Novel Plant-Associated Brevibacillus and Lysinibacillus Genomospecies Harbor a Rich Biosynthetic Potential of Antimicrobial Compounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Isolation, Growth Conditions and DNA Isolation
2.2. Genome Sequencing, Assembly and Annotation
2.3. Biocontrol Activity against Plant Pathogens and Plant Growth Promotion
2.4. Identification of Gene Clusters Involved in the Synthesis of Secondary Metabolites
2.5. Data Analysis
2.6. Gene Bank Accession Numbers of DNA Sequences
3. Results and Discussion
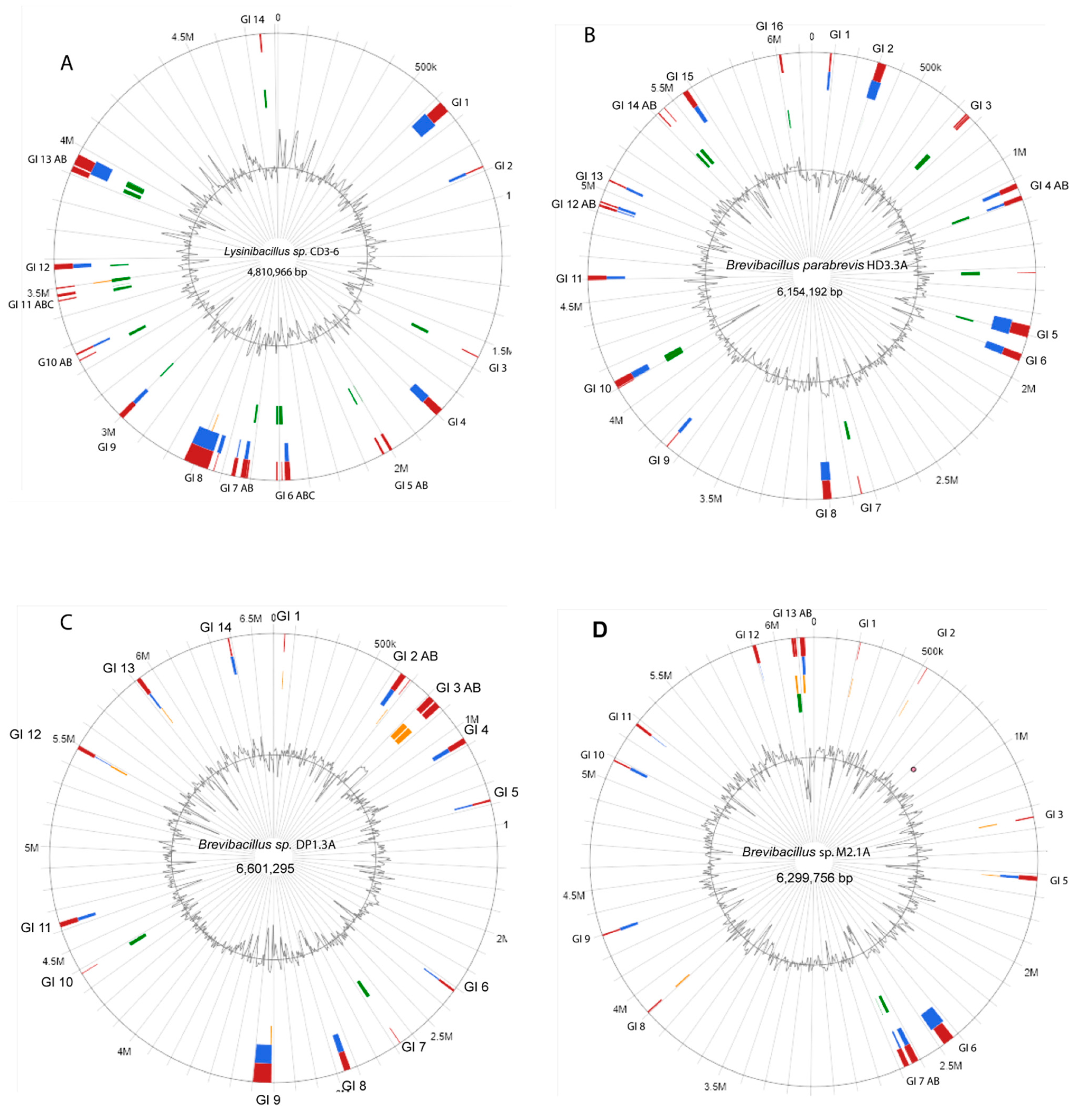
3.1. Resequencing of Selected Lysinibacillus and Brevibacillus Strains Revealed the Presence of Genomic Islands and Extrachromosomal Elements
| Genus | Lysini-bacillus | Brevibacillus | ||
|---|---|---|---|---|
| Strain | CD3-6 | HD3.3A | DP1.3A | M2.1A |
| Extrachrosomal elements (ECE) | ||||
