purine nucleotide biosynthetic enzymes in 8-Azaguanine resistant mutants of Bacillus subtilis. Agric. Biol. Chem. 1972, 36, 1511–1522. [Google Scholar] [CrossRef]
- Matsui, H.; Sato, K.; Enei, H.; Hirose, Y. Mutation of an inosine-producing strain of Bacillus subtilis to DL-methionine sulfoxide resistance for guanosine production. Appl. Environ. Microbiol. 1977, 34, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Sato, K.; Enei, H.; Hirose, Y. Production of guanosine by psicofuranine and decoyinine resistant mutants of Bacillus subtilis. Agric. Biol. Chem. 1979, 43, 1739–1744. [Google Scholar] [CrossRef]
- Asahara, T.; Mori, Y.; Zakataeva, N.P.; Livshits, V.A.; Yoshida, K.; Matsuno, K. Accumulation of gene-targeted Bacillus subtilis mutations that enhance fermentative inosine production. Appl. Microbiol. Biotechnol. 2010, 87, 2195–2207. [Google Scholar] [CrossRef]
- Lobanov, K.V.; Lopes, L.E.; Korol’kova, N.V.; Tyaglov, B.V.; Glazunov, A.V.; Shakulov, R.S.; Mironov, A.S. Reconstruction of purine metabolism in Bacillus subtilis to obtain the strain producer of AICAR: A new drug with a wide range of therapeutic applications. Acta Naturae 2011, 3, 79–89. [Google Scholar] [CrossRef]
- Lobanov, K.V.; Korol’kova, N.V.; Eremina, S.Y.; Lopes, L.É.; Mironov, A.S. Mutation analysis of the purine operon leader region in Bacillus subtilis. Russ. J. Genet. 2011, 47, 785–793. [Google Scholar] [CrossRef]
- Gusarov, I.I.; Kreneva, R.A.; Podcharniaev, D.A.; Iomantas, I.V.; Abalakina, E.G.; Stoĭnova, N.V.; Perumov, D.A.; Kozlov, I.I. Riboflavin biosynthetic genes in Bacillus amyloliquefaciens: Primary structure, organization and regulation of activity. Mol. Biol. 1997, 31, 446–453. [Google Scholar]
- Kil, Y.V.; Mironov, V.N.; Iyu, G.; Kreneva, R.A.; Perumov, D.A. Riboflavin operon of Bacillus subtilis: Unusual symmetric arrangement of the regulatory region. Mol. Gen. Genet. 1992, 233, 483–486. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, S.S.; Santa-Anna, S.; Anna, S.S.; Jiang, C.; Perkins, J. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 2001, 183, 7371–7380. [Google Scholar] [CrossRef]
- Gelfand, M.S.; Mironov, A.A.; Jomantas, J.; Kozlov, Y.I.; Perumov, D.A. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999, 15, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Cohenchalamish, S.; Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 2002, 99, 15908–15913. [Google Scholar] [CrossRef] [PubMed]
- Kreneva, R.A.; Perumov, D.A. Genetic mapping of regulatory mutations of Bacillus subtilis riboflavin operon. Mol. Gen. Genet. 1990, 222, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Solovieva, I.M.; Kreneva, R.A.; Leak, D.J.; Perumov, D.A. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology 1999, 145, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Higashitsuji, Y.; Angerer, A.; Berghaus, S.; Hobl, B.; Mack, M. RibR, a possible regulator of the Bacillus subtilis riboflavin biosynthetic operon, in vivo interacts with the 5’-untranslated leader of rib mRNA. FEMS Microbiol. Lett. 2010, 274, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Mack, M.; Van Loon, A.P.; Hohmann, H.P. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 1998, 180, 950–955. [Google Scholar] [CrossRef]
- Coquard, D.; Huecas, M.; Ott, M.; van Dijl, J.M.; van Loon, A.P.; Hohmann, H.P. Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: A point mutation in ribC results in riboflavin overproduction. Mol. Gen. Genet. 1997, 254, 81–84. [Google Scholar] [CrossRef]
- Zhao, G.; Dong, F.; Lao, X.; Zheng, H. Strategies to increase the production of biosynthetic riboflavin. Mol. Biotechnol. 2021, 63, 909–918. [Google Scholar] [CrossRef]
- Duan, Y.X.; Chen, T.; Chen, X.; Zhao, X. Overexpression of glucose-6-phosphate dehydrogenase enhances riboflavin production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2010, 85, 1907–1914. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, X.; Chen, T.; Shi, S.; Zhao, X. Over-expression of glucose dehydrogenase improves cell growth and riboflavin production in Bacillus subtilis. Biotechnol. Lett. 2006, 28, 1667–1672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, X.; Chen, X.; Li, M.; Wang, X. Enhancement of riboflavin production in Bacillus subtilis via in vitro and in vivo metabolic engineering of pentose phosphate pathway. Biotechnol. Lett. 2021, 43, 2209–2216. [Google Scholar] [CrossRef]
