Antimicrobial Resistance of Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella multocida Isolated from Romanian Swine Farms
Abstract
:1. Introduction
2. Materials and Methods
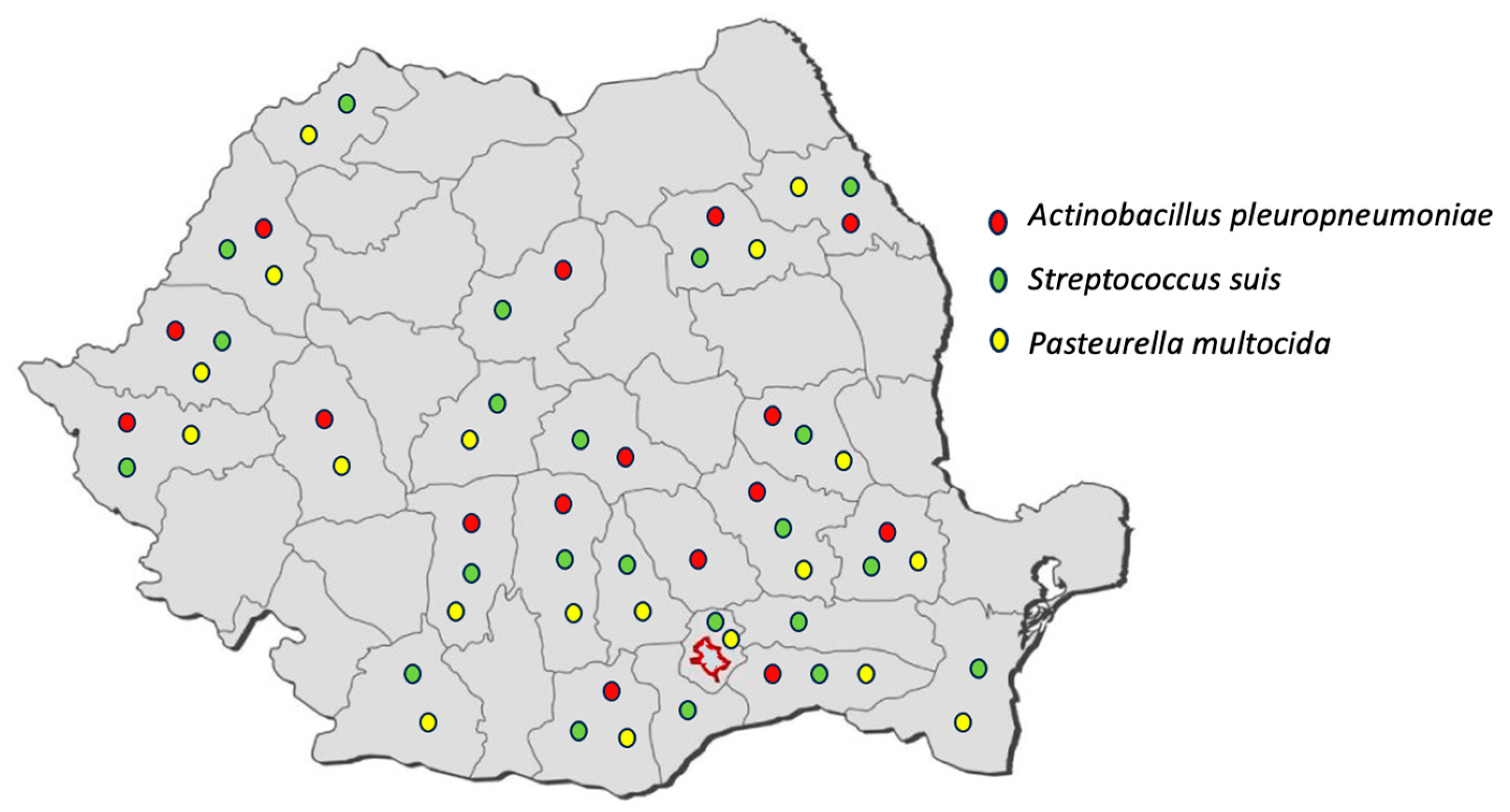
2.1. Sample Collection
2.2. Isolation and Identification
2.3. Antimicrobial Susceptibility Testing
3. Results
4. Discussion
4.1. Actinobacillus pleuropneumoniae
4.2. Streptococcus suis
4.3. Pasteurella multocia
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dayao, D.A.E.; Gibson, J.S.; Blackall, P.J.; Turni, C. Antimicrobial Resistance in Bacteria Associated with Porcine Respiratory Disease in Australia. Vet. Microbiol. 2014, 171, 232–235. [Google Scholar] [CrossRef] [PubMed]
- van Dixhoorn, I.D.E.; Te Beest, D.E.; Bolhuis, J.E.; Parmentier, H.K.; Kemp, B.; van Mourik, S.; Stockhofe-Zurwieden, N.; van Reenen, C.G.; Rebel, J.M.J. Animal-Based Factors Prior to Infection Predict Histological Disease Outcome in Porcine Reproductive and Respiratory Syndrome Virus- and Actinobacillus Pleuropneumoniae-Infected Pigs. Front. Vet. Sci. 2021, 8, 742877. [Google Scholar] [CrossRef] [PubMed]
- Petri, F.A.M.; Ferreira, G.C.; Arruda, L.P.; Malcher, C.S.; Storino, G.Y.; Almeida, H.M.D.S.; Sonalio, K.; Silva, D.G.D.; Oliveira, L.G.D. Associations between Pleurisy and the Main Bacterial Pathogens of the Porcine Respiratory Diseases Complex (PRDC). Animals 2023, 13, 1493. [Google Scholar] [CrossRef]
- Pepovich, R.; Nikolovn, B.; Genova, K.; Hristov, K.; Tfradjiiska-Hadjiolova, R.; Nikolova, E.; Stoimenov, G. The Comparative Therapeutic Efficacy of Antimicrobials in Pigs Infected with Mycoplasma Hyopneumoniae. Sci. Work. Ser. C. Vet. Med. 2016, LXII, 76–81. [Google Scholar]
- Sweeney, M.T.; Lindeman, C.; Johansen, L.; Mullins, L.; Murray, R.; Senn, M.K.S.; Bade, D.; Machin, C.; Kotarski, S.F.; Tiwari, R.; et al. Antimicrobial Susceptibility of Actinobacillus Pleuropneumoniae, Pasteurella Multocida, Streptococcus Suis, and Bordetella Bronchiseptica Isolated from Pigs in the United States and Canada, 2011 to 2015. J. Swine Health Prod. 2017, 25, 106–120. [Google Scholar] [CrossRef]
- Zimmerman, J.J.; Karriker, L.A.; Ramirez, A.; Schwartz, K.J.; Stevenson, G.W.; Zhang, J. (Eds.) Diseases of Swine, 11th ed.; Wiley-Blackwell/American Association of Swine Veterinarians: Hoboken, NJ, USA, 2019; pp. 396–397. [Google Scholar]
- Sargeant, J.M.; Bergevin, M.D.; Churchill, K.; Dawkins, K.; Deb, B.; Dunn, J.; Hu, D.; Moody, C.; O’Connor, A.M.; O’Sullivan, T.L.; et al. A Systematic Review of the Efficacy of Antibiotics for the Prevention of Swine Respiratory Disease. Anim. Health. Res. Rev. 2019, 20, 291–304. [Google Scholar] [CrossRef]
- Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2015 Annual Report; Government of Canada, Public Health Agency of Canada: Guelph, ON, Canada, 2017.