| CP085881 2485 bp | CP085875 61,606 bp ParM-like segregation | CP085877 44,610 bp ParM-like segregation | JABSUY020000003 19,434 bp ParA | |
| CP085878 10,996 bp Rep, RC replication, MobA/L | ||||
| CP085879 6349 bp | ||||
| Genomic features | ||||
| chromosome | CP085880 | CP085874 | CP085876 | JABSUY020000001 |
| Genome size (bp) | 4,810,966 | 6,154,192 | 6,601,295 | 6,172,625 |
| Replication origin (oriC) | 4,810,413…4,810,966 | 6,153,310…6,154,192 | 6,600,712…6,601,295 | 570,707…571,289 |
| G+C % | 37.1 | 52.1 | 47.4 | 47.4 |
| Number of genes (PGAP) | 4794 | 5819 | 6203 | 5956 |
| Genes coding (PGAP) | 4589 | 5570 | 5960 | 5728 |
| CDSs total (PGAP) | 5331 | 5661 | 6030 | 5783 |
| CDS core genome (EDGAR) | 2733 | 3044 | 2607 | 3295 |
| CDS pan genome (EDGAR) | 9100 | 9438 | 15,934 | 13,428 |
| CDS singletons (EDGAR) | 187 | 166 | 365 | 113 |