- You, J.; Du, Y.; Pan, X.; Zhang, X.; Yang, T.; Rao, Z. Increased production of riboflavin by coordinated expression of multiple genes in operons in Bacillus subtilis. ACS Synth. Biol. 2022, 11, 801–1810. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, T.; Ma, X.; Shen, Z.; Zhao, X. Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum. Bioresour. Technol. 2011, 102, 3934–3940. [Google Scholar] [CrossRef] [PubMed]
- Tannler, S.; Zamboni, N.C.; Aymerich, S.; Aymerich, S.; Sauer, U. Screening of Bacillus subtilis transposon mutants with altered riboflavin production. Metab. Eng. 2008, 10, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sun, Y.; Fu, S.; Xia, M.; Su, Y.; Liu, C.; Zhang, C.; Zhang, D. Improving the production of riboflavin by introducing a mutant ribulose 5-phosphate-3-epimerase gene in Bacillus subtilis. Front Bioeng. Biotechnol. 2021, 9, 704650. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Bai, L.; Wang, Z.; Shi, T.; Chen, T.; Zhao, X. Enhancement of riboflavin production by deregulating gluconeogenesis in Bacillus subtilis. World J. Microbiol. Biotechnol. 2014, 30, 1893–1900. [Google Scholar] [CrossRef]
- Goodwin, T.W.; Mcevoy, D. Studies on the biosynthesis of riboflavin 5. General factors controlling flavinogenesis in the yeast Candida flareri. Biochem. J. 1959, 71, 742–748. [Google Scholar] [CrossRef] [PubMed]
- Maclaren, J.A. The effects of certain purines and pyrimidines upon the production of riboflavin by Eremothecium ashbyii. J. Bacteriol. 1952, 63, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Santos, M.A.; Pompejus, M.; Revuelta, J. Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii. Appl. Environ. Microbiol. 2005, 71, 5743–5751. [Google Scholar] [CrossRef] [PubMed]
- Mateos, L.; Santos, A.A. Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a myb-related transcription factor in the fungus Ashbya gossypii. Appl. Environ. Microbiol. 2006, 72, 5052–5060. [Google Scholar] [CrossRef]
- Revuelta, J.L.; Santos, M.A.; Alberto, J. Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol. 2008, 8, 67. [Google Scholar]
- Xu, J.; Wang, C.; Ban, R. Improving riboflavin production by modifying related metabolic pathways in Bacillus subtilis. Lett. Appl. Microbiol. 2022, 74, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Chen, T.; Zhang, Z.; Chen, X.; Zhao, X. Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab. Eng. 2009, 11, 243–252. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Tang, W.; Zhang, D. Manipulation of purine metabolic networks for riboflavin production in Bacillus subtilis. ACS Omega 2020, 5, 29140–29146. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Wang, J.Y.; Zhao, X. Effect of riboflavin operon dosage on riboflavin productivity in Bacillus subtilis. J. Tianjin Univ. 2005, 11, 1–5. [Google Scholar]
- Hohmann, H.P.; Huembelin, M.; van Loon, A.P.; Schurter, W. Improved Riboflavin Production. European Patent EP0821063, 21 September 2006. [Google Scholar]
- Lehmann, M.; Degen, S.; Hohmann, H.P.; Wyss, M.; Bacher, A.; Schramek, N. Biosynthesis of riboflavin. Screening for an improved GTP cyclohydrolase II mutant. FEBS J. 2009, 276, 4119–4129. [Google Scholar] [CrossRef]
- Stouthamer, A.H.; Verseveld, H.W.V. Microbial energetics should be considered in manipulating metabolism for biotechnological purposes. Trends Biotechnol. 1987, 5, 149–155. [Google Scholar] [CrossRef]
- Dauner, M.; Sonderegger, M.; Hochuli, M.; Szyperski, T.; Wüthrich, K.; Hohmann, H.P.; Sauer, U.; Bailey, J.E. Intracellular carbon fluxes in riboflavin-producing Bacillus subtilis during growth on two-carbon substrate mixtures. Appl. Environ. Microbiol. 2002, 68, 1760–1771. [Google Scholar] [CrossRef]
- Sauer, U.; Hatzimanikatis, V.; Hohmann, H.P.; Manneberg, M.; van Loon, A.P.; Bailey, J.E. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl. Environ. Microbiol. 1996, 62, 3687–3696. [Google Scholar] [CrossRef]
