Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Cultivation
2.2. Microscopy
2.3. Physiological Characterization
2.4. 16S rRNA Gene Sequence Analysis
2.5. Genome Sequencing and Phylogenomics
2.6. Keratinase Activity Test
3. Results
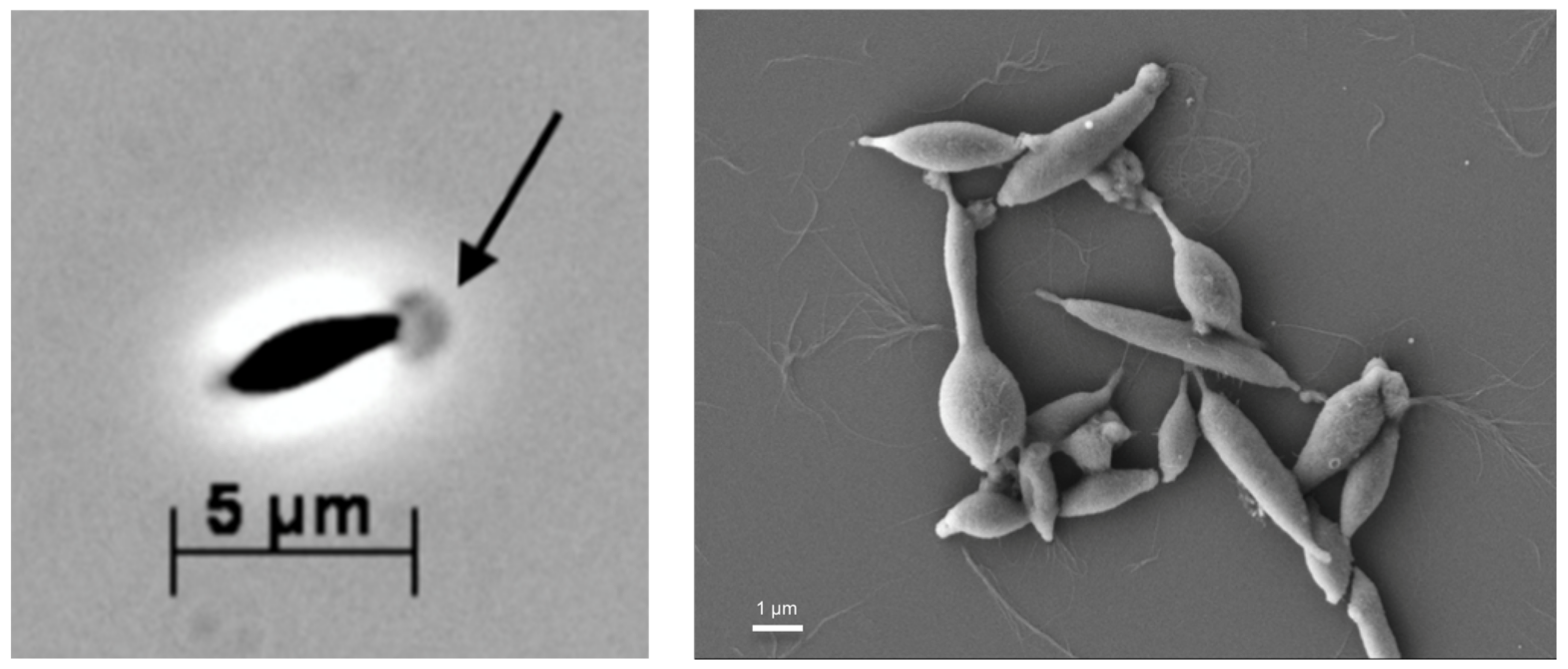
3.1. Microscopy and Morphology
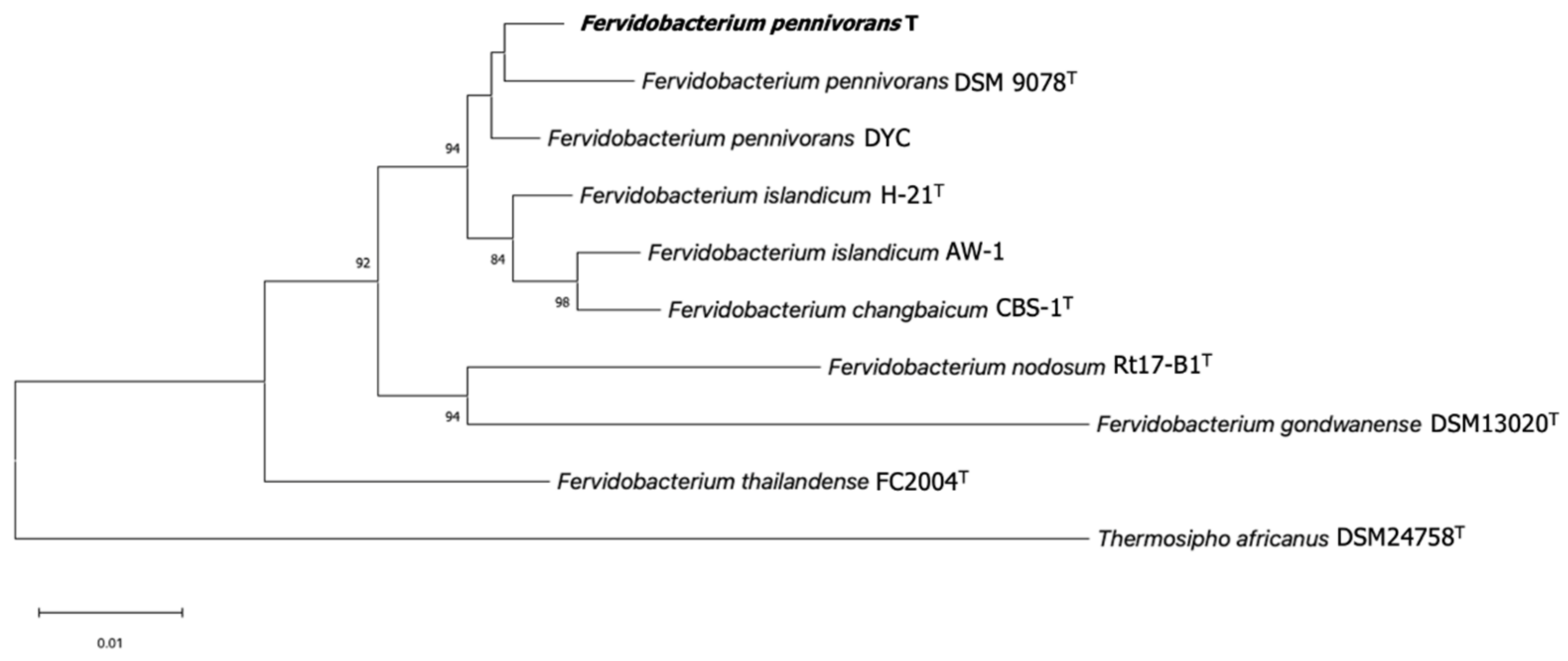
3.2. Phylogenetic Identification
3.3. Physiology
3.4. Feather Degradation
3.5. Genome Characteristics and Phylogenomics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frock, A.D.; Notey, J.S.; Kelly, R.M. The genus Thermotoga: Recent developments. Environ. Technol. 2010, 31, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Achenbachrichter, L.; Gupta, R.; Stetter, K.O.; Woese, C.R. Were the Original Eubacteria Thermophiles. Syst. Appl. Microbiol. 1987, 9, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, V.; Gupta, R.S. Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Anton. Leeuw. Int. J. G. 2015, 108, 1281. [Google Scholar] [CrossRef]
- Huber, R.; Langworthy, T.A.; Konig, H.; Thomm, M.; Woese, C.R.; Sleytr, U.B.; Stetter, K.O. Thermotoga-maritima Sp-Nov Represents a New Genus of Unique Extremely Thermophilic Eubacteria Growing up to 90-Degrees-C. Arch. Microbiol. 1986, 144, 324–333. [Google Scholar] [CrossRef]
- Gupta, R.S.; Bhandari, V. Phylogeny and molecular signatures for the phylum Thermotogae and its subgroups. Anton. Leeuw. Int. J. G. 2011, 100, 1–34. [Google Scholar] [CrossRef]
- Patel, B.K.C.; Morgan, H.W.; Daniel, R.M. Fervidobacterium nodosum Gen-Nov and Spec-Nov, a New Chemoorganotrophic, Caldoactive, Anaerobic Bacterium. Arch. Microbiol. 1985, 141, 63–69. [Google Scholar] [CrossRef]
- Andrews, K.T.; Patel, B.K.C. Fervidobacterium gondwanense sp. nov., a new thermophilic anaerobic bacterium isolated from nonvolcanically heated geothermal waters of the Great Artesian Basin of Australia. Int. J. Syst. Bacteriol. 1996, 46, 265–269. [Google Scholar] [CrossRef]
- Cai, J.; Wang, Y.; Liu, D.; Zeng, Y.; Xue, Y.; Ma, Y.; Feng, Y. Fervidobacterium changbaicum sp. nov., a novel thermophilic anaerobic bacterium isolated from a hot spring of the Changbai Mountains, China. Int. J. Syst. Evol. Microbiol. 2007, 57, 2333–2336. [Google Scholar] [CrossRef]
