Characterization of Five Psychrotolerant Alcanivorax spp. Strains Isolated from Antarctica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Sampling and Treatment
2.2. Bacterial Enrichments and Isolation of Hydrocarbon-Degrading Strains
2.3. Taxonomical Characterization of Hydrocarbon-Degrading Isolates
2.4. Analyses on Alcanivorax spp. Strains
2.4.1. Partial 16S rRNA, alk-B1, and P450 Gene Sequencing
2.4.2. Physiological Characterization
2.4.3. Evaluation of Biosurfactant Production and Emulsification Activity
3. Results
3.1. Isolation and Taxonomical Characterization of Hydrocarbon-Degrading Isolates
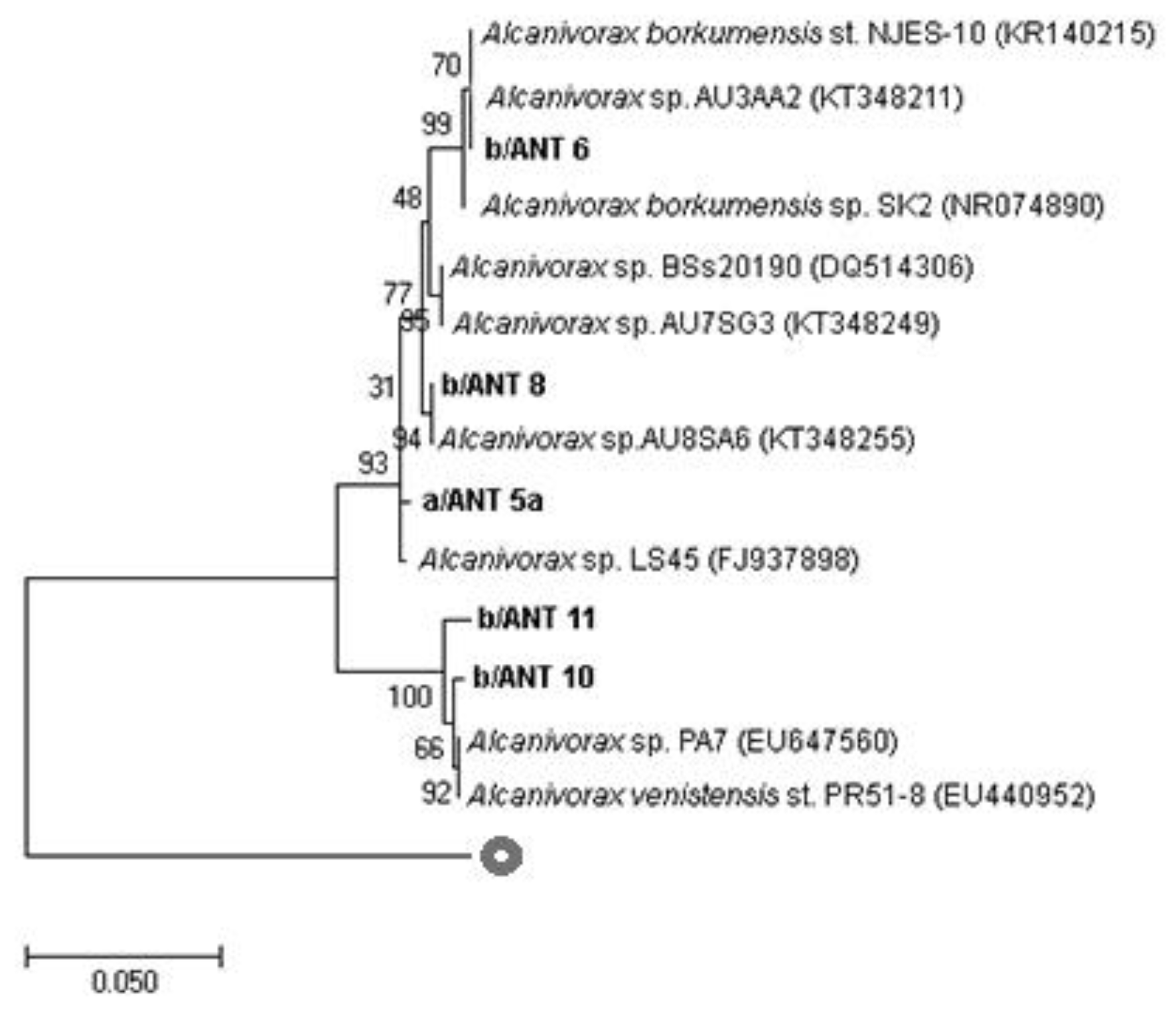
3.2. Analyses on Alcanivorax spp. Strains
3.3. Partial 16S rRNA, alk-B1, and P450 Gene Sequencing
3.4. Physiological Characterization
3.5. Biosurfactant Production and Emulsification Activity
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fingas, M. The Basics of Oil Spill Cleanup; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Xue, J.; Yu, Y.; Bai, Y.; Wang, L.; Wu, Y. Marine Oil-Degrading Microorganisms and Biodegradation Process of Petroleum Hydrocarbon in Marine Environments: A Review. Curr. Microbiol. 2015, 71, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Michaud, L.; Lo Giudice, A.; Saitta, M.; De Domenico, M.; Bruni, V. The biodegradation efficiency on diesel oil by two psychrotrophic Antarctic marine bacteria during a two-month-long experiment. Mar. Pollut. Bull. 2004, 49, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Radovic’, J.R.; Aeppli, C.; Nelson, R.K.; Jimenez, N.; Reddy, C.M.; Bayona, J.M.; Albaigés, J. Assessment of photochemical processes in marine oil spill fingerprinting. Mar. Pollut. Bull. 2014, 79, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Hazen, T.C.; Dubinsky, E.A.; DeSantis, T.Z.; Andersen, G.L.; Piceno, Y.M.; Singh, N.; Jansson, J.K.; Probst, A.; Borglin, S.E.; Fortney, J.L.; et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 2010, 330, 204–208. [Google Scholar] [CrossRef] [Green Version]
- Guibert, L.M.; Loviso, C.L.; Marcos, M.S.; Commendatore, M.G.; Dionisi, H.M.; Lozada, M. Alkane Biodegradation Genes from Chronically Polluted Subantarctic Coastal Sediments and Their Shifts in Response to Oil Exposure. Microb. Ecol. 2012, 64, 605–616. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Giuliano, L.; Gentile, G.; Crisafi, E.; Chernikova, T.N.; Abraham, W.R.; Lunsdorf, H.; Timmis, K.N.; Golyshin, P.N. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal seawater. Int. J. Syst. Evol. Microbiol. 2003, 53, 779–785. [Google Scholar] [CrossRef]
- Gentile, G.; Bonasera, V.; Amico, C.; Giuliano, L.; Yakimov, M. Shewanella sp. GA-22, a psychrophilic hydrocarbonoclastic Antarctic bacterium producing polyunsaturated fatty acids. J. Appl. Microbiol. 2003, 95, 1124–1133. [Google Scholar] [CrossRef]