- Bush, E. Swine 2012 Part II: Reference of Swine Health and Health Management in the United States; USDA: Washington, DC, USA, 2016. [Google Scholar]
- Vanni, M.; Merenda, M.; Barigazzi, G.; Garbarino, C.; Luppi, A.; Tognetti, R.; Intorre, L. Antimicrobial Resistance of Actinobacillus Pleuropneumoniae Isolated from Swine. Vet. Microbiol. 2012, 156, 172–177. [Google Scholar] [CrossRef]
- De Jong, A.; Thomas, V.; Simjee, S.; Moyaert, H.; El Garch, F.; Maher, K.; Morrissey, I.; Butty, P.; Klein, U.; Marion, H.; et al. Antimicrobial Susceptibility Monitoring of Respiratory Tract Pathogens Isolated from Diseased Cattle and Pigs across Europe: The VetPath Study. Vet. Microbiol. 2014, 172, 202–215. [Google Scholar] [CrossRef]
- Yoo, A.N.; Cha, S.B.; Shin, M.K.; Won, H.K.; Kim, E.H.; Choi, H.W.; Yoo, H.S. Serotypes and Antimicrobial Resistance Patterns of the Recent Korean Actinobacillus Pleuropneumoniae Isolates. Vet. Rec. 2014, 174, 223. [Google Scholar] [CrossRef]
- Pageaut, H.; Lacouture, S.; Lehoux, M.; Marois-Créhan, C.; Segura, M.; Gottschalk, M. Interactions of Mycoplasma Hyopneumoniae and/or Mycoplasma Hyorhinis with Streptococcus Suis Serotype 2 Using In Vitro Co-Infection Models with Swine Cells. Pathogens 2023, 12, 866. [Google Scholar] [CrossRef]
- Petrocchi-Rilo, M.; Gutiérrez-Martín, C.-B.; Pérez-Fernández, E.; Vilaró, A.; Fraile, L.; Martínez-Martínez, S. Antimicrobial Resistance Genes in Porcine Pasteurella Multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern. Antibiotics 2020, 9, 614. [Google Scholar] [CrossRef] [PubMed]
- Bourély, C.; Cazeau, G.; Jouy, E.; Haenni, M.; Madec, J.-Y.; Jarrige, N.; Leblond, A.; Gay, E. Antimicrobial Resistance of Pasteurella Multocida Isolated from Diseased Food-Producing Animals and Pets. Vet. Microbiol. 2019, 235, 280–284. [Google Scholar] [CrossRef] [PubMed]
- El Garch, F.; de Jong, A.; Simjee, S.; Moyaert, H.; Klein, U.; Ludwig, C.; Marion, H.; Haag-Diergarten, S.; Richard-Mazet, A.; Thomas, V.; et al. Monitoring of Antimicrobial Susceptibility of Respiratory Tract Pathogens Isolated from Diseased Cattle and Pigs across Europe, 2009–2012: VetPath Results. Vet. Microbiol. 2016, 194, 11–22. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI Standard VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Pall, E.; Spinu, M.; Sandru, C.D.; Duca, G.; Suatean, M.I.; Szafta, A.-A.; Olah, D.; Vasiu, A. A Comparison of Antibiotic Resistance and Multiple Antibiotic Resistance Index in Wild Boars from Covasna and Cluj Counties. Sci. Works. Ser. C Vet. Med. 2022, LXVIII, 94–99. [Google Scholar]