- Friedrich, A.B.; Antranikian, G. Keratin Degradation by Fervidobacterium pennavorans, a Novel Thermophilic Anaerobic Species of the Order Thermotogales. Appl. Environ. Microbiol. 1996, 62, 2875–2882. [Google Scholar] [CrossRef]
- Huber, R.; Woese, C.R.; Langworthy, T.A.; Kristjansson, J.K.; Stetter, K.O. Fervidobacterium-Islandicum sp.-nov., a New Extremely Thermophilic Eubacterium Belonging to the Thermotogales. Arch. Microbiol. 1990, 154, 105–111. [Google Scholar] [CrossRef]
- Kanoksilapatham, W.; Pasomsup, P.; Keawram, P.; Cuecas, A.; Portillo, M.C.; Gonzalez, J.M. Fervidobacterium thailandense sp. nov., an extremely thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2016, 66, 5023–5027. [Google Scholar] [CrossRef]
- Podosokorskaya, O.A.; Merkel, A.Y.; Kolganova, T.V.; Chernyh, N.A.; Miroshnichenko, M.L.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Fervidobacterium riparium sp. nov., a thermophilic anaerobic cellulolytic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2011, 61, 2697–2701. [Google Scholar] [CrossRef]
- Gerday, C.; Glansdorff, N.; American Society for Microbiology. Physiology and Biochemistry of Extremophiles; ASM Press: Washington, DC, USA, 2007; p. xvi. 429p. [Google Scholar]
- Parry, D.A.; Crewther, W.G.; Fraser, R.D.; MacRae, T.P. Structure of alpha-keratin: Structural implication of the amino acid sequences of the type I and type II chain segments. J. Mol. Biol. 1977, 113, 449–454. [Google Scholar] [CrossRef]
- Shavandi, A.; Silva, T.H.; Bekhit, A.A.; Bekhit, A.E.A. Keratin: Dissolution, extraction and biomedical application. Biomater. Sci. 2017, 5, 1699–1735. [Google Scholar] [CrossRef]
- Lee, Y.J.; Dhanasingh, I.; Ahn, J.S.; Jin, H.S.; Choi, J.M.; Lee, S.H.; Lee, D.W. Biochemical and structural characterization of a keratin-degrading M32 carboxypeptidase from Fervidobacterium islandicum AW-1. Biochem. Biophys. Res. Commun. 2015, 468, 927–933. [Google Scholar] [CrossRef]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial decomposition of keratin in nature—A new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef]