- Cai, Q.; Zhang, B.; Chen, B.; Zhu, Z.; Lin, W.; Cao, T. Screening of biosurfactant producers from petroleum hydrocarbon contaminated sources in cold marine environments. Mar. Pollut. Bull. 2014, 86, 402–410. [Google Scholar] [CrossRef]
- Jain, D.K.; Collins-Thompson, D.L.; Lee, H.; Trevors, J.T. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Meth. 1991, 13, 271–279. [Google Scholar] [CrossRef]
- Gentile, G.; Bonsignore, M.; Santisi, S.; Catalfamo, M.; Giuliano, L.; Genovese, L.; Yakimov, M.M.; Denaro, R.; Genovese, M.; Cappello, S. Biodegradation potentiality of psychrophilic bacterial strain Oleispira antarctica RB-8T. Mar. Pollut. Bull. 2016, 105, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Silveira, C.B.; Thompson, F. The family Alcanivoraceae. In The Prokaryotes—Gammaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F.., Eds.; Springer: Berlin, Germany, 2014; pp. 59–67. [Google Scholar]
- Crisafi, F.; Giuliano, L.; Yakimov, M.M.; Azzaro, M.; Denaro, R. Isolation and degradation potential of a cold-adapted oil/PAH-degrading marine bacterial consortium from Kongsfjorden (Arctic region). Rend. Lincei-Sci. Fis. 2016, 27, 261–270. [Google Scholar] [CrossRef]
- Schneiker, S.; Dos Santos, V.A.; Bartels, D.; Bekel, T.; Brecht, M.; Buhrmester, J.; Chernikova, T.N.; Denaro, R.; Ferrer, M.; Gertler, C.; et al. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 2006, 24, 997–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabirova, J.S.; Becker, A.; Lunsdorf, H.; Nicaud, J.M.; Timmis, K.N.; Golyshin, P.N. Transcriptional profiling of the marine oil-degrading bacterium Alcanivorax borkumensis during growth on n-alkanes. FEMS Microbiol. Lett. 2011, 319, 160–168. [Google Scholar] [CrossRef] [Green Version]
- Yakimov, M.M.; Golyshin, P.N.; Lang, S.; Moore, E.R.B.; Abraham, W.R.; Lünsdorf, H.; Timmis, K.N. Alcanivorax borkumensis gen. now, sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 1998, 48, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, E.; Bellicanta, G.S.; Pellizari, V.H. New alk genes detected in Antarctic marine sediments. Environ. Microbiol. 2009, 11, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Goordial, J.; Davila, A.; Lacelle, D.; Pollard, W.; Marinova, M.M.; Greer, C.W.; DiRuggiero, J.; McKay, C.P.; Whyte, L.G. Nearing the cold-arid limits of microbial life in permafrost of an upper dry valley, Antarctica. ISME J. 2016, 10, 1613–1624. [Google Scholar] [CrossRef]
- Dyksterhouse, S.E.; Gray, J.P.; Herwig, R.P.; Lara, J.C.; Staley, J.T. Cycloclasticus pugetii gen. nov., sp. nov., an Aromatic Hydrocarbon-Degrading Bacterium from Marine Sediments. Int. J. Syst. Evol. Microbiol. 1995, 45, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Zhang, Y.J.; Yu, Y.; Li, H.J.; Gao, Z.M.; Chen, X.L.; Chen, B.; Zhang, Y.Z. Neptunomonas antarctica sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2010, 60, 1958–1961. [Google Scholar] [CrossRef]
