Effect of Parthenium hysterophorus L. Invasion on Soil Microbial Communities in the Yellow River Delta, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site
2.2. Sampling Collection, Determination of Soil Physical and Chemical Properties
2.3. High-Throughput Sequencing of Soil Microbe
2.4. Data Analysis
3. Results
3.1. Soil Physical and Chemical Properties
3.2. Alpha Diversity Index Difference Analysis among Different Groups
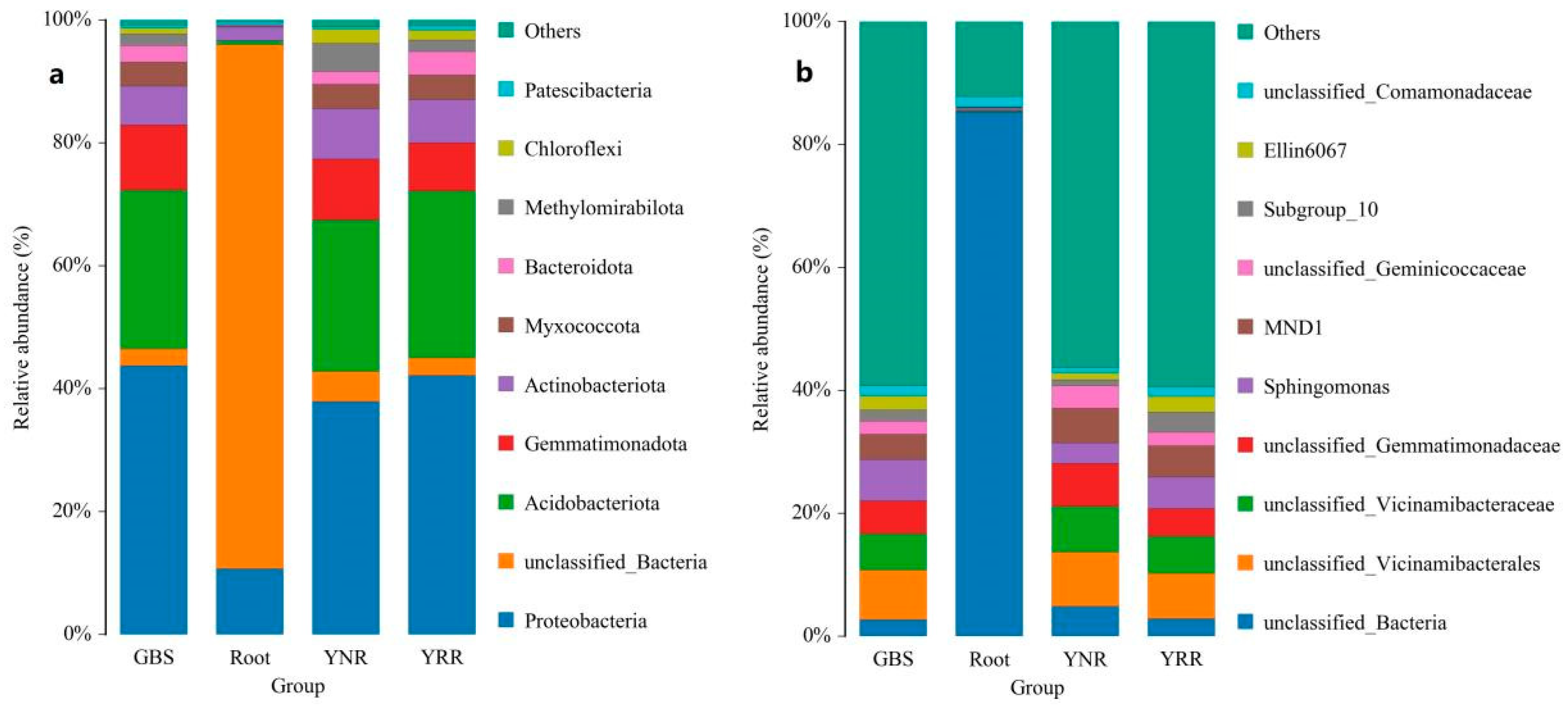
3.3. OTU Abundance Analysis
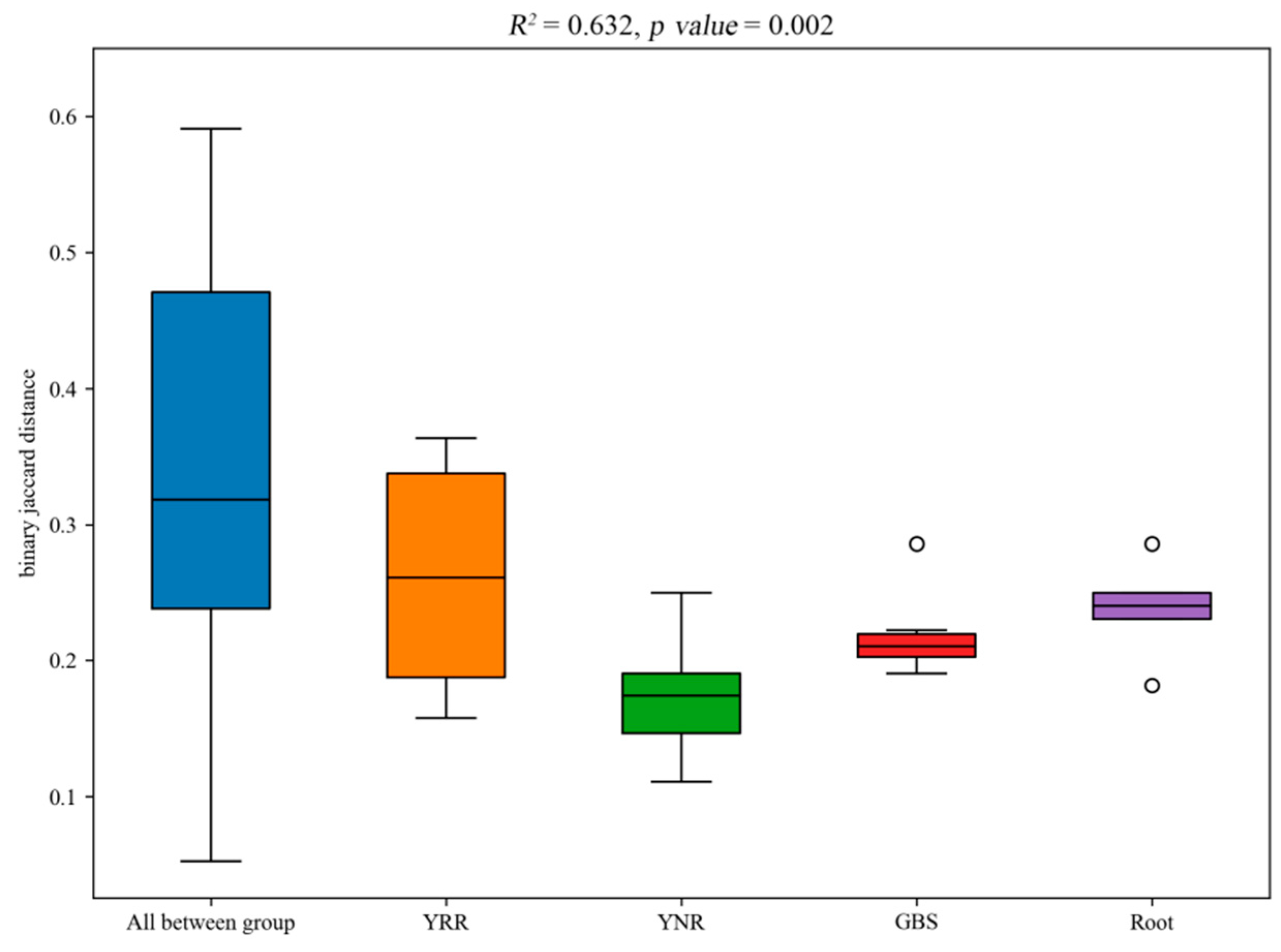
3.4. Soil Microbial Community Structure Analysis
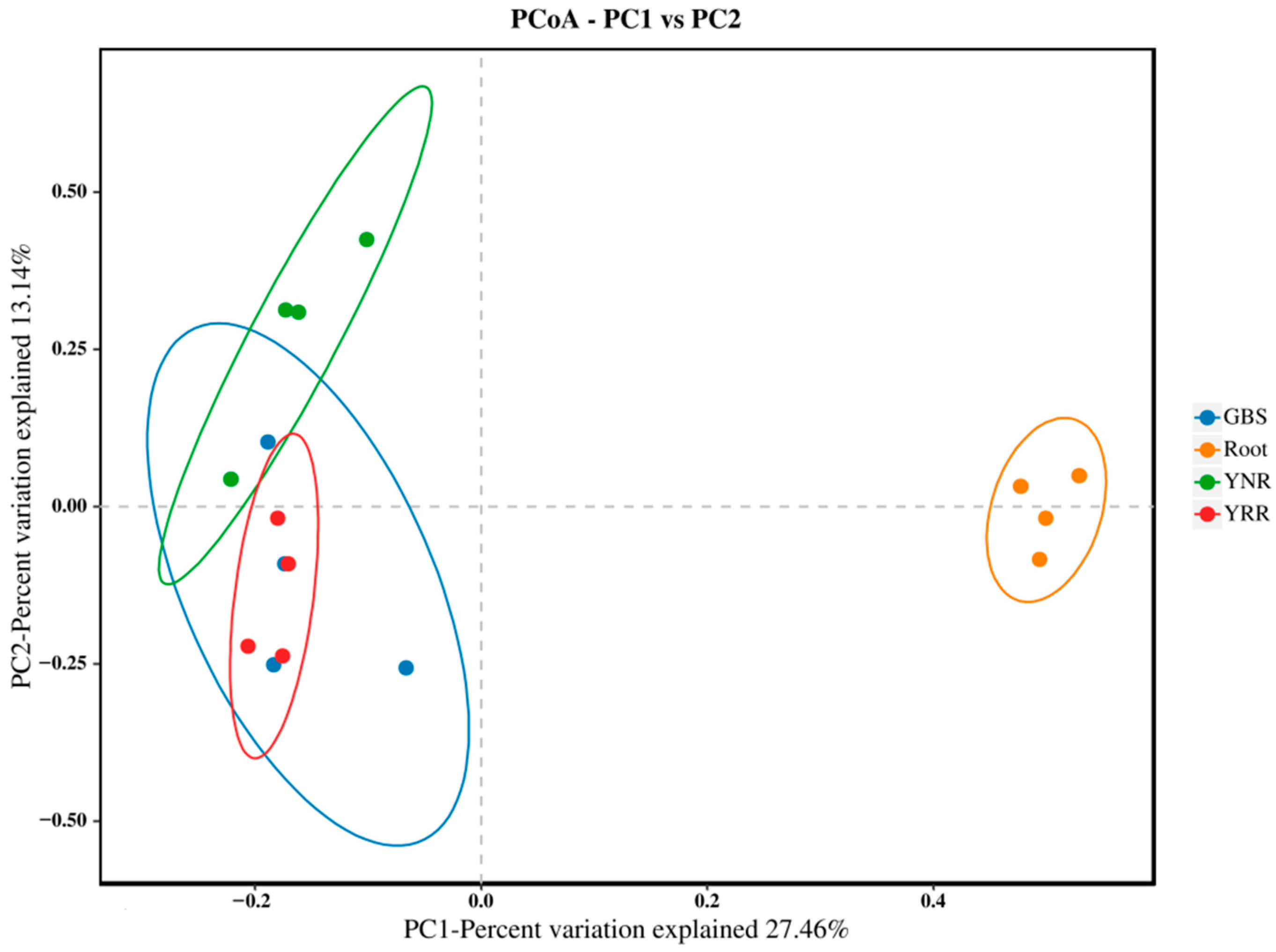
3.5. PCoA Analysis
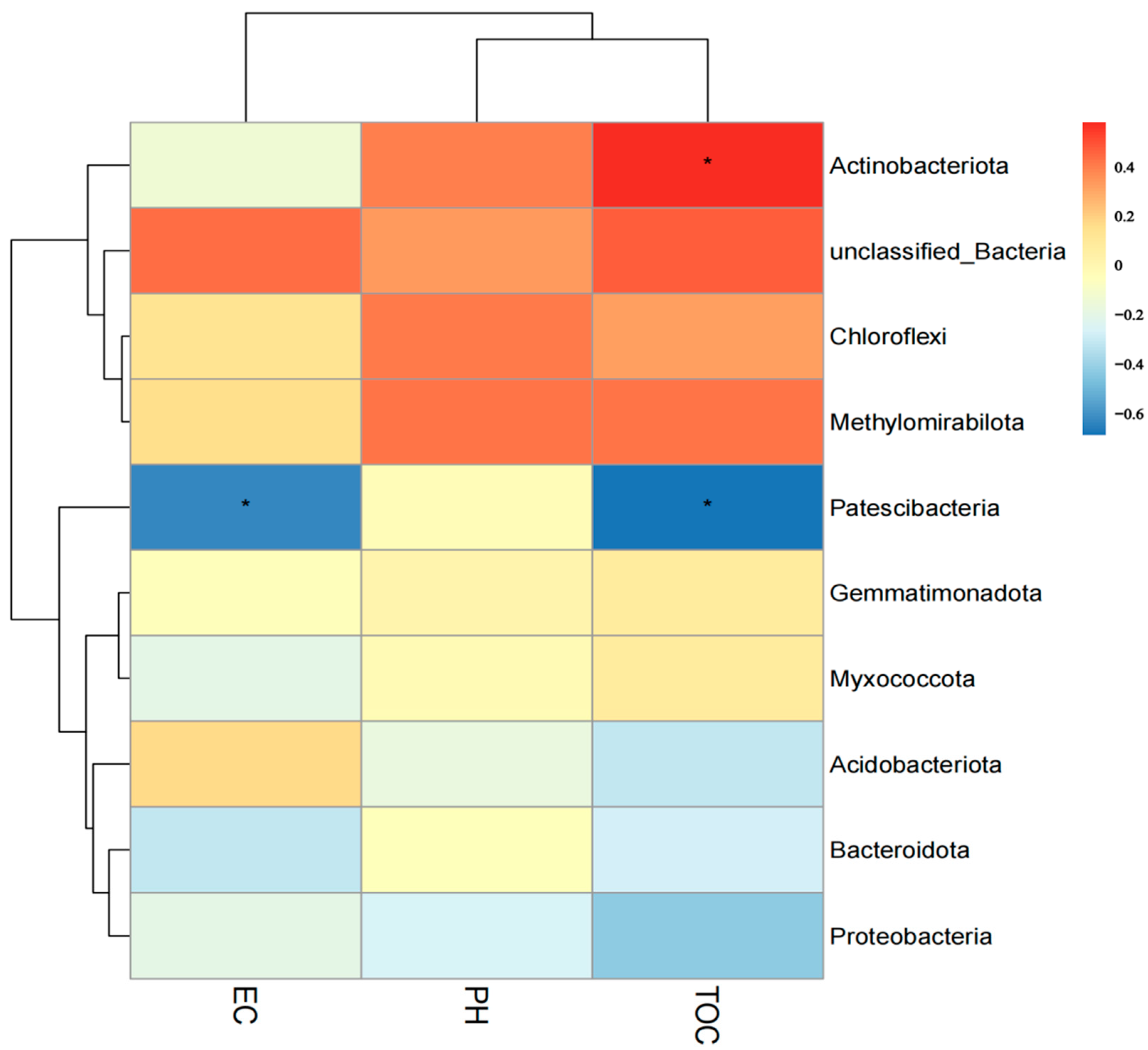