- Qiu, J.; Wilkens, C.; Barrett, K.; Meyer, A.S. Microbial enzymes catalyzing keratin degradation: Classification, structure, function. Biotechnol. Adv. 2020, 44, 107607. [Google Scholar] [CrossRef]
- Li, Q. Progress in Microbial Degradation of Feather Waste. Front. Microbiol. 2019, 10, 2717. [Google Scholar] [CrossRef]
- Laba, W.; Zarowska, B.; Chorazyk, D.; Pudlo, A.; Piegza, M.; Kancelista, A.; Kopec, W. New keratinolytic bacteria in valorization of chicken feather waste. AMB Express 2018, 8, 9. [Google Scholar] [CrossRef]
- Tesfaye, T.; Sithole, B.; Ramjugernath, D.; Chunilall, V. Valorisation of chicken feathers: Characterisation of chemical properties. Waste Manag. 2017, 68, 626–635. [Google Scholar] [CrossRef]
- Williams, C.M.; Lee, C.G.; Garlich, J.D.; Shih, J.C.H. Evaluation of a Bacterial Feather Fermentation Product, Feather-Lysate, as a Feed Protein. Poultry. Sci. 1991, 70, 85–94. [Google Scholar] [CrossRef]
- Papadopoulos, M.C. Effect of Processing on High-Protein Feedstuffs—A Review. Biol. Waste 1989, 29, 123–138. [Google Scholar] [CrossRef]
- Conners, S.B.; Mongodin, E.F.; Johnson, M.R.; Montero, C.I.; Nelson, K.E.; Kelly, R.M. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. Fems. Microbiol. Rev. 2006, 30, 872–905. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Tello, E.; Fardeau, M.L.; Thomas, P.; Ramirez, F.; Casalot, L.; Cayol, J.L.; Garcia, J.L.; Ollivier, B. Petrotoga mexicana sp nov., a novel thermophilic, anaerobic and xylanolytic bacterium isolated from an oil-producing well in the Gulf of Mexico. Int. J. Syst. Evol. Micr. 2004, 54, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Dzhuraeva, M.M.; Margaryan, A.A. (Eds.) Microbial Diversity of High-Altitude Geothermal Springs in Tajikistan; Springer: Singapore, 2021; Volume 32. [Google Scholar]
- Miller, T.L.; Wolin, M.J. Serum Bottle Modification of Hungate Technique for Cultivating Obligate Anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [CrossRef]