- Trumpower, B.L.; Gennis, R.B. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: The enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu. Rev. Biochem. 1994, 63, 675–716. [Google Scholar] [CrossRef]
- Zamboni, N.; Mouncey, N.; Hohmann, H.P.; Sauer, U. Reducing maintenance metabolism by metabolic engineering of respiration improves riboflavin production by Bacillus subtilis. Metab. Eng. 2003, 5, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, T.; Chen, X.; Zhao, X. Redirection electron flow to high coupling efficiency of terminal oxidase to enhance riboflavin biosynthesis. Appl. Microbiol. Biotechnol. 2006, 73, 374–383. [Google Scholar] [CrossRef]
- Duan, Y.X. Metabolic Engineering of Producing Riboflavin Strain B. subtilis PY. Ph.D. Thesis, Tianjin University, Tianjin, China, 2009. [Google Scholar]
- Zhu, Y.; Chen, X.; Chen, T.; Zhao, X. Enhancement of riboflavin production by overexpression of acetolactate synthase in a pta mutant of Bacillus subtilis. FEMS Microbiol. Lett. 2007, 266, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Kreneva, R.A.; Gel’Fand, M.S.; Mironov, A.A.; Iomantas, I.; Kozlov, I.; Mironov, A.S.; Perumov, D.A. Study of the phenotypic occurrence of ura gene inactivation in Bacillus subtilis. Genetika 2000, 36, 1166–1168. [Google Scholar]
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic. Acids Res. 2002, 30, 3141–3151. [Google Scholar] [CrossRef]
- Hemberger, S.; Pedrolli, D.B.; Stolz, J.; Vogl, C.; Lehmann, M.; Mack, M. RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains. BMC Biotechnol. 2011, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Maarten, V.D.J.; Michael, H. Bacillus subtilis: From soil bacterium to super-secreting cell factory. Microb. Cell Fact. 2013, 12, 1–6. [Google Scholar]
- Peters, J.M.; Colavin, A.; Shi, H.; Czarny, T.L.; Larson, M.H.; Wong, S.; Hawkins, J.S.; Lu, C.H.S.; Koo, B.; Marta, E.; et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 2016, 165, 1493–1506. [Google Scholar] [CrossRef]
- Westbrook, A.W.; Moo-Young, M.; Chou, C.P. Development of a CRISPR-Cas9 tool kit for comprehensive engineering of Bacillus subtilis. Appl. Environ. Microbiol. 2016, 82, 4876–4895. [Google Scholar] [CrossRef]
- Welsch, N.; Homuth, G.; Schweder, T. Stepwise optimization of a low-temperature Bacillus subtilis expression system for "difficult to express" proteins. Appl. Microbiol. Biotechnol. 2015, 99, 6363–6376. [Google Scholar] [CrossRef]
- Yang, S.; Kang, Z.; Cao, W.; Du, G.; Chen, J. Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J. Biotechnol. 2015, 219, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, J.; Du, G.; Chen, J.; Liu, L. Metabolic engineering of Bacillus subtilis fueled by systems biology: Recent advances and future directions. Biotechnol. Adv. 2016, 35, 20–30. [Google Scholar] [CrossRef]
- Oh, Y.K.; Palsson, B.O.; Park, S.M.; Schilling, C.H.; Mahadevan, R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J. Biol. Chem. 2007, 282, 28791–28799. [Google Scholar] [CrossRef]
- Henry, C.S.; Zinner, J.F.; Cohoon, M.P.; Stevens, R.L. iBsu1103: A new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol. 2009, 10, R69. [Google Scholar] [CrossRef]
- Hao, T.; Han, B.; Ma, H.; Fu, J.; Wang, H.; Wang, Z.; Tang, B.; Chen, T.; Zhao, X. In silico metabolic engineering of Bacillus subtilis for improved production of riboflavin, Egl-237, (R, R)-2,3-butanediol and isobutanol. Mol. Biosyst. 2013, 9, 2034–2044. [Google Scholar] [CrossRef]
- Buescher, J.M.; Liebermeister, W.; Jules, M.; Uhr, M.; Muntel, J.; Botella, E.; Hessling, B.; Kleijn, R.J.; Le Chat, L.; Lecointe, F.; et al. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 2012, 335, 1099–1103. [Google Scholar] [CrossRef]
- Nicolas, P.; Mäder, U.; Dervyn, E.; Uhr, M.; Muntel, J.; Botella, E.; Hessling, B.; Kleijn, R.J.; Le Chat, L.; Lecointe, F.; et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 2012, 335, 1103–1106. [Google Scholar] [CrossRef]
- Revuelta, J.L.; Ledesma-Amaro, R.; Lozano-Martinez, P.; Díaz-Fernández, D.; Buey, R.M.; Jiménez, A. Bioproduction of riboflavin: A bright yellow history. J. Ind. Microbiol. Biotechnol. 2017, 44, 659–665. [Google Scholar] [CrossRef]