3.6. Correlation between Soil Bacterial Species and Soil Physicochemical Properties
3.7. LEfSe Analysis and Random Forest Analyses
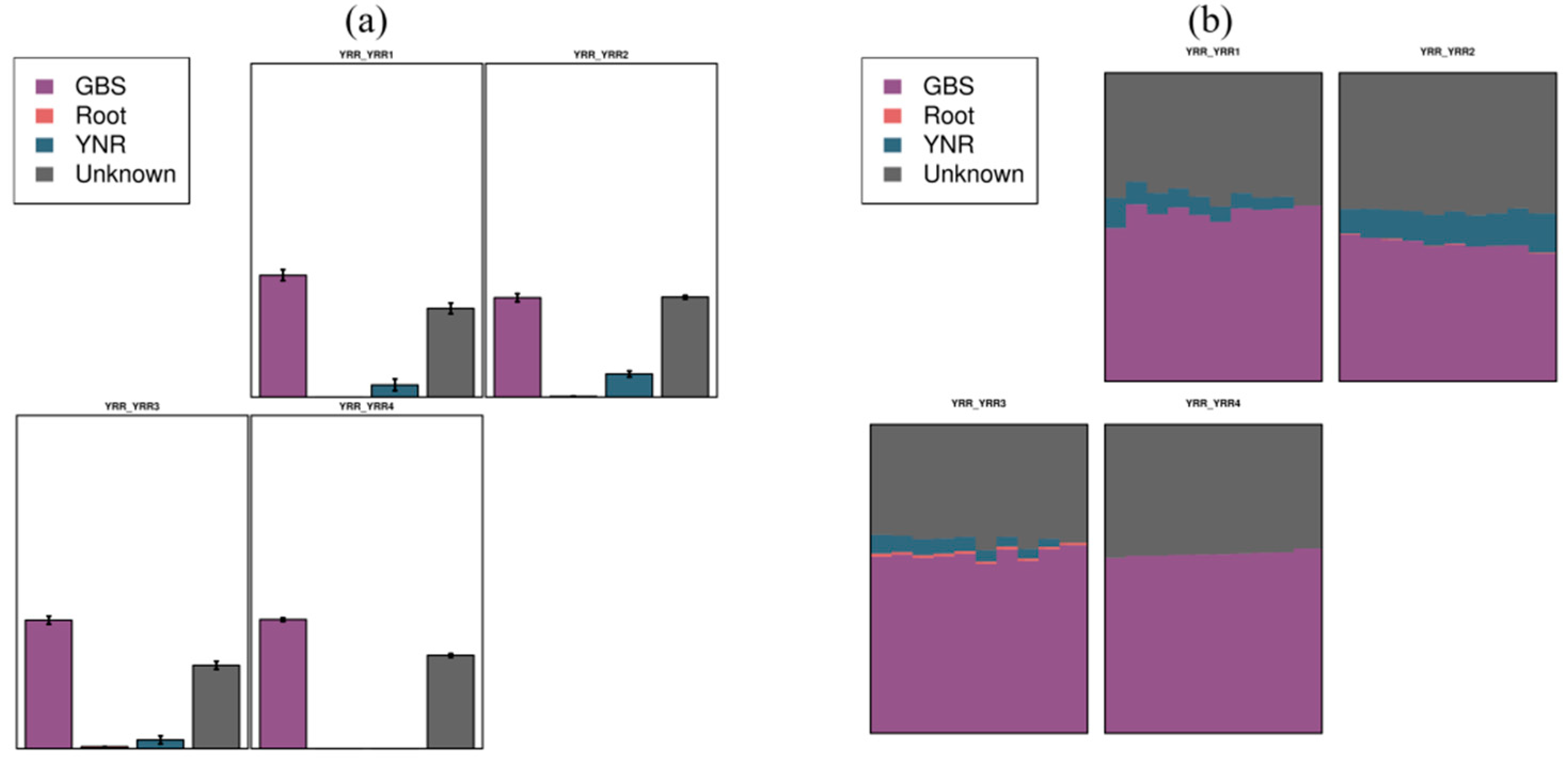
3.8. SourceTracker Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rai, P.K. Paradigm of plant invasion: Multifaceted review on sustainable management. Environ. Monit. Assess. 2015, 187, 759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, X.B.; Luo, Y.M. An overview of ecohydrology of the Yellow River delta wetland. Ecohydrol. Hydrobiol. 2016, 16, 39–44. [Google Scholar] [CrossRef]
- Shang, S.; Hu, S.X.; Liu, X.X.; Zang, Y.; Chen, J.; Gao, N.; Li, L.Y.; Wang, J.; Liu, L.X.; Xu, J.K.; et