| Number of RNAs (PGAP) | 149 | 173 | 168 | 166 |
| rRNAs (5S, 16S, 23S, PGAP) | 37 | 38 | 41 | 41 |
| tRNAs (PGAP) | 112 | 130 | 127 | 106 |
| ncRNAs (PGAP) | 5 | 5 | 5 | 5 |
| Pseudo genes (PGAP) | 51 | 76 | 70 | 55 |
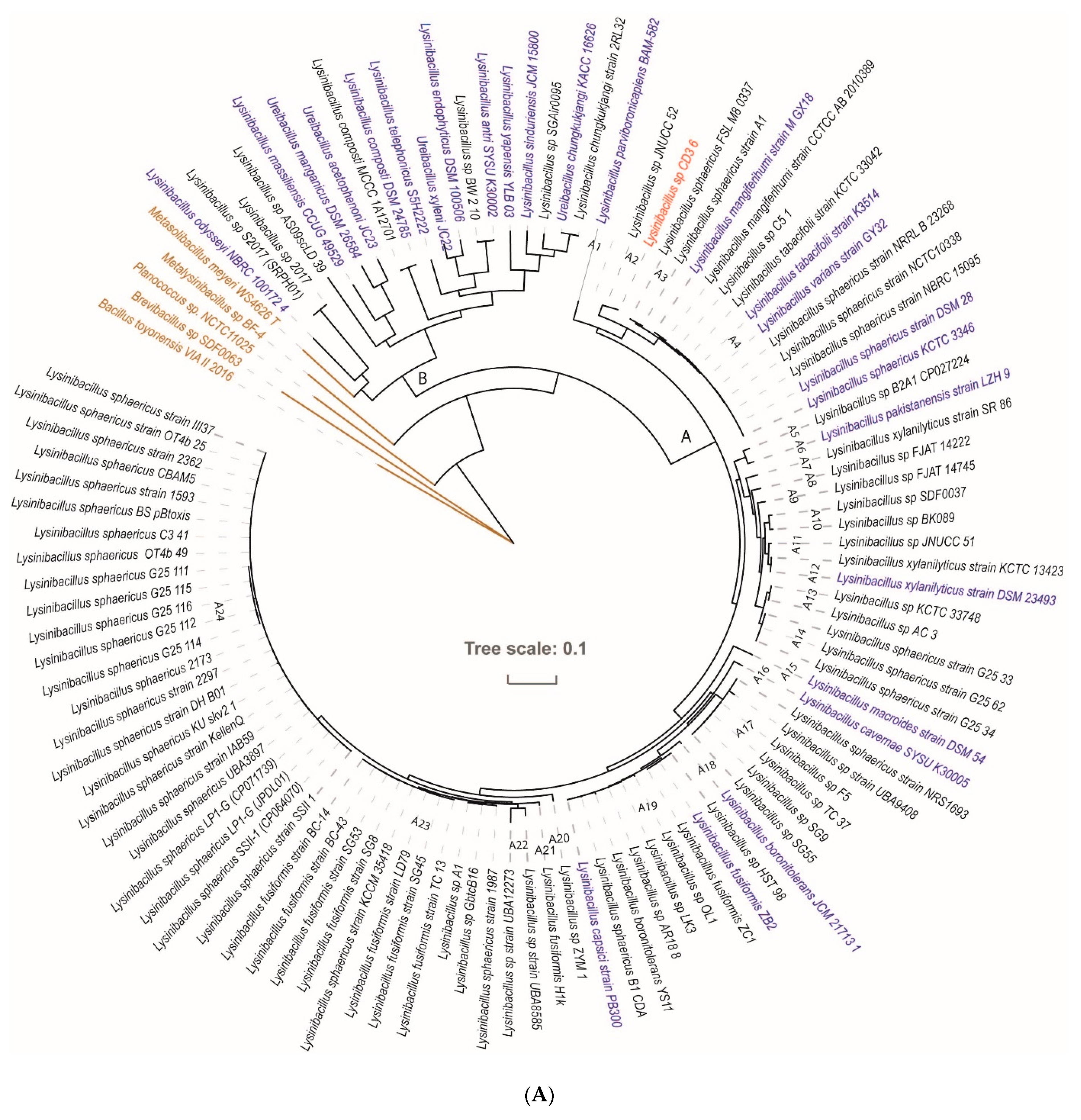
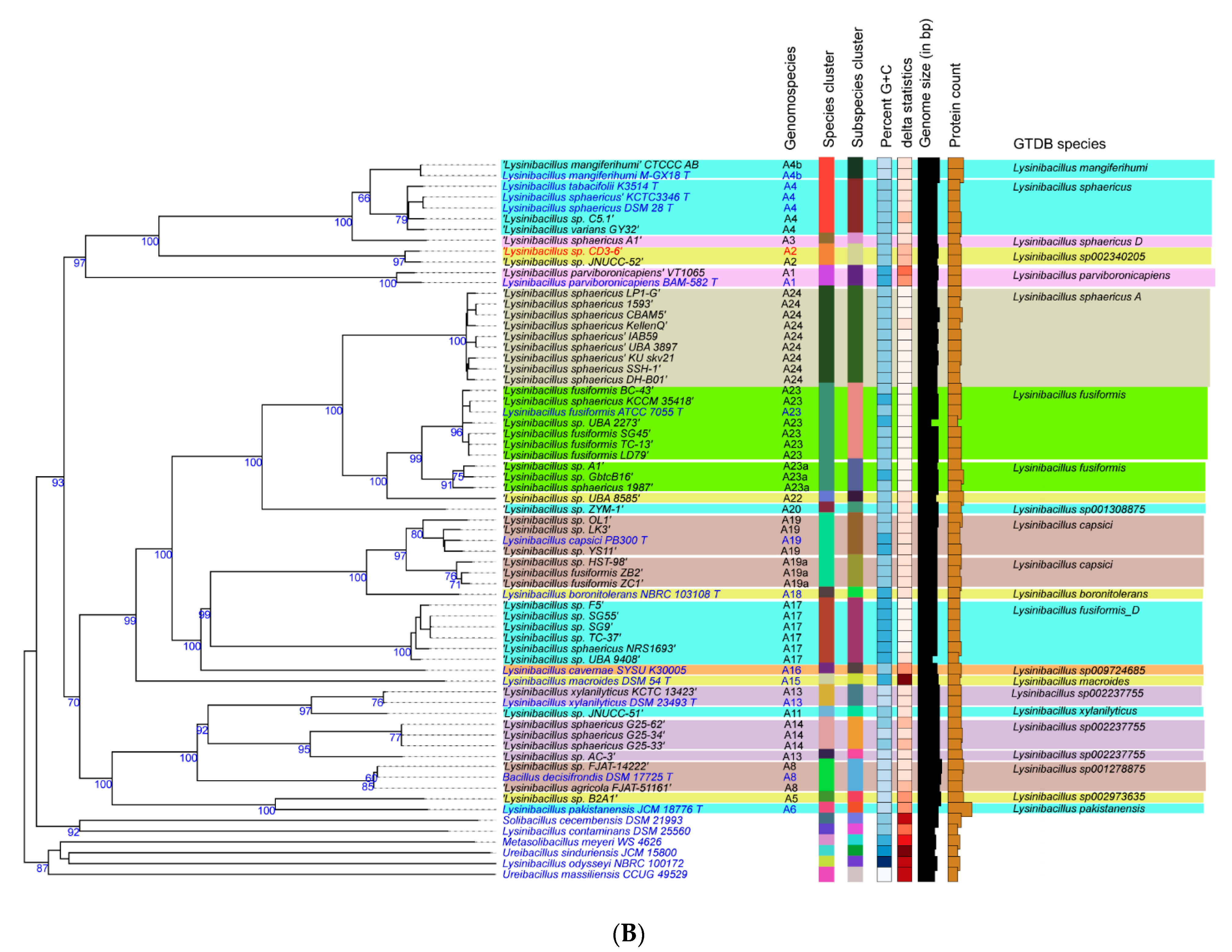
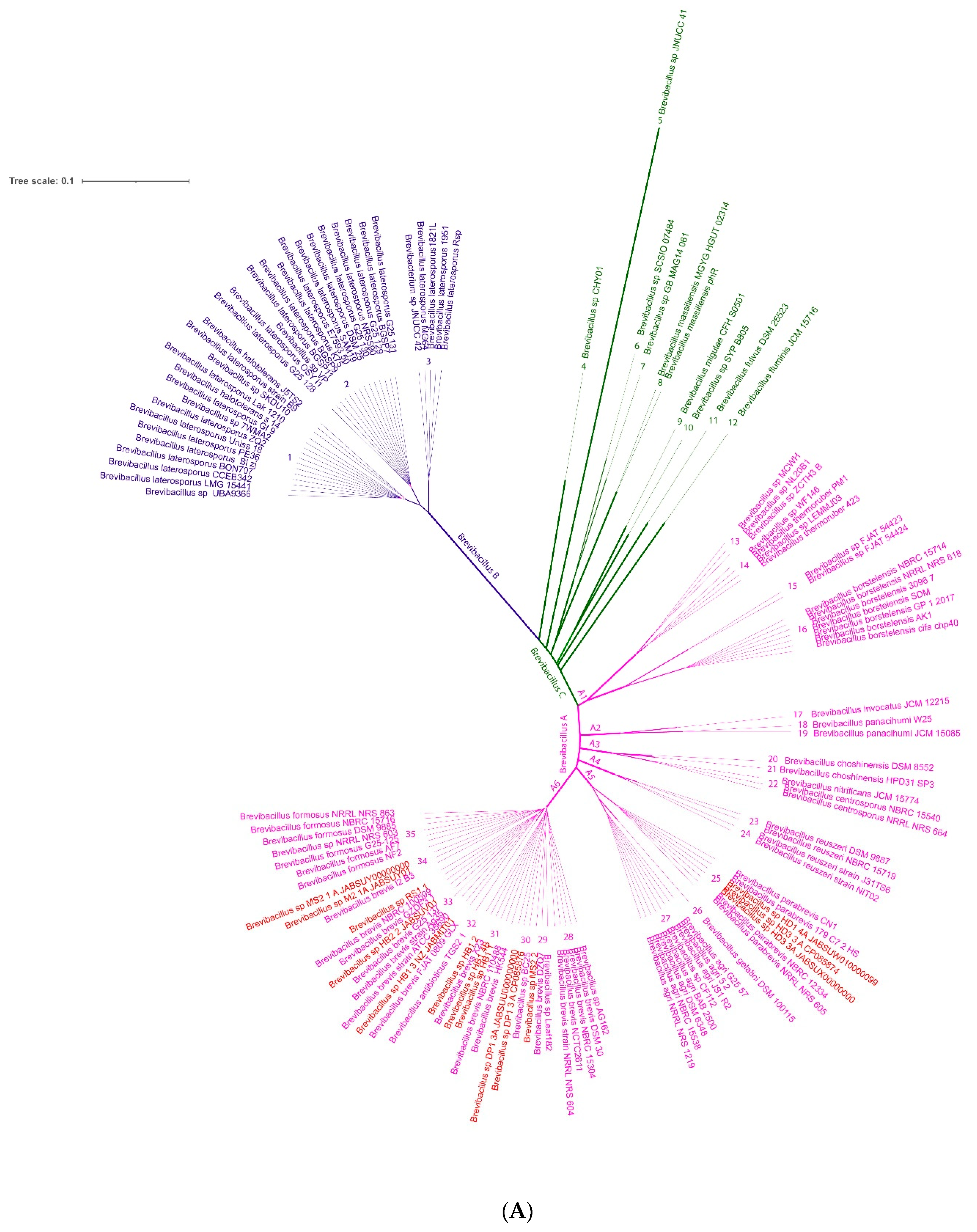
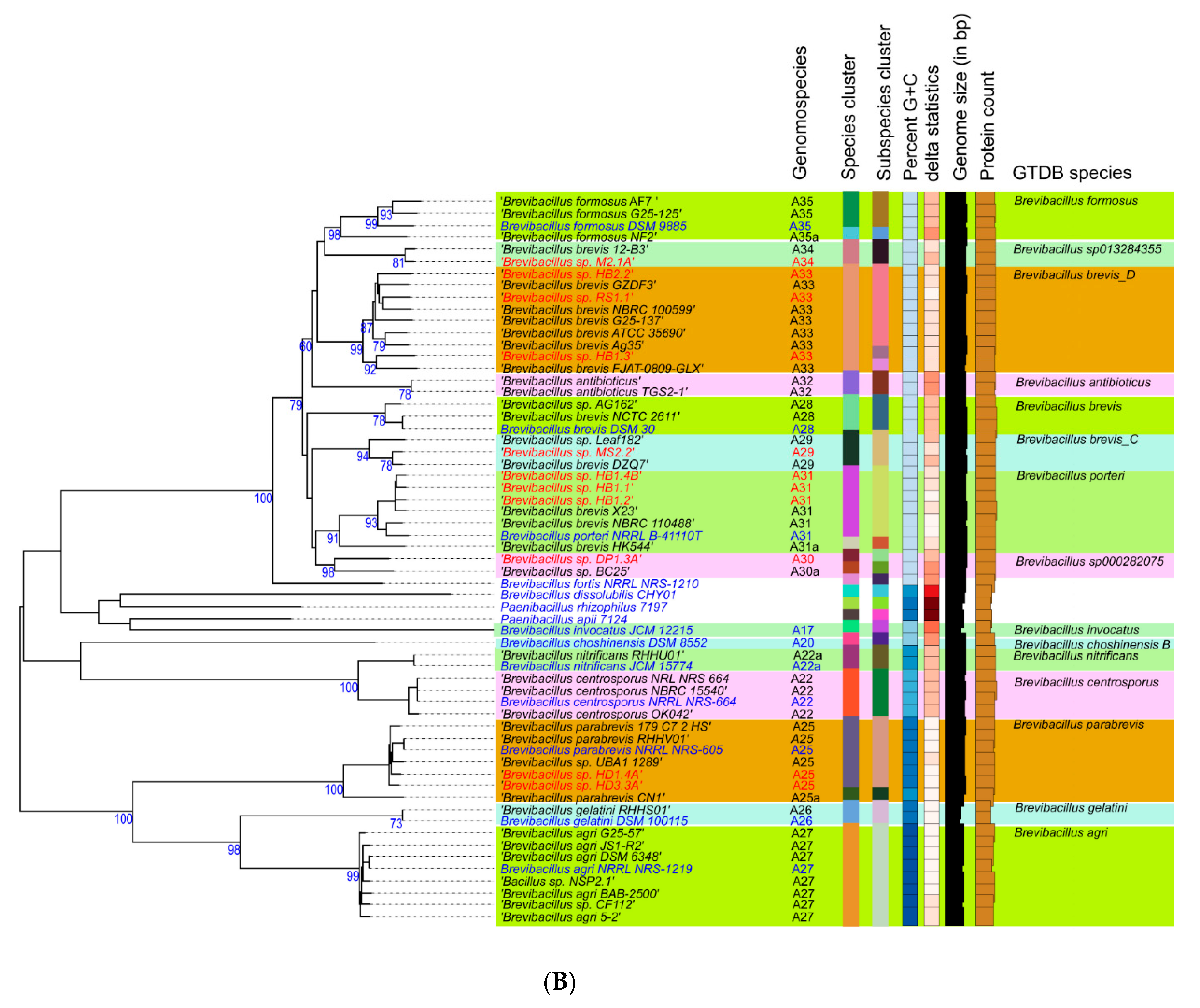
3.2. Taxonomic Evaluation of Lysinibacillus and Brevibacillus Strains Revealed Novel Genomospecies
3.2.1. Lysinibacillus CD3-6 Forms a Distinct Genomospecies Together with Lysinibacillus JNUCC-52
3.2.2. Novel GS Were Assigned for Plant-Associated Brevibacillus Isolates
- Strains HD1.4A and HD3.3A represented the Brevibacillus parabrevis species cluster (GS A5-25). They only shared ANIb values of ≤85% with the other group A Brevibacillus clusters.