- Portis, E.; Lindeman, C.; Johansen, L.; Stoltman, G. Antimicrobial Susceptibility of Porcine Pasteurella Multocida, Streptococcus Suis, and Actinobacillus Pleuropneumoniae from the United States and Canada, 2001 to 2010. J. Swine Health Prod. 2013, 21, 30–41. [Google Scholar]
- Vilaró, A.; Novell, E.; Enrique-Tarancón, V.; Balielles, J.; Vilalta, C.; Martinez, S.; Fraile Sauce, L.J. Antimicrobial Susceptibility Pattern of Porcine Respiratory Bacteria in Spain. Antibiotics 2020, 9, 402. [Google Scholar] [CrossRef]
- Holmer, I.; Salomonsen, C.M.; Jorsal, S.E.; Astrup, L.B.; Jensen, V.F.; Høg, B.B.; Pedersen, K. Antibiotic Resistance in Porcine Pathogenic Bacteria and Relation to Antibiotic Usage. BMC Vet. Res. 2019, 15, 449. [Google Scholar] [CrossRef]
- Xu, X.; Li, J.; Huang, P.; Cui, X.; Li, X.; Sun, J.; Huang, Y.; Ji, Q.; Wei, Q.; Bao, G.; et al. Isolation, Identification and Drug Resistance Rates of Bacteria from Pigs in Zhejiang and Surrounding Areas during 2019–2021. Vet. Sci. 2023, 10, 502. [Google Scholar] [CrossRef]
- Kucerova, Z.; Hradecka, H.; Nechvatalova, K.; Nedbalcova, K. Antimicrobial Susceptibility of Actinobacillus Pleuropneumoniae Isolates from Clinical Outbreaks of Porcine Respiratory Diseases. Vet. Microbiol. 2011, 150, 203–206. [Google Scholar] [CrossRef]
- Pascu, C.; Costinar, L.; Herman, V. Antibiotic Resistance of Actinobacillus Pleuropneumoniae Strains in the Period 2016–2022 in Romania. Rom. Rev. Vet. Med. 2022, 32, 40–46. [Google Scholar]
- Kamimura, S.; Sameshima, T.; Ito, H. Serovar and Antimicrobial Resistance Profiles of Actinobacillus Pleuropneumoniae Isolated in Japan from 2006 to 2011. JARQ 2016, 50, 73–77. [Google Scholar] [CrossRef]
- de Jong, A.; Morrissey, I.; Rose, M.; Temmerman, R.; Klein, U.; Simjee, S.; El Garch, F. Antimicrobial Susceptibility among Respiratory Tract Pathogens Isolated from Diseased Cattle and Pigs from Different Parts of Europe. J. Appl. Microbiol. 2023, 134, lxad132. [Google Scholar] [CrossRef] [PubMed]
- van Hout, J.; Heuvelink, A.; Gonggrijp, M. Monitoring of Antimicrobial Susceptibility of Streptococcus Suis in the Netherlands, 2013–2015. Vet. Microbiol. 2016, 194, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, J.; Wang, J.; Restif, O.; Holmes, M.A.; Mather, A.E.; Weinert, L.A.; Wileman, T.M.; Thomson, J.R.; Langford, P.R.; Wren, B.W.; et al. Patterns of Antimicrobial Resistance in Streptococcus Suis Isolates from Pigs with or without Streptococcal Disease in England between 2009 and 2014. Vet. Microbiol. 2017, 207, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-H.; Moon, D.-C.; Lee, Y.J.; Hyun, B.-H.; Lim, S.-K. Antimicrobial Resistance of Pasteurella Multocida Strains Isolated from Pigs between 2010 and 2016. Vet. Rec. Open 2018, 5, e000293. [Google Scholar] [CrossRef]