- Rong, J.C.; Liu, M.; Li, Y.; Sun, T.Y.; Xie, B.B.; Shi, M.; Chen, X.L.; Qin, Q.L. Insight into the genome sequence of a sediment-adapted marine bacterium Neptunomonas antarctica S3-22T from Antarctica. Mar. Genom. 2016, 2, 29–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, D.; Shi, Z.; Shen, X.; He, Y.; Sun, M.; Iuo, Q.; Wang, Q. Isolation, identification and alkane hydroxylase genes detection of a marine diesel-degrading bacterial strain (F9). Afr. J. Microbiol. Res. 2013, 7, 2794–2802. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. Molecular Evolutionary Genetics Analysis version 7.0 for big datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, S.G.; Krieg, N.R.; Sneath, P.H.A.; Stanley, J.T.; Williams, S.T. Bergey’s Manual of Determinate Bacteriology; Williams and Wilkins: New York, NY, USA, 1998. [Google Scholar]
- Maneerat, S.; Phetrong, K. Isolation of biosurfactant-producing marine bacteria and characteristics of selected biosurfactant. J. Sci. Technol. 2007, 29, 781–791. [Google Scholar]
- Maia, M.; Capão, A.; Procópio, L. Biosurfactant produced by oil-degrading Pseudomonas putida AM-b1 strain with potential for microbial enhanced oil recovery. Bioremediat. J. 2019, 23, 302–310. [Google Scholar] [CrossRef]
- Li, C.Q.; Liu, W.C.; Zhu, P.; Yang, J.L.; Cheng, K.D. Phylogenetic Diversity of Bacteria Associated with the Marine Sponge Gelliodes carnosa collected from the Hainan Island Coastal Waters of the South China Sea. Microb. Ecol. 2011, 62, 800–812. [Google Scholar] [CrossRef]
- Catania, V.; Santisi, S.; Signa, G.; Vizzini, S.; Mazzola, A.; Cappello, S.; Yakimov, M.M.; Quatrini, P. Intrinsic bioremediation potential of a chronically polluted marine coastal area. Mar. Pollut. Bull. 2015, 99, 138–149. [Google Scholar] [CrossRef]
- Olivera, N.L.; Nievas, M.L.; Lozada, M.; del Prado, G.; Dionisi, H.M.; Faustino Siñeriz, F. Isolation and characterization of biosurfactant-producing Alcanivorax strains: Hydrocarbon accession strategies and alkane hydroxylase gene analysis. Res. Microbiol. 2009, 160, 19–26. [Google Scholar] [CrossRef]
- Yuan, J.; Lai, Q.; Sun, F.; Zheng, T.; Shao, Z. The diversity of PAH-degrading bacteria in a deep-seawater column above the Southwest Indian Ridge. Front. Microbiol. 2015, 6, 853. [Google Scholar] [CrossRef] [Green Version]
- Lai, Q.; Wang, J.; Gu, L.; Zheng, T.; Shao, Z. Alcanivorax marinus sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 2013, 63, 4428–4432. [Google Scholar] [CrossRef]
- Park, C.; Park, W. Survival and Energy Producing Strategies of Alkane Degraders Under Extreme Conditions and Their Biotechnological Potential. Front. Microbiol. 2018, 9, 1081. [Google Scholar] [CrossRef] [PubMed]
- Gran-Scheuch, A.; Fuentes, E.; Bravo, D.M.; Jiménez, J.C.; Pérez-Donoso, J.M. Isolation and Characterization of Phenanthrene Degrading Bacteria from Diesel Fuel-Contaminated Antarctic Soils. Front. Microbiol. 2017, 8, 1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aislabie, J.; Foght, J.; Saul, D. Aromatic hydrocarbon-degrading bacteria from soil near Scott Base, Antarctica. Polar Biol. 2000, 23, 183–188. [Google Scholar] [CrossRef]