- The Brevibacillus porteri species cluster (GS A6-31) was formed by HB1.1, HB 1.2, and HB1.4B. Their ANIb-values (92–93%), when compared with the other “brevis” group strains, were below the species cut-off level (95–96%) recommended as the ANI criterion for interspecies identity.
- The same was true for HB1.3, HB2.2, and RS1.1 forming together with other genomes GS A6-33. The cluster was designated according to the GTDB classification, as Brevibacillus brevis D and did not contain a type strain.
- Strain MS2.2 formed together with Leaf182 the GS A6-29, designated as Brevibacillus brevis C. Their ANI values were found below 93% when compared with the most related clusters of the A6-branch.
- Brevibacillus sp. DP1.3A shared a common cluster (A6-30) with the genome of Brevibacillus sp. BC25. The GTDB classification of this cluster was s_Brevibacillus sp. 000282075.
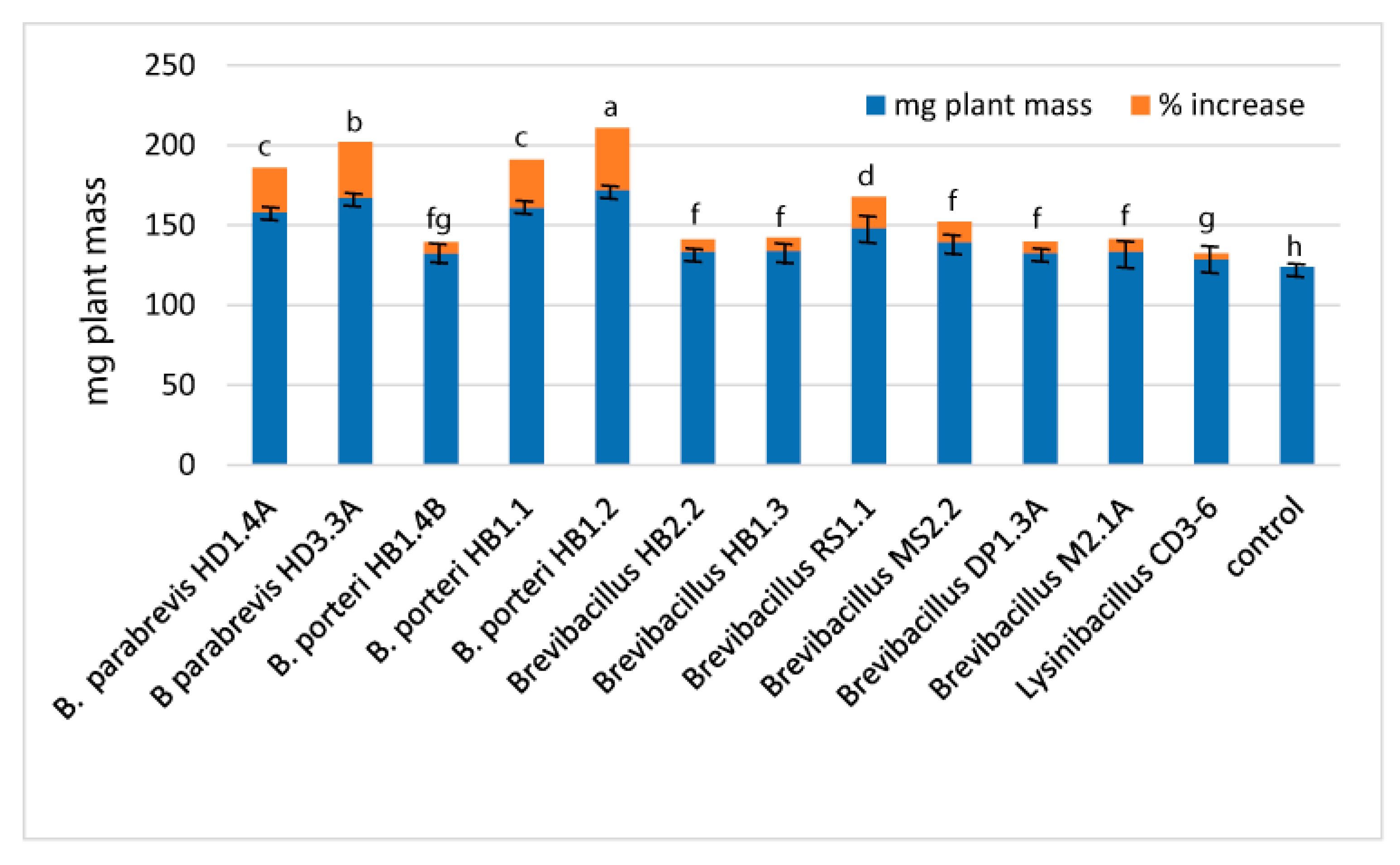
3.3. Plant-Associated Brevibacilli Promote Plant Growth and Suppress Plant Pathogens
3.3.1. Antagonistic Activity against Gram-Positive and Gram-Negative Bacteria
3.3.2. Antifungal Activity
3.3.3. Nematicidal Activity
3.3.4. Plant Growth Promotion
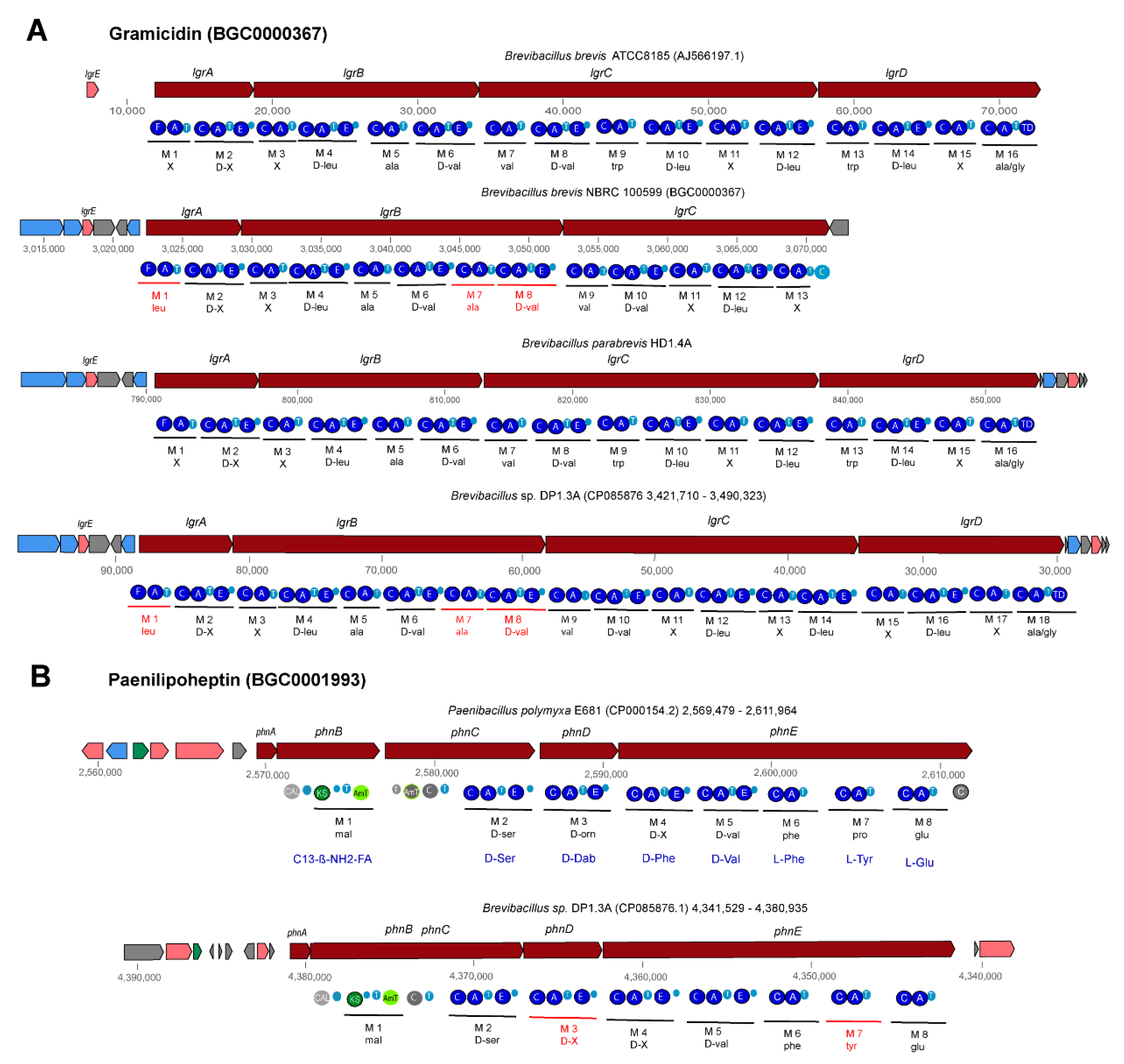
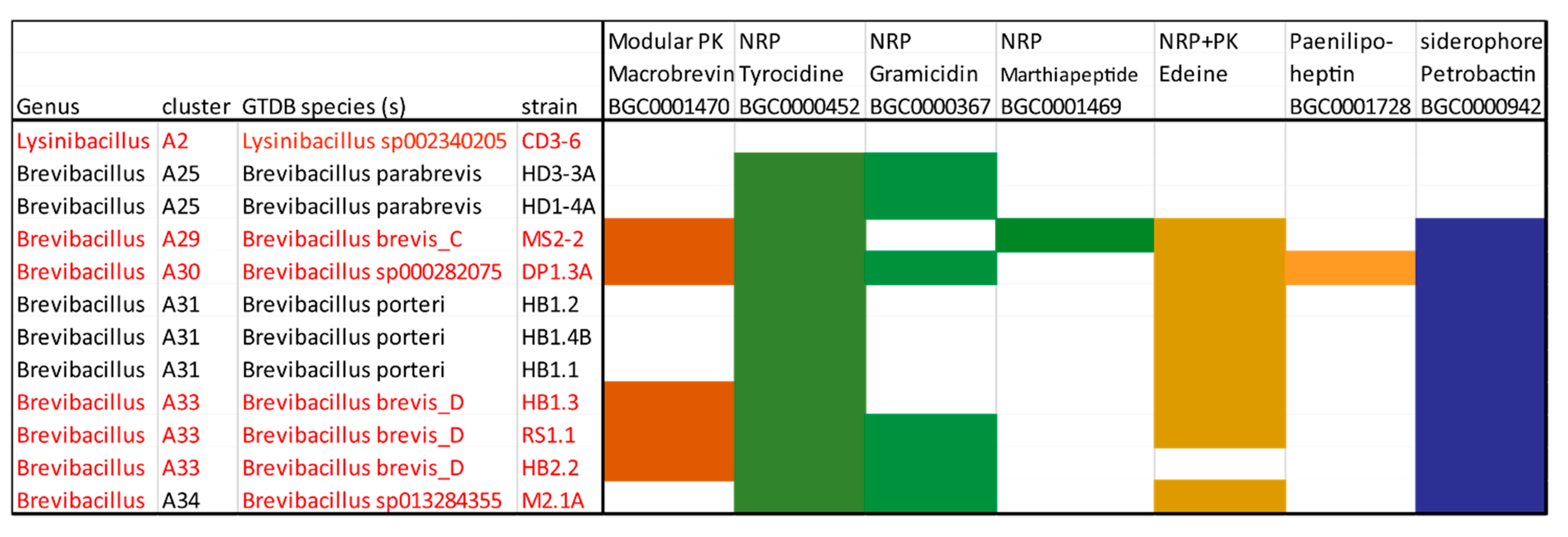
3.4. Genome Mining for Putative Natural Product Biosynthesis Gene Clusters
3.4.1. Gene Clusters Encoding Modular and Nonmodular Polyketides
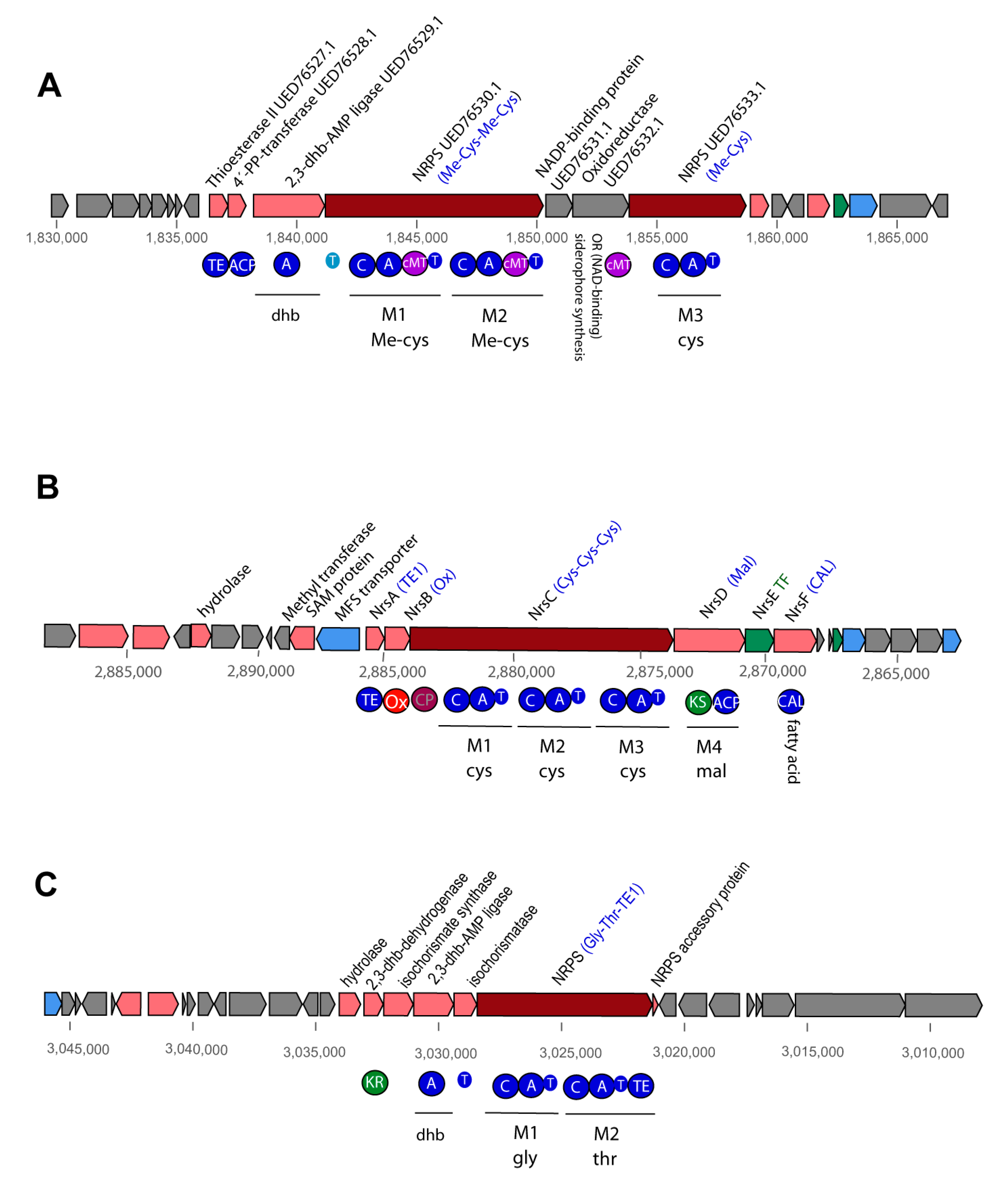
3.4.2. Non-Ribosomal-Synthesized Antimicrobial Peptides (NRP)
3.4.3. Gene Clusters Encoding PK-NRP Hybrids
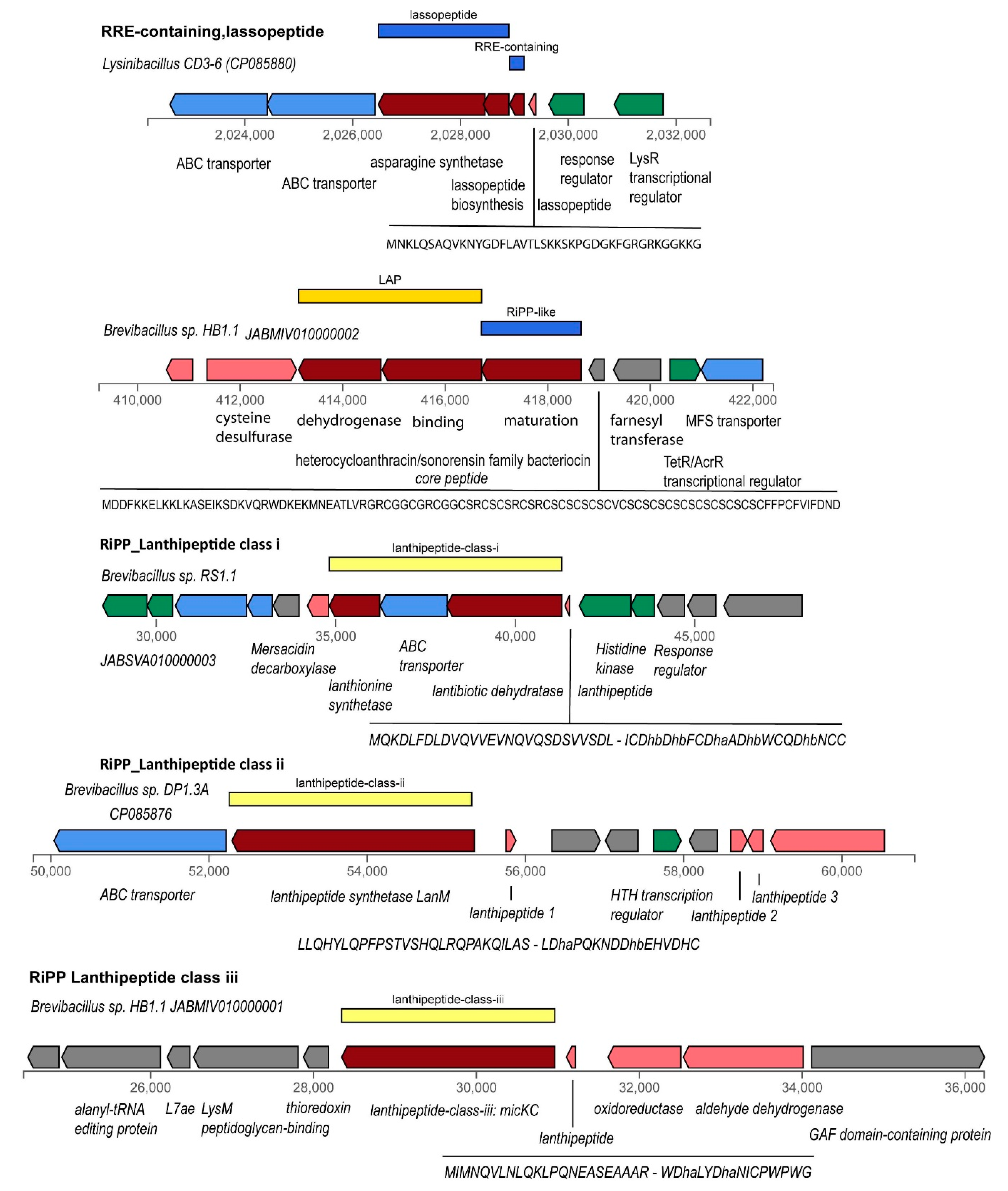
3.4.4. Gene Clusters Representing RiPPs, and Bacteriocins
3.4.5. Gene Cluster Involved in Synthesis of Siderophores and Other BGCs
3.4.6. Uncharacterized NRP and PK-NRP Hybrid Gene Clusters in Brevibacilli
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tam, L.T.T.; Jähne, J.; Luong, P.T.; Thao, L.T.P.; Chung, L.T.K.; Schneider, A.; Blumenscheit, C.; Lasch, P.; Schweder, T.; Borriss, R. Draft genome sequences of 59 endospore-forming Gram-positive bacteria associated with crop plants grown in Vietnam. Microbiol. Resour. Announc. 2020, 9, e01154-20. [Google Scholar] [CrossRef] [PubMed]
- Blumenscheit, C.; Jähne, J.; Schneider, A.; Blom, J.; Schweder, T.; Lasch, P.; Borriss, R. Genome sequence data of Bacillus velezensis BP1.2A and BT2.4. Data Brief 2022, 41, 107978. [Google Scholar] [CrossRef] [PubMed]
- Shida, O.; Takagi, H.; Kadowaki, K.; Komagata, K. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int. J. Syst. Bacteriol. 1996, 46, 939–946, Erratum in Int. J. Syst. Bacteriol. 1997, 47, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Edwards, S.G.; Seddon, B. Mode of antagonism of Brevibacillus brevis against Botrytis cinerea in vitro. J. Appl. Microbiol. 2001, 91, 652–659. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, B.X.; Zhang, Z.; Shen, W.F.; Yang, C.H.; Yu, J.Q.; Zhao, Y.H. Survival of the biocontrol agents Brevibacillus brevis ZJY-1 and Bacillus subtilis ZJY-116 on the spikes of barley in the field. J. Zhejiang Univ. Sci. B 2005, 8, 770–777. [Google Scholar] [CrossRef] [Green Version]