al. Effects of Spartina alterniflora invasion on the community structure and diversity of wetland soil bacteria in the Yellow River Delta. Ecol. Evol. 2022, 12, e8905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.L.; Bai, J.H.; Jia, J.; Wang, W.; Wang, X.; Zhao, Q.Q.; Lu, Q.Q. Shifts of soil microbial community composition along a short-term invasion chronosequence of Spartina alterniflora in a Chinese estuary. Sci. Total Environ. 2019, 657, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bibi, K.; Ullah, R. Distribution pattern and ecological determinants of an invasive plant Parthenium hysterophorus L., in Malakand division of Pakistan. J. Mt. Sci. 2020, 17, 1670–1683. [Google Scholar] [CrossRef]
- Adkins, S.; Shabbir, A. Biology, ecology and management of the invasive parthenium weed (Parthenium hysterophorus L.). Pest Manag. Sci. 2014, 70, 1023–1029. [Google Scholar] [CrossRef]
- Gao, X.; Li, M.; Gao, Z.; Zhang, J.; Liu, Y.; Cao, A. Seed germination characteristics and clone reproductive capacity of Parthenium hysterophorus L. Ecol. Environ. Sci. 2013, 22, 100–104. [Google Scholar]
- Liu, F.D.; Mo, X.; Kong, W.J.; Song, Y. Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the Yellow River Delta, China. Sci. Total Environ. 2020, 740, 140144. [Google Scholar] [CrossRef]