- Cappello, S.; Caruso, G.; Bergami, E.; Macrì, A.; Venuti, V.; Majolino, D.; Corsi, I. New insights into the structure and function of the prokaryotic communities colonizing plastic debris collected in King George Island (Antarctica): Preliminary observations from two plastic fragments. J. Hazard. Mater. 2021, 414, 125586. [Google Scholar] [CrossRef] [PubMed]
- Zappalà, G.; Caruso, G.; Denaro, R.; Crisafi, F.; Monticelli, L.S. Ecological and molecular approach to the assessment of oil pollution: A comparative study between two coastal marine (Mediterranean and Patagonian) ecoregions. WIT Trans. Ecol. Environ. 2020, 242, 85–96. [Google Scholar] [CrossRef]
- Dastgheib, S.M.M.; Amoozegar, M.A.; Khajeh, K.; Ventosa, A. A halotolerant Alcanivorax sp. strain with potential application in saline soil remediation. Appl. Microbiol. Biotechnol. 2011, 90, 305–312. [Google Scholar] [CrossRef]
- Liu, Y.C.; Li, L.Z.; Wu, Y.; Tian, W.; Zhang, L.P.; Xu, L.; Shen, Q.R.; Shen, B. Isolation of an alkane-degrading Alcanivorax sp. strain 2B5 and cloning of the alkB gene. Bioresour. Technol. 2010, 101, 310–316. [Google Scholar] [CrossRef]
- Golyshin, P.N.; Harayama, S.; Timmis, K.N.; Yakimov, M.M. Family II. Alcanivoraceae fam. nov. In Bergey’s Manual of Systematic Bacteriology: Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria; Springer: New York, NY, USA, 2007. [Google Scholar]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef]
- Campbell, I.B.; Claridge, G.G.C.; Campbell, D.I.; Balks, M.R. The soil environment of the McMurdo Dry Valleys, Antarctica. In Ecosystem Dynamics in a Polar Desert. The McMurdo Dry Valleys, Antarctica; Priscu, J.C., Ed.; American Geophysical Union: Washington, DC, USA, 1998; pp. 297–322. [Google Scholar]
- Kube, M.; Chernikova, T.N.; Al-Ramahi, Y.; Beloqui, A.; Lopez-Cortez, N.; Guazzaroni, M.E.; Heipieper, H.J.; Klages, S.; Kotsyurbenko, O.R.; Langer, I.; et al. Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat. Commun. 2013, 4, 2156. [Google Scholar] [CrossRef] [Green Version]
- Habib, S.; Ahmad, S.A.; Johari, W.L.; Shukor, M.Y.; Alias, S.A.; Khalil, K.A.; Yasid, N.A. Evaluation of conventional and response surface level optimisation of n-dodecane (n-C12) mineralisation by psychrotolerant strains isolated from pristine soil at Southern Victoria Island, Antarctica. Microb. Cell Factories 2018, 17, 44. [Google Scholar] [CrossRef] [Green Version]
- Whyte, L.G.; Schultz, A.; van Beilen, J.B.; Luz, A.P.; Pellizari, V.; Labbé, D.; Greer, C.W. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 2002, 41, 141–150. [Google Scholar] [CrossRef] [PubMed]




| Selected Isolate | No. of Identical Sequences | Closest Species | GenBank No. | Sequence Similarity (%) | Sequence Size (bp) | ONR7a | ONR7a/ Na-Acetate | Marine Agar |