- Li, C.P.; Shi, W.C.; Wu, D.; Tian, R.M.; Wang, B.; Lin, R.S.; Zhou, B.; Gao, Z. Biocontrol of potato common scab by Brevibacillus laterosporus BL12 is related to the reduction of pathogen and changes in soil bacterial community. Biol. Control 2021, 153, 104496. [Google Scholar] [CrossRef]
- Yang, X.; Yousef, A.E. Antimicrobial peptides produced by Brevibacillus spp.: Structure, classification and bioactivity: A mini review. World J. Microbiol. Biotechnol. 2018, 34, 57. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Vogel, C.M.; Ueoka, R.; Schäfer, M.; Ryffel, F.; Müller, D.B.; Probst, S.; Kreuzer, M.; Piel, J.; Vorholt, J.A. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat. Microbiol. 2018, 3, 909–919. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1117–1125. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 10, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Dieckmann, M.A.; Beyvers, S.; Nkouamedjo-Fankep, R.C.; Hanel, P.H.G.; Jelonek, L.; Blom, J.; Goesmann, A. EDGAR3.0: Comparative genomics and phylogenomics on a scalable infrastructure. Nucleic Acids Res. 2021, 49, W185–W192. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, X.; Luo, H.; Fan, Z.; Luo, J.; He, S.; Yue, H.; Zhang, P.; Chen, R. BioCircos.js: An interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics 2016, 32, 1740–1742. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Sardà Carbasse, J.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, J.A.; Fleming, J.T. Basic culture methods. In Caenorhabditis Elegans: Modern Biological Analysis of an Organism; Epstein, H.F., Shakes, D.C., Eds.; Academic: San Diego, CA, USA, 1995; pp. 3–29. [Google Scholar]
- Liu, Z.; Budiharjo, A.; Wang, P.; Shi, H.; Fang, J.; Borriss, R.; Zhang, K.; Huang, X. The highly modified microcin peptide plantazolicin is associated with nematicidal activity of Bacillus amyloliquefaciens FZB42. Appl. Microbiol. Biotechnol. 2013, 97, 10081–10090. [Google Scholar] [CrossRef]
- Hooper, D.J.; Hallmann, J.; Subbotin, S.A. Methods for extraction, processing and detection of plant and soil nematodes. In Plant Parasitic Nematodes in Subtropical and Tropical Agriculture; Luc, M., Sikora, R., Bridge, J., Eds.; CAB International: Wallingford, UK, 2005; pp. 53–86. [Google Scholar]
- Bridge, J.; Page, S.L.J. Estimation of Root Knot Nematode Infestation Levels in Roots Using a Rating Chart. Trop. Pest Manag. 1980, 26, 296–298. [Google Scholar] [CrossRef]
- Budiharjo, A.; Chowdhury, S.P.; Dietel, K.; Beator, B.; Dolgova, O.; Fan, B.; Bleiss, W.; Ziegler, J.; Schmid, M.; Hartmann, A.; et al. Transposon mutagenesis of the plant-associated Bacillus amyloliquefaciens ssp. plantarum FZB42 revealed that the nfrA and RBAM17410 genes are involved in plant-microbe-interactions. PLoS ONE 2014, 9, e98267. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Makarova, K.S.; Grishin, N.V.; Koonin, E.V. The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics 2006, 22, 2581–2584. [Google Scholar] [CrossRef] [Green Version]