- Mashzhan, A.; Javier-Lopez, R.; Kistaubayeva, A.; Savitskaya, I.; Birkeland, N.K. Metagenomics and Culture-Based Diversity Analysis of the Bacterial Community in the Zharkent Geothermal Spring in Kazakhstan. Curr. Microbiol. 2021, 78, 2926–2934. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Neron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing, G.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- DeBoy, R.T.; Mongodin, E.F.; Emerson, J.B.; Nelson, K.E. Chromosome evolution in the Thermotogales: Large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 2006, 188, 2364–2374. [Google Scholar] [CrossRef]
- Xu, Z.; Puranik, R.; Hu, J.; Xu, H.; Han, D. Complete genome sequence of Thermotoga sp. strain RQ7. Stand. Genom. Sci. 2017, 12, 62. [Google Scholar] [CrossRef]





| Characteristic | F. pennivorans subsp. keratinolyticus T | F. pennivorans DSM9078 | F. thailandense FC2004 | F. nodosum Rt17-B1 | F. islandicum H21 | F. gondwanense AB39 | F. changbaicum CBS-1 |
|---|---|---|---|---|---|---|---|
| Isolation Source | Tajikistan | Azores Islands, Portugal | Thailand | New Zealand | Iceland | Geothermal artesian basin, Australia | China |
| Cell Size (µm) | 0.5 × 2–20 | 0.5 × 2–20 | 0.5–0.6 × 1.1–30 | 0.5–0.55 × 1–2.5 | 0.6 × 1–4 | 0.5–0.6 × 4–40 | 0.5–0.6 × 1–8 |
| Temperature Range (°C) Optimum | 55–75 | 50–80 | 60–88 | 47–80 | 50–80 | >45 to <80 | 55–90 |
| 65 | 70 | 78–80 | 70 | 65 | 65–68 | 75–80 | |
| pH Range Optimum | 6.5–7.5 | 5.5–8.0 | 6.0–8.5 | 6.0–8.0 | 6.0–8.0 | 6.0–8.0 | 6.3–8.5 |
| 6.5 | 6.5 | 7.5–8 | 7 | 7.2 | 7 | 7.5 | |
| NaCl Range (g/L) Optimum | 0–30 | 0–40 | 0–5 | <10 | <10 | 0–6 | 0–10 |
| 3 | 4 | 0–1.0 | ≤1 (NR) | 2 | 1 | 0 | |
| Generation Time (min) | 150 | 126 | 85 | 105 | 150 | 79 | 99 |
| Utilization of | |||||||
| Glucose | + | + | + | + | + | + | + |
| Sucrose | slow | + | + | + | + | slow | + |
| Lactose | + | − | − | + | − | + | + |
| Arabinose | − | − | − | slow | + | − | − |
| Galactose | + | + | − | + | + | slow | + |
| Mannose | slow | + | − | + | + | + | − |
| Sorbitol | + | slow | − | + | + | − | + |
| Mannitol | − | slow | − | slow | − | slow | − |
| Starch | slow | + | + | + | + | + | + |
| Cellulose | + | − | − | − | + | − | − |
| Peptone | + | + | + | + | + | + | + |