- Yang, W.; Yan, Y.E.; Jiang, F.; Leng, X.; Cheng, X.L.; An, S.Q. Response of the soil microbial community composition and biomass to a short-term Spartina alterniflora invasion in a coastal wetland of eastern China. Plant Soil 2016, 408, 443–456. [Google Scholar] [CrossRef]
- Jiang, S.; Xing, Y.J.; Liu, G.C.; Hu, C.Y.; Wang, X.C.; Yan, G.Y.; Wang, Q.G. Changes in soil bacterial and fungal community composition and functional groups during the succession of boreal forests. Soil Biol. Biochem. 2021, 161, 108393. [Google Scholar] [CrossRef]
- Qu, Z.L.; Liu, B.; Ma, Y.; Xu, J.; Sun, H. The response of the soil bacterial community and function to forest succession caused by forest disease. Funct. Ecol. 2020, 34, 2548–2559. [Google Scholar] [CrossRef]
- Zhou, B.H.; Liu, Z.W.; Yang, G.; He, H.; Liu, H.J. Microbial activity and diversity in the rhizosphere soil of the invasive species Zizania latifolia in the wetland of Wuchang Lake, China. Mar. Freshw. Res. 2020, 71, 1702–1713. [Google Scholar] [CrossRef]
- Shang, S.; Li, L.Y.; Zhang, Z.W.; Zang, Y.; Chen, J.; Wang, J.; Wu, T.; Xia, J.B.; Tang, X.X. The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta. Microorganisms 2022, 10, 1722. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 (vol 37, pg 852, 2019). Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- IBM Corp. IBM SPSS Statistics for Windows, Version 22.0; IBM Corp: Armonk, NY, USA, 2013.