- Min, Y.N.; Tabuchi, A.; Womble, D.D.; Rownd, R.H. Transcription of the stability operon of IncFII plasmid NR1. J. Bacteriol. 1991, 173, 2378–2384. [Google Scholar] [CrossRef]
- Marsin, S.; Forterre, P. A rolling circle replication initiator protein with a nucleotidyl-transferase activity encoded by the plasmid pGT5 from the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 1998, 6, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Monzingo, A.F.; Ozburn, A.; Xia, S.; Meyer, R.J.; Robertus, J.D. The structure of the minimal relaxase domain of MobA at 2.1 A resolution. J. Mol. Biol. 2007, 366, 165–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free. Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Baxter, J.C.; Waples, W.G.; Funnell, B.E. Nonspecific DNA binding by P1 ParA determines the distribution of plasmid partition and repressor activities. J. Biol. Chem. 2020, 295, 17298–17309. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.J.; Luo, H.; Gao, F. Ori-Finder 2022: A Comprehensive Web Server for Prediction and Analysis of Bacterial Replication Origins. Genom. Proteom. Bioinform. Biorxiv 2022. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, C.A. Lysinibacillus mangiferihumi, Lysinibacillus tabacifolii and Lysinibacillus varians are later heterotypic synonyms of Lysinibacillus sphaericus. Int. J. Syst. Evol. Microbiol. 2019, 69, 2958–2962. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory friendly classification with the Genome Taxonomy Database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.A.; Hugenholtz, P. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arie, T. Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J. Pestic. Sci. 2019, 44, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Chi, N.M.; Thu, P.Q.; Nam, H.B.; Quang, D.Q.; Phong, L.V.; Van, N.D.; Trang, T.T.; Kien, T.T.; Tam, T.T.T.; Dell, B. Management of Phytophthora palmivora disease in Citrus reticulata with chemical fungicides. J. Gen. Plant Pathol. 2020, 86, 494–502. [Google Scholar] [CrossRef]
- Eisenback, J.D.; Triantaphyllou, H.H. Root-knot nematodes: Meloidogyne species and races. In Manual of Agricultural Nematology; Nickle, W.R., Ed.; Marcell Dekker: New York, NY, USA, 1991; pp. 191–274. [Google Scholar]
- Hussey, P.S.; Barker, K.R. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis. Rep. 1973, 57, 1025–1028. [Google Scholar]
- Anastasiadis, I.A.; Giannakou, I.O.; Prophetou-Athanasiadou, D.A.; Gowen, S.R. The combined effect of the application of a biocontrol agent Paecilomyces lilacinus, with various practices for the control of root-knot nematodes. Crop Prot. 2008, 27, 352–361. [Google Scholar] [CrossRef]
- Engelbrecht, G.; Claassens, S.; Mienie, M.S.; Fourie, H. Filtrates of mixed Bacillus spp. inhibit second-stage juvenile motility of root-knot nematodes. Rhizosphere 2022, 22, 100528. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [Green Version]
