Ethanol-Producing Enterocloster bolteae Is Enriched in Chronic Hepatitis B-Associated Gut Dysbiosis: A Case–Control Culturomics Study
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Design
2.2. Study Population
2.3. High-Throughput Culturomics Approach
2.4. Method of 16S Ribosomal DNA Gene Amplification and Sequencing
2.5. Measurement of Ethanol Production by Strains Enriched in Chronic HBV Samples
2.6. Bioinformatic Analysis
2.7. Statistical Analysis
3. Results
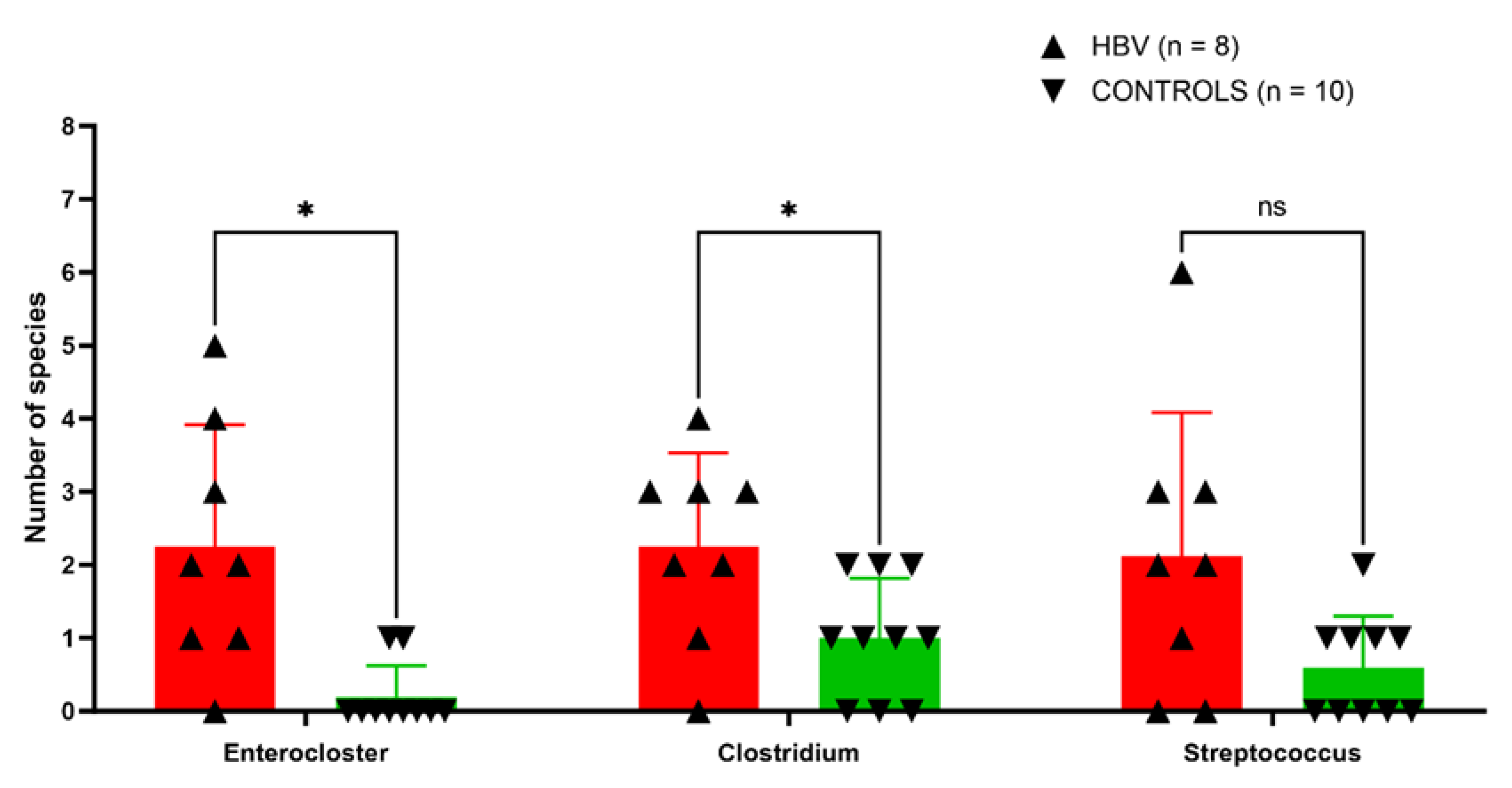
3.1. Altered Diversity in Chronic HBV Samples by Culturomics
3.2. Diversity Assessed by Metagenomics
3.3. Missing Repertoire in Patients with Chronic HBV Infection
3.4. Ethanol Quantification Produced by Enterocloster Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, L.S.Y.; Covert, E.; Wilson, E.; Kottilil, S. Chronic Hepatitis B Infection: A Review. JAMA 2018, 319, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.S.; Huang, J.L.W.; George, J.; Huang, J.; Leung, C.; Eslam, M.; Chan, H.L.Y.; Ng, S.C. The changing epidemiology of liver diseases in the Asia-Pacific region. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, J.; Wang, L.; Wu, X.; Chen, Y.; Piao, H.; Lu, L.; Jiang, W.; Xu, Y.; Feng, B.; et al. New classification of liver biopsy assessment for fibrosis in chronic hepatitis B patients before and after treatment. Hepatology 2017, 65, 1438–1450. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.-R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, Y.L.; Yang, Y.Y.; Zhang, N.P.; Niu, C.; Shen, X.Z.; Wu, J. Novel approaches to intervene gut microbiota in the treatment of chronic liver diseases. FASEB J. 2021, 35, e21871. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Tang, R.; Li, B.; Ma, X.; Schnabl, B.; Tilg, H. Gut microbiome, liver immunology, and liver diseases. Cell. Mol. Immunol. 2021, 18, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, V.; Di Gregorio, V.; Iebba, V.; Giusto, M.; Schippa, S.; Merli, M.; Thalheimer, U. Microbiota and the gut-liver axis: Bacterial translocation, inflammation and infection in cirrhosis. World J. Gastroenterol. 2014, 20, 16795–16810. [Google Scholar] [CrossRef]