- Wang, G.; Shi, T.; Chen, T.; Wang, X.; Wang, Y.; Liu, D.; Guo, J.; Fu, J.; Feng, L.; Wang, Z.; et al. Integrated whole-genome and transcriptome sequence analysis reveals the genetic characteristics of a riboflavin-overproducing Bacillus subtilis. Metab. Eng. 2018, 48, 138–149. [Google Scholar] [CrossRef]
- You, J.; Yang, C.; Pan, X.; Hu, M.; Du, Y.; Osire, T.; Yang, T.; Rao, Z. Metabolic engineering of Bacillus subtilis for enhancing riboflavin production by alleviating dissolved oxygen limitation. Bioresour. Technol. 2021, 333, 125228. [Google Scholar] [CrossRef]
- Barbaupiednoir, E.; de Keersmaecker, S.; Wuyts, V.; Pirovano, W.; Costessi, A.; Philipp, P.; Roosens, N.H. Genome sequence of EU-unauthorized genetically modified Bacillus subtilis strain 2014-3557 overproducing riboflavin, isolated from a Vitamin B2 80% feed additive. Genome Announc. 2015, 3, e00214-15. [Google Scholar] [PubMed]
- Barbau-Piednoir, E.; de Keersmaecker, S.; Delvoye, M.; Gau, C.; Philipp, P.; Roosens, N.H. Use of next generation sequencing data to develop a qPCR method for specific detection of EU-unauthorized genetically modified Bacillus subtilis overproducing riboflavin. BMC Biotechnol. 2015, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Paracchini, V.; Petrillo, M.; Reiting, R.; Angers-Loustau, A.; Wahler, D.; Stolz, A.; Schönig, B.; Matthies, A.; Bendiek, J.; Meinel, D.M.; et al. Molecular characterization of an unauthorized genetically modified Bacillus subtilis production strain identified in a vitamin B2 feed additive. Food Chem. 2017, 230, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hu, W.; Wang, Z.; Chen, T. Rational engineering of Escherichia coli for high-level production of riboflavin. J. Agric. Food Chem. 2021, 69, 12241–12249. [Google Scholar] [CrossRef]
| Gene | Function | Optimum pH * | Optimum Temperature * | Km Value (μM) * | Cofactor | EC Number |
|---|---|---|---|---|---|---|
| ribG | Diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino) uracil reductase | 8.0 | 37 | -/0.005 | -/ NADPH | EC:3.5.4.26/ 1.1.1.93 |
| ribB | riboflavin synthase | 7.4 | 37 | 0.010–0.030 | - | EC:2.5.1.9 |
| ribA | GTP cyclohydrolase II/3,4-dihydroxy-2-butanone-4-phosphate synthase | 8.5/8.0 | 37 | 0.031–0.112/ 0.116–0.181 | - | EC:3.5.4.25/ 4.1.99.12 |
| ribH | 6,7-dimethyl-8-ribityllumazine synthase | 7.0 | 37 | 0.130 | - | EC:2.5.1.78 |
| ribT | unknown | - | - | - | - | - |
| ribC | riboflavin kinase/FAD synthetase | 8.5/- | 52/- | 0.180/- | ATP | EC:2.7.1.26/ 2.7.7.2 |
| ribR | riboflavin kinase | 8.5 | 52 | 0.180 | ATP | EC:2.7.1.26 |
| Screening Drugs | Analogue | Reaction Mechanism | Strain Background | Guanosine Improvement | Reference |
|---|---|---|---|---|---|
| 8-azaguanine | Guanine | Deregulation of the feedback of PRPP amidotransferase | RDA-16 | 1.6–1.8 | [35] |
| Methionine sulfoxide | Purine | Enzyme activity of 5′ -nuclease decreased and enhance XMP synthesis from IMP | AG169 | 1.455 | [36] |
| Psicofuranine | Purine | Improve enzyme activity of GMP synthetase | GP-1 | 1.325 | [37] |
| Decoyinine | Purine | Enhance GMP synthesis from XMP | MG-1 | 1.509 | [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhang, Q.; Qi, X.; Gao, H.; Wang, M.; Guan, H.; Yu, B. Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review. Microorganisms 2023, 11, 164. https://doi.org/10.3390/microorganisms11010164
Liu Y, Zhang Q, Qi X, Gao H, Wang M, Guan H, Yu B. Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review. Microorganisms. 2023; 11(1):164. https://doi.org/10.3390/microorganisms11010164
Chicago/Turabian StyleLiu, Yang, Quan Zhang, Xiaoxiao Qi, Huipeng Gao, Meng Wang, Hao Guan, and Bo Yu. 2023. "Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review" Microorganisms 11, no. 1: 164. https://doi.org/10.3390/microorganisms11010164
APA StyleLiu, Y., Zhang, Q., Qi, X., Gao, H., Wang, M., Guan, H., & Yu, B. (2023). Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review. Microorganisms, 11(1), 164. https://doi.org/10.3390/microorganisms11010164