- Tracanna, V.; de Jong, A.; Medema, M.H.; Kuipers, O.P. Mining prokaryotes for antimicrobial compounds: From diversity to function. FEMS Microbiol. Rev. 2017, 41, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, L.; Wang, Y.; Zhang, C.; Liu, T.; Duan, C.; Bian, X.; Guo, Z.; Long, Q.; Tang, Y.; et al. Enhancement of edeine production in Brevibacillus brevis X23 via in situ promoter engineering. Microb. Biotechnol. 2022, 15, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Vater, J.; Herfort, S.; Doellinger, J.; Weydmann, M.; Borriss, R.; Lasch, P. Genome Mining of the Lipopeptide Biosynthesis of Paenibacillus polymyxa E681 in Combination with Mass Spectrometry: Discovery of the Lipoheptapeptide Paenilipoheptin. ChemBioChem 2018, 19, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Khosla, C.; Gokhale, R.S.; Jacobsen, J.R.; Cane, D.E. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 1999, 68, 219–253. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef]
- Funa, N.; Ohnishi, Y.; Fujii, I.; Shibuya, M.; Ebizuka, Y.; Horinouchi, S. A new pathway for polyketide synthesis in microorganisms. Nature 1999, 400, 897–899. [Google Scholar] [CrossRef]
- Nakano, C.; Ozawa, H.; Akanuma, G.; Funa, N.; Horinouchi, S. Biosynthesis of aliphatic polyketides by type III polyketide synthase and methyltransferase in Bacillus subtilis. J. Bacteriol. 2009, 191, 4916–4923. [Google Scholar] [CrossRef] [Green Version]
- Finking, R.; Marahiel, M.A. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- Mootz, H.D.; Marahiel, M.A. The tyrocidine biosynthesis operon of Bacillus brevis: Complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 1997, 179, 6843–6850. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, M.; Rautenbach, M.; Vosloo, J.A.; Siersma, T.; Aisenbrey, C.H.M.; Zaitseva, E.; Laubscher, W.E.; van Rensburg, W.; Behrends, J.C.; Bechinger, B.; et al. The Multifaceted Antibacterial Mechanisms of the Pioneering Peptide Antibiotics Tyrocidine and Gramicidin S. mBio 2018, 9, e00802-18. [Google Scholar] [CrossRef] [Green Version]
- Kessler, N.; Schuhmann, H.; Morneweg, S.; Linne, U.; Marahiel, M.A. The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J. Biol. Chem. 2004, 279, 7413–7419. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Huang, H.; Chen, Y.; Tan, J.; Song, Y.; Zou, J.; Tian, X.; Hua, Y.; Ju, J. Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 2012, 75, 2251–2255. [Google Scholar] [CrossRef]
- Kurylo-Borowska, Z.; Szer, W. Inhibition of bacterial DNA synthesis by edeine. Effect on Escherichia coli mutants lacking DNA polymerase I. Biochim. Biophys. Acta 1972, 287, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Westman, E.L.; Yan, M.; Waglechner, N.; Koteva, K.; Wright, G.D. Self resistance to the atypical cationic antimicrobial peptide edeine of Brevibacillus brevis Vm4 by the N-acetyltransferase EdeQ. Chem Biol. 2013, 20, 983–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, B.J.; Hudson, G.A.; Dunbar, K.L.; Mitchell, D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 2015, 11, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Kloosterman, A.M.; Shelton, K.E.; van Wezel, G.P.; Medema, M.H.; Mitchell, D.A. RRE-Finder: A Genome-Mining Tool for Class-Independent RiPP Discovery. mSystems 2020, 5, e00267-20. [Google Scholar] [CrossRef]