- Ladoşi, I.; Păpuc, T.A.; Ladoşi, D. The Impact of African Swine Fever (ASF) on Romanian Pig Meat Production: A Review. Acta Vet. 2023, 73, 1–12. [Google Scholar] [CrossRef]
- Mader, R.; Damborg, P.; Amat, J.-P.; Bengtsson, B.; Bourély, C.; Broens, E.M.; Busani, L.; Crespo-Robledo, P.; Filippitzi, M.-E.; Fitzgerald, W.; et al. Building the European Antimicrobial Resistance Surveillance Network in Veterinary Medicine (EARS-Vet). Eurosurveillance 2021, 26, 2001359. [Google Scholar] [CrossRef]
- Mader, R.; Muñoz Madero, C.; Aasmäe, B.; Bourély, C.; Broens, E.M.; Busani, L.; Callens, B.; Collineau, L.; Crespo-Robledo, P.; Damborg, P.; et al. Review and Analysis of National Monitoring Systems for Antimicrobial Resistance in Animal Bacterial Pathogens in Europe: A Basis for the Development of the European Antimicrobial Resistance Surveillance Network in Veterinary Medicine (EARS-Vet). Front. Microbiol. 2022, 13, 838490. [Google Scholar] [CrossRef]
- Lazar, C.I.; Duca, G.; Sandru, C.D.; Olah, D.; Spinu, M.; Pall, E.; Cerbu, C.; Giurgiu, O.; Potarniche, A.; Vasiu, A. Antibiotic Profile of Bacteria Isolated from the Skin Surface from Extensively Raised Swine. AGROLIFE 2022, 11, 80–84. [Google Scholar] [CrossRef]
- Vasiu, A.; Pall, E.; Spinu, M.; Zablau, S.D.; Ungureanu, E.; Suatean, M.I.; Brudasca, G.F.; Olah, D.I. Evaluation of Bacterial Involvement in an Episode of Neonatal Calf Diarrhea. AGROLIFE 2022, 11, 239–243. [Google Scholar] [CrossRef]
- Baraitareanu, S.; Vidu, L.; Stefan, G.; Mihai, B.; Mihai, R.; Militaru, I.S.; Birtoiu, D.; Nastase, V.; Catana, M.C.; Constantin, T.; et al. The Development of Dairy Farm Level Multi-Actor Teams Targeting Reduced Antibiotic Use in Romania. Sci. Work. Ser. C Vet. Med. 2021, LXVII, 35–42. [Google Scholar]
- Pană, B.C. Strengthening Romania’s Health System to Address Antimicrobial Resistance: EVIPNet Evidence Brief for Policy, Number 6; World Health Organization, Regional Office for Europe: København, Danmark, 2020. [Google Scholar]
| Title 1 | 2017 No. Isolates (%) | 2018 No. Isolates (%) | 2019 No. Isolates (%) | 2020 No. Isolates (%) | 2021 No. Isolates (%) | 2022 No. Isolates (%) | Total |
|---|---|---|---|---|---|---|---|
| APP * | 13 (10) | 14 (11) | 10 (7) | 15 (12) | 30 (23) | 55 (43) | 137 |
| Streptococcus suis | 21 (10) | 24 (12) | 14 (14) | 31 (15) | 55 (27) | 62 (30) | 207 |
| Pasteurella multocida | 9 (7) | 18 (15) | 8 (14) | 20 (17) | 36 (30) | 30 (25) | 121 |
| The Antimicrobial | Tablet Concentration (μg) | Susceptible (mm) | Intermediate (mm) | Resistant (mm) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| APP | S. suis | P. multocida | APP | S. suis | P. multocida | APP | S. suis | P. multocida | ||