|---|---|---|---|---|---|---|---|---|
| Alcanivorax | ||||||||
| a/ANT5a | 1 | Alcanivorax sp. LS45 | FJ937898 | 99 | 670 | − | + | − |
| b/ANT6 | 2 | Alcanivorax sp. AU3AA2 | KT348211 | 100 | 721 | − | + | − |
| b/ANT8 | 1 | Alcanivorax sp. AU8SA6 | KT348255 | 99 | 560 | − | + | − |
| b/ANT10 | 1 | Alcanivorax sp. PA7 | EU647560 | 99 | 632 | − | + | − |
| b/ANT11 | 1 | A. venustensis strain PR51-8 | EU440952 | 99 | 650 | − | + | − |
| Oleispira | ||||||||
| a/ANT1a | 4 | Oleispira antarctica RB8 | FO203512 | 99 | 634 | − | − | − |
| a/ANT4a | 10 | Oleispira lenta strain. DFH11 | NR108293 | 99 | 534 | − | − | − |
| Pseudoalteromonas | ||||||||
| a/ANT2a | 6 | Pseudoalteromonas sp. 204Z-3 | GU584162 | 99 | 623 | + | + | + |
| a/ANT2d | 5 | Pseudoalteromonas sp. KJF2-15 | JQ800018 | 100 | 660 | + | + | + |
| a/ANT3 | 1 | Pseudoalteromonas sp. H2-49 | KM979170 | 99 | 701 | + | + | + |
| Marinomonas | ||||||||
| b/ANT9 | 1 | Marinomonas sp. KJF11-23 | JQ800192 | 99 | 637 | + | + | + |
| Halomonas | ||||||||
| b/ANT12a | 8 | Halomonas sp. whb35 | FJ444981 | 100 | 739 | + | + | + |
| Vibrio | ||||||||
| b/ANT15a | 2 | Vibrio artabrorum | FN667877 | 99 | 732 | + | + | + |
| Isolate | Drop-Collapse | Oil Spreading (cm) | E24 Test (%) | ||||
|---|---|---|---|---|---|---|---|
| T7 | T20 | T7 | T20 | T7 | T20 | ||
| 4 °C | a/ANT5a | - | - | 1.0 ± 0.1 | 0.2 ± 0.1 | - | - |
| b/ANT6 | - | - | 1.5 ± 0.1 | 0.4 ± 0.1 | - | 14.3 ± 0.5 | |
| b/ANT8 | - | - | 1.0 ± 0.1 | - | - | - | |
| b/ANT10 | - | - | 0.8 ± 0.1 | 0.3 ± 0.1 | - | - | |
| b/ANT11 | - | - | 1.0 ± 0.1 | 0.4 ± 0.1 | - | - | |
| 25 °C | a/ANT5a | - | - | 2.0 ± 0.1 | 0.2 ± 0.1 | 58.6 ± 0.5 | 15.5 ± 0.5 |
| b/ANT6 | - | - | 1.9 ± 0.1 | 0.3 ± 0.1 | 75.2 ± 0.5 | 27.6 ± 0.5 | |
| b/ANT8 | - | - | 1.0 ± 0.1 | - | 62.6 ± 0.5 | 3.4 ± 0.5 | |
| / | b/ANT10 | - | - | 1.0 ± 0.1 | 0.2 ± 0.1 | 60.0 ± 0.5 | 3.4 ± 0.5 |
| b/ANT11 | - | - | 0.7 ± 0.1 | 0.5 ± 0.1 | 5.5 ± 0.5 | 3.4 ± 0.5 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cappello, S.; Corsi, I.; Patania, S.; Bergami, E.; Azzaro, M.; Mancuso, M.; Genovese, M.; Lunetta, A.; Caruso, G. Characterization of Five Psychrotolerant Alcanivorax spp. Strains Isolated from Antarctica. Microorganisms 2023, 11, 58. https://doi.org/10.3390/microorganisms11010058
Cappello S, Corsi I, Patania S, Bergami E, Azzaro M, Mancuso M, Genovese M, Lunetta A, Caruso G. Characterization of Five Psychrotolerant Alcanivorax spp. Strains Isolated from Antarctica. Microorganisms. 2023; 11(1):58. https://doi.org/10.3390/microorganisms11010058
Chicago/Turabian StyleCappello, Simone, Ilaria Corsi, Sabrina Patania, Elisa Bergami, Maurizio Azzaro, Monique Mancuso, Maria Genovese, Alessia Lunetta, and Gabriella Caruso. 2023. "Characterization of Five Psychrotolerant Alcanivorax spp. Strains Isolated from Antarctica" Microorganisms 11, no. 1: 58. https://doi.org/10.3390/microorganisms11010058