| Feather Hydrolysis | + | + | + | − | − | − | − |
| DNA G + C Content (mol%) | 39 | 38.9 | 45.8 | 33.7 | 41 | 35 | 31.9 |
| F. pennivorans T | F. pennivorans DSM9078 | F. pennivorans DYC | F. islandicum AW-1 | F. thailandense FC2004 | F. nodosum Rt17-B1 | F. changbaicum CBS-1 | F. gondwanense 13020 | |
|---|---|---|---|---|---|---|---|---|
| Country of Origin | Tajikistan | Portugal (Azores) | New Zealand | Indonesia | Thailand | New Zealand | China | Australia |
| Year of Isolation | 2021 | 1999 | 2016 | 2004 | 2016 | 1985 | 2007 | 1996 |
| Genome Size (bps) | 2,002,515 | 2,166,381 | 2,061,852 | 2,237,377 | 2,040,210 | 1,948,941 | 2,266,449 | 2,145,239 |
| N° CDS with Protein | 1875 | 1973 | 1893 | 2055 | 1870 | 1796 | 2038 | 1975 |
| N° RNAs | 57 | 57 | 58 | 56 | 54 | 58 | 57 | 54 |
| N° rRNAs (23S-16S-5S) | 2 Feburary 2002 | 3, 1, 2 | 2, 2, 2 | 2, 2, 2 | 1, 3, 1 | 2, 2, 2 | 2, 2, 2 | 1, 3, 1 |
| % GC | 39 | 38.9 | 38.9 | 40.7 | 45.8 | 3 | 40.7 | 39.7 |
| CRISPR Clusters | 3 | 4 | 3 | 2 | 8 | 2 | 2 | 1 |
| NCBI Accession Number | CP050868 | CP003260.1 | CP011393.1 | CP014334.2 | NZ_LWAF00000000.1 | CP000771.1 | NZ_CP026721.1 | NZ_FRDJ00000000.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javier-Lopez, R.; Mandolini, E.; Dzhuraeva, M.; Bobodzhanova, K.; Birkeland, N.-K. Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms 2023, 11, 22. https://doi.org/10.3390/microorganisms11010022
Javier-Lopez R, Mandolini E, Dzhuraeva M, Bobodzhanova K, Birkeland N-K. Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms. 2023; 11(1):22. https://doi.org/10.3390/microorganisms11010022
Chicago/Turabian StyleJavier-Lopez, Rubén, Edoardo Mandolini, Munavvara Dzhuraeva, Khursheda Bobodzhanova, and Nils-Kåre Birkeland. 2023. "Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile" Microorganisms 11, no. 1: 22. https://doi.org/10.3390/microorganisms11010022
APA StyleJavier-Lopez, R., Mandolini, E., Dzhuraeva, M., Bobodzhanova, K., & Birkeland, N.-K. (2023). Fervidobacterium pennivorans subsp. keratinolyticus subsp. nov., a Novel Feather-Degrading Anaerobic Thermophile. Microorganisms, 11(1), 22. https://doi.org/10.3390/microorganisms11010022