| β-lactamase | ||||||||||
| Penicillin (P) | 10 | ≥17 | ≥24 | ≥17 | 14–16 | – | 14–16 | ≤13 | – | ≤13 |
| Ampicillin (AM) | 10 | ≥17 | ≥24 | ≥17 | 14–16 | – | 14–16 | ≤13 | – | ≤13 |
| Amoxicillin (AMX) | 25 | ≥17 | ≥24 | ≥17 | 14–16 | – | 14–16 | ≤13 | – | ≤13 |
| Amoxicillin + clavulanic acid (AMC) | 20/10 | ≥18 | ≥18 | ≥18 | 14–17 | 14–17 | 14–17 | ≤13 | ≤13 | ≤13 |
| Ceftiofur (CEF) | 30 | ≥21 | ≥21 | ≥21 | 18–22 | 18–20 | 18–20 | ≤17 | ≤17 | ≤17 |
| Cefquinome (CEQ) | 30 | ≥33 | ≥22 | ≥22 | 20–21 | 20–21 | 20–21 | ≤19 | ≤19 | ≤19 |
| Quinolones | ||||||||||
| Enrofloxacin (ENR) | 5 | ≥23 | ≥23 | ≥23 | 19–22 | 19–22 | 19–22 | ≤18 | ≤18 | ≤18 |
| Marbofloxacin (MAR) | 5 | ≥20 | ≥20 | ≥20 | 15–19 | 15–19 | 15–19 | ≤14 | ≤14 | ≤14 |
| Macrolides | ||||||||||
| Erythromycin (E) | 15 | – | ≥21 | – | – | 16–20 | – | – | ≤15 | – |
| Tilmicosin (TIL) | 15 | ≥11 | – | ≥11 | – | – | – | ≤10 | – | ≤10 |
| Tetracyclines | ||||||||||
| Oxytetracycline (OT) | 30 | ≥15 | ≥23 | ≥15 | 12–14 | 19–22 | 12–14 | ≤11 | ≤18 | ≤11 |
| Tetracycline (TE) | 30 | ≥15 | ≥23 | ≥15 | 12–14 | 19–22 | 12–14 | ≤11 | ≤18 | ≤11 |
| Doxycycline (DOXY) | 30 | ≥15 | ≥23 | ≥15 | 12–14 | 19–22 | 12–14 | ≤11 | ≤18 | ≤11 |
| Amphenicols | ||||||||||
| Florfenicol (FFC) | 30 | ≥22 | ≥22 | ≥22 | 19–21 | 19–21 | 19–21 | ≤18 | ≤18 | ≤18 |
| Aminoglycosides | ||||||||||
| Streptomycin (S) | 10 | ≥15 | – | ≥15 | 14 | – | 14 | ≤13 | – | ≤13 |
| Gentamycin (GM) | 10 | ≥16 | – | ≥16 | 13–15 | – | 13–15 | ≤12 | – | ≤12 |
| Neomicin (N) | 30 | – | – | ≥17 | – | – | 16 | – | – | ≤15 |
| Apramycin (AP) | 15 | – | – | ≥20 | – | – | 19–16 | – | – | ≤15 |
| Pleuromutilin deriv. | ||||||||||
| Tiamulin (TIA) | 30 | ≥9 | – | – | – | – | – | ≤8 | – | – |
| Sulphonamides | ||||||||||
| Trimethoprim – Sulfamethoxazole (SxT) | 1.25/ 23.75 | ≥16 | ≥19 | ≥16 | 11–15 | 16–18 | 11–15 | ≤10 | ≤15 | ≤10 |
| The Antimicrobial | No. of Isolates Tested | A. pleuropneumoniae | No. of Isolates Tested | S. suis | No. of Isolates Tested | P. multocida | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S % (No.) | I % (No.) | R % (No.) | S % (No.) | I % (No.) | R % (No.) | S % (No.) | I % (No.) | R % (No.) | ||||
| Amoxicillin | 133 | 95 (126) | 2 (2) | 4 (5) | 207 | 95 (197) | 1 (2) | 4 (8) | 119 | 97 (115) | 2 (2) | 2 (2) |
| Ampicillin | 136 | 94 (128) | 1 (1) | 5 (7) | 206 | 95 (196) | 0 (1) | 4 (9) | 121 | 96 (116) | 1 (1) | 3 (4) |
| Amoxicillin + clavulanic acid | 136 | 99 (135) | 1 (1) | 0 (0) | 206 | 98 (201) | 2 (4) | 0 (1) | 121 | 98 (119) | 1 (1) | 1 (1) |
| Penicillin | 128 | 95 (122) | 0 (0) | 5 (6) | 207 | 92 (191) | 0 (1) | 7 (15) | 120 | 97 (116) | 0 (0) | 3 (4) |