- Liu, Y.; Zhao, H. Variable importance-weighted Random Forests. Quant. Biol. (Beijing China) 2017, 5, 338–351. [Google Scholar] [CrossRef]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef]
- Rai, P.K.; Singh, J.S. Invasive alien plant species: Their impact on environment, ecosystem services and human health. Ecol. Indic. 2020, 111, 106020. [Google Scholar] [CrossRef]
- Wagner, V.; Vecera, M.; Jimenez-Alfaro, B.; Pergl, J.; Lenoir, J.; Svenning, J.C.; Pysek, P.; Agrillo, E.; Biurrun, I.; Campos, J.A.; et al. Alien plant invasion hotspots and invasion debt in European woodlands. J. Veg. Sci. 2021, 32, e13014. [Google Scholar] [CrossRef]
- te Beest, M.; Stevens, N.; Olff, H.; van der Putten, W.H. Plant-soil feedback induces shifts in biomass allocation in the invasive plant Chromolaena odorata. J. Ecol. 2009, 97, 1281–1290. [Google Scholar] [CrossRef]
- de la Pena, E.; Rodriguez-Echeverria, S.; van der Putten, W.H.; Freitas, H.; Moens, M. Mechanism of control of root-feeding nematodes by mycorrhizal fungi in the dune grass Ammophila arenaria. New Phytol. 2006, 169, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Lai, I.L.; Chang, S.C. Changes in Soil Microbial Community and Carbon Flux Regime across a Subtropical Montane Peatland-to-Forest Successional Series in Taiwan. Forests 2022, 13, 958. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; An, S.Q.; Zhao, H.; Xu, L.Q.; Qiao, Y.J.; Cheng, X.L. Impacts of Spartina alterniflora invasion on soil organic carbon and nitrogen pools sizes, stability, and turnover in a coastal salt marsh of eastern China. Ecol. Eng. 2016, 86, 174–182. [Google Scholar] [CrossRef]
- Copetta, A.; Lingua, G.; Berta, G. Effects of three AM fungi on growth, distribution of glandular hairs, and essential oil production in Ocimum basilicum L. var. Genovese. Mycorrhiza 2006, 16, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Liu, Y.J.; Brunel, C.; van Kleunen, M. Soil-microorganism-mediated invasional meltdown in plants. Nat. Ecol. Evol. 2020, 4, 1612–1621. [Google Scholar] [CrossRef]




| Groups | EC (ms/cm) | pH | TOC (g/kg) |
|---|---|---|---|
| YRR | 2.16 ± 0.56a | 7.78 ± 0.07ab | 18.33 ± 0.90b |
| YNR | 2.14 ± 0.19a | 7.90 ± 0.07a | 47.08 ± 7.41a |
| GBS | 1.87 ± 0.24a | 7.69 ± 0.02b | 25.74 ± 7.68ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, S.; Zhang, Z.; Zhao, L.; Liu, L.; Shi, D.; Xu, H.; Zhang, H.; Xie, W.; Zhao, F.; Zhou, Z.; et al. Effect of Parthenium hysterophorus L. Invasion on Soil Microbial Communities in the Yellow River Delta, China. Microorganisms 2023, 11, 18. https://doi.org/10.3390/microorganisms11010018
Shang S, Zhang Z, Zhao L, Liu L, Shi D, Xu H, Zhang H, Xie W, Zhao F, Zhou Z, et al. Effect of Parthenium hysterophorus L. Invasion on Soil Microbial Communities in the Yellow River Delta, China. Microorganisms. 2023; 11(1):18. https://doi.org/10.3390/microorganisms11010018
Chicago/Turabian StyleShang, Shuai, Zaiwang Zhang, Liping Zhao, Longxiang Liu, Dongli Shi, Hui Xu, Hanjie Zhang, Wenjun Xie, Fengjuan Zhao, Zhihao Zhou, and et al. 2023. "Effect of Parthenium hysterophorus L. Invasion on Soil Microbial Communities in the Yellow River Delta, China" Microorganisms 11, no. 1: 18. https://doi.org/10.3390/microorganisms11010018
APA StyleShang, S., Zhang, Z., Zhao, L., Liu, L., Shi, D., Xu, H., Zhang, H., Xie, W., Zhao, F., Zhou, Z., Xu, J., & Wang, J. (2023). Effect of Parthenium hysterophorus L. Invasion on Soil Microbial Communities in the Yellow River Delta, China. Microorganisms, 11(1), 18. https://doi.org/10.3390/microorganisms11010018