- Valdés-Stauber, N.; Scherer, S. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl. Environ. Microbiol. 1994, 60, 3809–3814. [Google Scholar] [CrossRef] [Green Version]
- Hegemann, J.D.; Süssmuth, R.D. Matters of class: Coming of age of class III and IV lanthipeptides. RSC Chem. Biol. 2020, 1, 110–127. [Google Scholar] [CrossRef]
- Anthony, T.; Chellappa, G.S.; Rajesh, T.; Gunasekaran, P. Functional analysis of a putative holin-like peptide-coding gene in the genome of Bacillus licheniformis AnBa9. Arch. Microbiol. 2010, 192, 51–56. [Google Scholar] [CrossRef]
- Aunpad, R.; Panbangred, W. Evidence for two putative holin-like peptides encoding genes of Bacillus pumilus strain WAPB4. Curr. Microbiol. 2012, 64, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Janes, B.K.; Passalacqua, K.D.; Pfleger, B.F.; Bergman, N.H.; Liu, H.; Håkansson, K.; Somu, R.V.; Aldrich, C.C.; Cendrowski, S.; et al. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J. Bacteriol. 2007, 189, 1698–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagan, A.K.; Carlson, P.E., Jr.; Hanna, P.C. Flying under the radar: The non-canonical biochemistry and molecular biology of petrobactin from Bacillus anthracis. Mol. Microbiol. 2016, 102, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Koppisch, A.T.; Dhungana, S.; Hill, K.K.; Boukhalfa, H.; Heine, H.S.; Colip, L.A.; Romero, R.B.; Shou, Y.; Ticknor, L.O.; Marrone, B.L.; et al. Petrobactin is produced by both pathogenic and non-pathogenic isolates of the Bacillus cereus group of bacteria. Biometals 2008, 21, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhou, H.; Dai, G.; Zhong, G.; Huo, L.; Li, A.; Liu, Y.; Yang, M.; Ravichandran, V.; Zheng, Z.; et al. Characterization of a Cryptic NRPS Gene Cluster in Bacillus velezensis FZB42 Reveals a Discrete Oxidase Involved in Multithiazole Biosynthesis. ACS Catal. 2022, 12, 3371–3381. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jähne, J.; Le Thi, T.T.; Blumenscheit, C.; Schneider, A.; Pham, T.L.; Le Thi, P.T.; Blom, J.; Vater, J.; Schweder, T.; Lasch, P.; et al. Novel Plant-Associated Brevibacillus and Lysinibacillus Genomospecies Harbor a Rich Biosynthetic Potential of Antimicrobial Compounds. Microorganisms 2023, 11, 168. https://doi.org/10.3390/microorganisms11010168
Jähne J, Le Thi TT, Blumenscheit C, Schneider A, Pham TL, Le Thi PT, Blom J, Vater J, Schweder T, Lasch P, et al. Novel Plant-Associated Brevibacillus and Lysinibacillus Genomospecies Harbor a Rich Biosynthetic Potential of Antimicrobial Compounds. Microorganisms. 2023; 11(1):168. https://doi.org/10.3390/microorganisms11010168
Chicago/Turabian StyleJähne, Jennifer, Thanh Tam Le Thi, Christian Blumenscheit, Andy Schneider, Thi Luong Pham, Phuong Thao Le Thi, Jochen Blom, Joachim Vater, Thomas Schweder, Peter Lasch, and et al. 2023. "Novel Plant-Associated Brevibacillus and Lysinibacillus Genomospecies Harbor a Rich Biosynthetic Potential of Antimicrobial Compounds" Microorganisms 11, no. 1: 168. https://doi.org/10.3390/microorganisms11010168