| Tetracycline | 123 | 76 (94) | 6 (7) | 18 (22) | 205 | 20 (41) | 5 (11) | 75 (153) | 106 | 85 (90) | 1 (1) | 14 (15) |
| Trimethoprim - Sulfamethoxazole | 137 | 91 (125) | 1 (2) | 7 (12) | 206 | 84 (174) | 2 (4) | 14 (28) | 118 | 93 (110) | 2 (2) | 5 (6) |
| Ceftiofur | 135 | 99 (134) | 1 (1) | 0 (0) | 204 | 99 (202) | 0 (1) | 0 (1) | 119 | 98 (117) | 0 (0) | 2 (2) |
| Oxytetracycline | 137 | 76 (104) | 4 (6) | 20 (27) | 203 | 22 (44) | 6 (12) | 72 (147) | 109 | 84 (92) | 3 (3) | 13 (14) |
| Enrofloxacin | 137 | 92 (126) | 3 (4) | 5 (7) | 207 | 83 (171) | 9 (19) | 8 (17) | 114 | 96 (109) | 4 (5) | 0 (0) |
| Marbofloxacin | 132 | 92 (122) | 3 (4) | 5 (6) | 206 | 91 (188) | 3 (7) | 5 (11) | 119 | 99 (118) | 0 (0) | 1 (1) |
| Florfenicol | 136 | 93 (128) | 3 (4) | 3 (4) | 191 | 97 (186) | 1 (2) | 2 (3) | 108 | 97 (105) | 0 (0) | 3 (3) |
| Cefquinome | 107 | 100 (107) | 0 (0) | 0 (0) | 180 | 93 (167) | 4 (7) | 3 (6) | 105 | 92 (97) | 4 (4) | 4 (4) |
| Doxycycline | 127 | 83 (106) | 6 (7) | 11 (14) | 199 | 29 (58) | 19 (38) | 52 (103) | 109 | 90 (98) | 6 (6) | 5 (5) |
| Erythromycin | 3 | 67 (2) | 0 (0) | 33 (1) | 196 | 42 (83) | 7 (14) | 51 (99) | 4 | 75 (3) | 25 (1) | 0 (0) |
| Gentamycin | 133 | 35 (47) | 20 (26) | 45 (60) | 1 | 100 (1) | 0 (0) | 0 (0) | 115 | 95 (83) | 9 (8) | 10 (11) |
| Tilmicosin | 132 | 58 (76) | 3 (4) | 39 (52) | 47 | 23 (11) | 4 (2) | 72 (34) | 119 | 62 (74) | 8 (10) | 29 (35) |
| Streptomycin | 131 | 18 (24) | 5 (7) | 77 (101) | 54 | 0 (0) | 2 (1) | 98 (53) | 113 | 35 (39) | 3 (3) | 63 (71) |
| Tiamulin | 20 | 60 (12) | 10 (2) | 30 (6) | 7 | 57 (4) | 0 (0) | 43 (3) | 8 | 38 (3) | 0 (0) | 63 (5) |
| Apramycin | – | – | – | – | – | – | – | – | 18 | 50 (9) | 11 (2) | 39 (7) |
| Neomicin | 4 | 100 (4) | 0 (0) | 0 (0) | 2 | 50 (1) | 0 (0) | 50 (1) | 102 | 62 (63) | 19 (19) | 20 (20) |
| The Antimicrobial | No. of Resistant Isolates | 2017 | 2018 | 2019 | 2020 | 2021 | 2022 |
|---|---|---|---|---|---|---|---|
| % (No.) | % (No.) | % (No.) | % (No.) | % (No.) | % (No.) | ||
| Amoxicillin | 5 | 0 (0) | 0 (0) | 0 (0) | 27 (4) | 3 (1) | 0 (0) |
| Ampicillin | 7 | 8 (1) | 0 (0) | 0 (0) | 27 (4) | 0 (0) | 4 (2) |
| Amoxicillin + clavulanic acid | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Penicillin | 6 | 0 (0) | 0 (0) | 0 (0) | 27 (4) | 4 (1) | 2 (1) |
| Tetracycline | 22 | 0 (0) | 0 (0) | 20 (2) | 40 (6) | 27 (8) | 15 (6) |
| Trimethoprim–Sulfamethoxazole | 12 | 0 (0) | 14 (2) | 10 (1) | 20 (3) | 7 (2) | 7 (4) |
| Ceftiofur | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Oxytetracycline | 27 | 1 (8) | 1 (7) | 1 (10) | 7 (47) | 8 (27) | 9 (16) |
| Enrofloxacin | 7 | 0 (0) | 0 (0) | 0 (0) | 13 (2) | 7 (2) | 5 (3) |
| Marbofloxacin | 6 | 0 (0) | 0 (0) | 0 (0) | 13 (2) | 3 (1) | 6 (3) |
| Florfenicol | 4 | 0 (0) | 0 (0) | 0 (0) | 7 (1) | 7 (2) | 2 (1) |
| Cefquinome | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Doxycycline | 14 | 0 (0) | 0 (0) | 0 (0) | 13 (2) | 15 (4) | 16 (8) |
| Erythromycin | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 100 (1) |
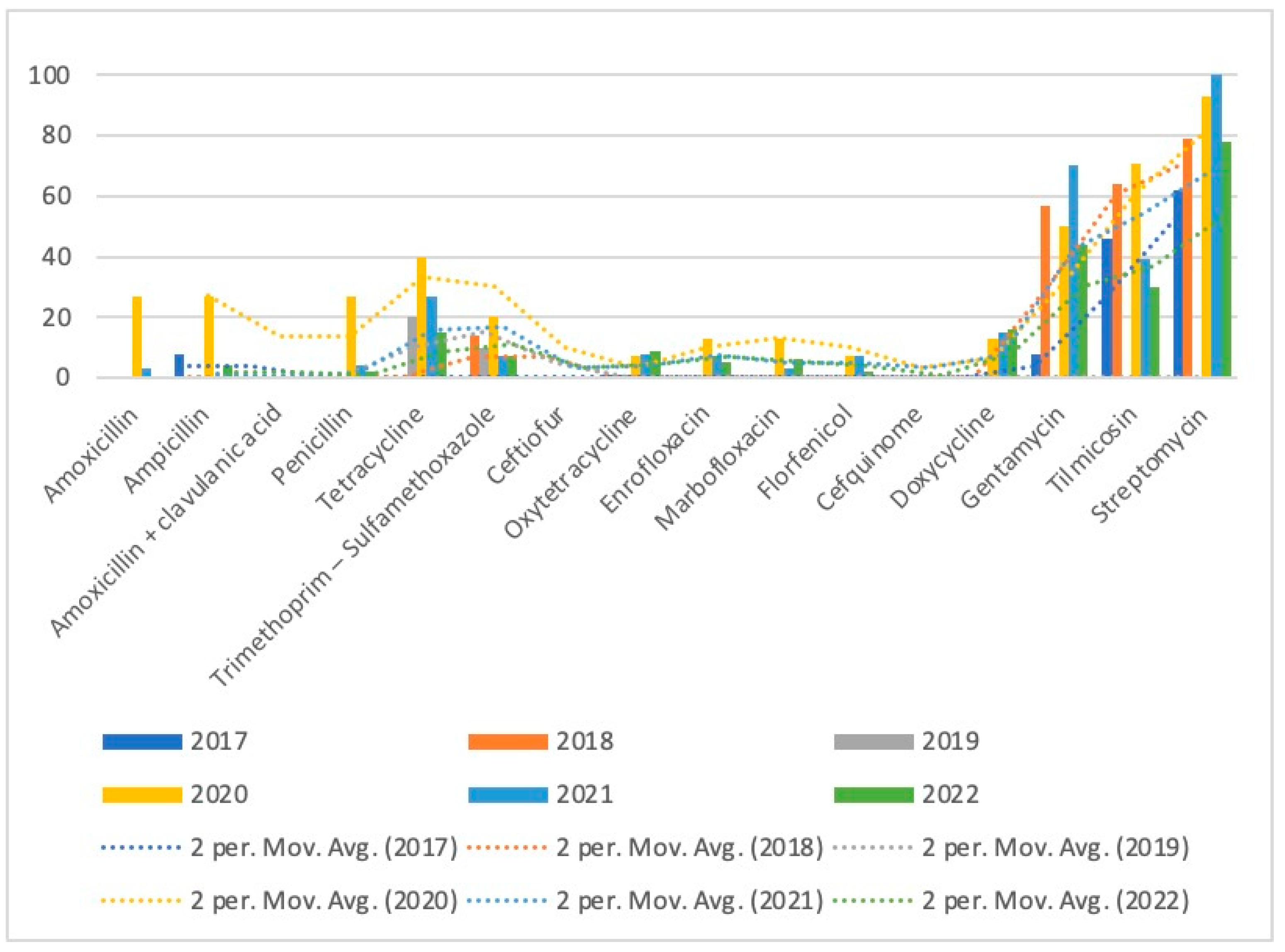
| Gentamycin | 60 | 8 (1) | 57 (8) | 0 (0) | 50 (7) | 70 (21) | 44 (23) |
| Tilmicosin | 52 | 46 (6) | 64 (9) | 0 (0) | 71 (10) | 39 (11) | 30 (16) |
| Streptomycin | 101 | 62 (8) | 79 (11) | 0 (0) | 93 (14) | 100 (28) | 78 (40) |
| Tiamulin | 6 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 25 (1) | 71 (5) |
| Neomicin | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| The Antimicrobial | No. of Resistant Isolates | 2017 | 2018 | 2019 | 2020 | 2021 | 2022 |
|---|---|---|---|---|---|---|---|
| % (No.) | % (No.) | % (No.) | % (No.) | % (No.) | % (No.) | ||
| Amoxicillin | 8 | 5 (1) | 0 (0) | 7 (1) | 0 (0) | 4 (2) | 6 (4) |
| Ampicillin | 9 | 0 (0) | 17 (4) | 0 (0) | 0 (0) | 4 (2) | 5 (3) |
| Amoxicillin + clavulanic acid | 1 | 0 (0) | 4 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Penicillin | 15 | 5 (1) | 17 (4) | 7 (1) | 6 (2) | 5 (3) | 6 (4) |
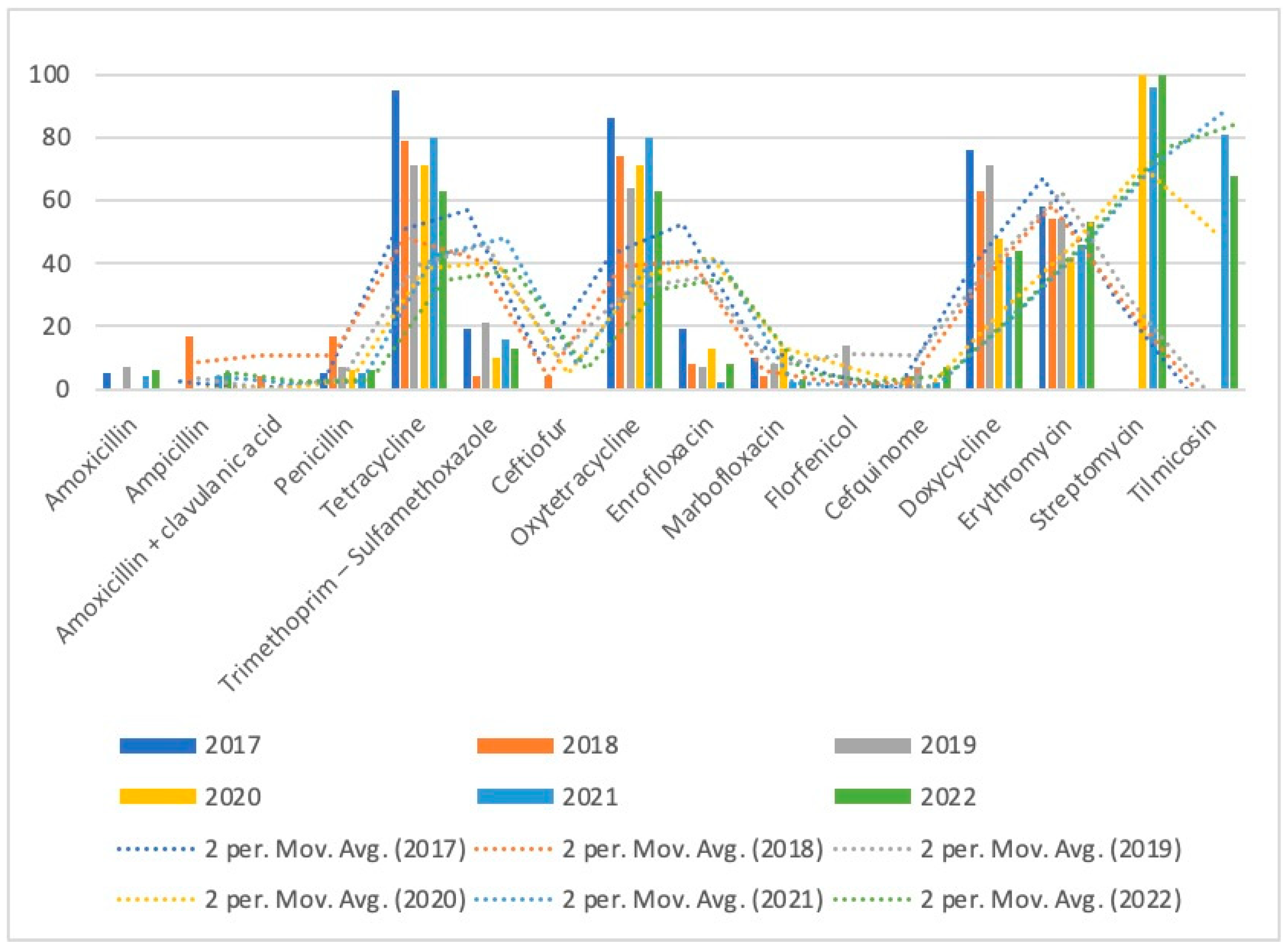
| Tetracycline | 153 | 95 (20) | 79 (19) | 71 (10) | 71 (22) | 80 (44) | 63 (38) |
| Trimethoprim–Sulfamethoxazole | 28 | 19 (4) | 4 (1) | 21 (3) | 10 (3) | 16 (9) | 13 (8) |
| Ceftiofur | 1 | 0 (0) | 4 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Oxytetracycline | 147 | 86 (18) | 74 (17) | 64 (9) | 71 (22) | 80 (43) | 63 (38) |
| Enrofloxacin | 17 | 19 (4) | 8 (2) | 7 (1) | 13 (4) | 2 (1) | 8 (5) |
| Marbofloxacin | 11 | 10 (2) | 4 (1) | 8 (1) | 13 (4) | 2 (1) | 3 (2) |
| Florfenicol | 3 | 0 (0) | 0 (0) | 14 (2) | 0 (0) | 0 (0) | 2 (1) |
| Cefquinome | 6 | 0 (0) | 4 (1) | 7 (1) | 0 (0) | 2 (1) | 7 (3) |
| Doxycycline | 103 | 76 (16) | 63 (15) | 71 (10) | 48 (15) | 42 (22) | 44 (25) |
| Erythromycin | 99 | 58 (11) | 54 (13) | 54 (7) | 42 (11) | 46 (24) | 53 (33) |
| Streptomycin | 53 | 0 (0) | 0 (0) | 0 (0) | 100 (1) | 96 (24) | 100 (28) |
| Tiamulin | 3 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 33 (1) | 50 (2) |
| Tilmicosin | 34 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 81 (13) | 68 (21) |
| Neomicin | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 50 (1) | 0 (0) |
| Gentamycin | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| The Antimicrobial | No. of Resistant Isolates | 2017 | 2018 | 2019 | 2020 | 2021 | 2022 |
|---|---|---|---|---|---|---|---|
| % (No.) | % (No.) | % (No.) | % (No.) | % (No.) | % (No.) | ||
| Amoxicillin | 2 | 0 (0) | 6 (1) | 0 (0) | 0 (0) | 0 (0) | 4 (1) |
| Ampicillin | 4 | 0 (0) | 6 (1) | 13 (1) | 0 (0) | 0 (0) | 7 (2) |
| Amoxicillin + clavulanic acid | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (1) |
| Penicillin | 4 | 11 (1) | 6 (1) | 0 (0) | 0 (0) | 3 (1) | 3 (1) |
| Tetracycline | 15 | 11 (1) | 11 (2) | 25 (2) | 18 (2) | 22 (7) | 4 (1) |
| Trimethoprim–Sulfamethoxazole | 6 | 11 (1) | 0 (0) | 29 (2) | 10 (2) | 3 (1) | 0 (0) |
| Ceftiofur | 2 | 0 (0) | 0 (0) | 0 (0) | 5 (1) | 0 (0) | 3 (1) |
| Oxytetracycline | 14 | 22 (2) | 0 (0) | 29 (2) | 18 (2) | 19 (7) | 3 (1) |
| Enrofloxacin | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Marbofloxacin | 1 | 0 (0) | 0 (0) | 0 (0) | 5 (1) | 0 (0) | 0 (0) |
| Florfenicol | 3 | 0 (0) | 11 (2) | 13 (1) | 0 (0) | 0 (0) | 0 (0) |
| Cefquinome | 4 | 0 (0) | 6 (1) | 38 (3) | 0 (0) | 0 (0) | 0 (0) |
| Doxycycline | 5 | 0 (0) | 0 (0) | 14 (1) | 6 (1) | 9 (3) | 0 (0) |
| Erythromycin | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
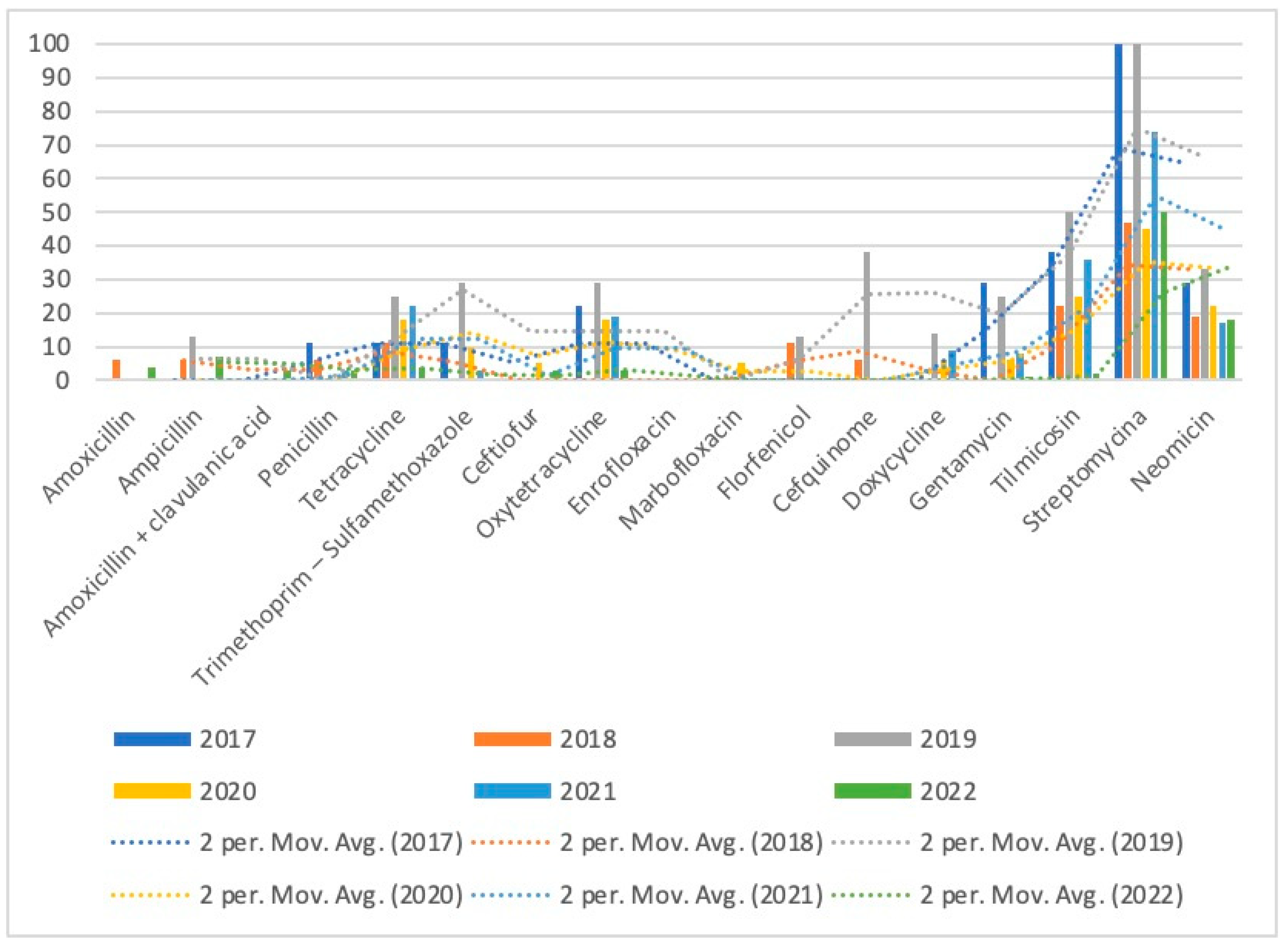
| Gentamycin | 11 | 29 (2) | 0 (0) | 25 (2) | 6 (1) | 8 (3) | 11 (3) |
| Tilmicosin | 35 | 38 (3) | 22 (4) | 50 (4) | 25 (5) | 36 (13) | 21 (6) |
| Streptomycin | 71 | 100 (7) | 47 (8) | 100 (8) | 45 (9) | 74 (26) | 50 (13) |
| Neomicin | 20 | 29 (2) | 19 (3) | 33 (2) | 22 (2) | 17 (6) | 18 (5) |
| Apramycin | 7 | 0 (0) | 100 (1) | 0 (0) | 67 (2) | 0 (0) | 29 (4) |
| Tiamulin | 5 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 50 (2) | 100 (3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siteavu, M.I.; Drugea, R.I.; Pitoiu, E.; Ciobotaru-Pirvu, E. Antimicrobial Resistance of Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella multocida Isolated from Romanian Swine Farms. Microorganisms 2023, 11, 2410. https://doi.org/10.3390/microorganisms11102410
Siteavu MI, Drugea RI, Pitoiu E, Ciobotaru-Pirvu E. Antimicrobial Resistance of Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella multocida Isolated from Romanian Swine Farms. Microorganisms. 2023; 11(10):2410. https://doi.org/10.3390/microorganisms11102410
Chicago/Turabian StyleSiteavu, Madalina Iulia, Roxana Ionela Drugea, Elena Pitoiu, and Emilia Ciobotaru-Pirvu. 2023. "Antimicrobial Resistance of Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella multocida Isolated from Romanian Swine Farms" Microorganisms 11, no. 10: 2410. https://doi.org/10.3390/microorganisms11102410
APA StyleSiteavu, M. I., Drugea, R. I., Pitoiu, E., & Ciobotaru-Pirvu, E. (2023). Antimicrobial Resistance of Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella multocida Isolated from Romanian Swine Farms. Microorganisms, 11(10), 2410. https://doi.org/10.3390/microorganisms11102410