- Zeuzem, S. Gut-liver axis. Int. J. Colorectal Dis. 2000, 15, 59–82. [Google Scholar] [CrossRef]
- McCuskey, R.S. Sinusoidal endothelial cells as an early target for hepatic toxicants. Clin. Hemorheol. Microcirc. 2006, 34, 5–10. [Google Scholar]
- Sun, Z.; Huang, C.; Shi, Y.; Wang, R.; Fan, J.; Yu, Y.; Zhang, Z.; Zhu, K.; Li, M.; Ni, Q.; et al. Distinct Bile Acid Profiles in Patients with Chronic Hepatitis B Virus Infection Reveal Metabolic Interplay Between Host, Virus and Gut Microbiome. Front. Med. 2021, 8, 708495. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, L.; Wang, H.; Cai, W.; Xie, Q. Modulation of bile acid profile by gut microbiota in chronic hepatitis B. J. Cell. Mol. Med. 2020, 24, 2573–2581. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xie, Y.; Zhou, F.; Zhang, B.; Wu, J.; Yang, L.; Xu, S.; Stedtfeld, R.; Chen, Q.; Liu, J.; et al. Featured Gut Microbiomes Associated With the Progression of Chronic Hepatitis B Disease. Front. Microbiol. 2020, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Chen, S.; Fu, Y.; Wu, W.; Chen, T.; Chen, J.; Yang, B.; Ou, Q. Gut microbiota dysbiosis in patients with hepatitis B virus-induced chronic liver disease covering chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. J. Viral Hepat. 2020, 27, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.A.; Lv, F.; Wang, R.; Chang, Y.; Zhao, Y.; Cui, X.; Li, H.; Yang, S.; Li, S.; Zhao, X.; et al. Potential role of intestinal microflora in disease progression among patients with different stages of Hepatitis B. Gut Pathogens. 2020, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Yan, X.; Zou, D.; Yang, Z.; Wang, X.; Liu, W.; Wang, S.; Li, X.; Han, J.; Huang, L.; et al. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Ayling, M.; Clark, M.D.; Leggett, R.M. New approaches for metagenome assembly with short reads. Brief. Bioinform. 2020, 21, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Tidjani Alou, M.; Million, M.; Traore, S.I.; Mouelhi, D.; Khelaifia, S.; Bachar, D.; Caputo, A.; Delerce, J.; Brah, S.; Alhousseini, D.; et al. Gut Bacteria Missing in Severe Acute Malnutrition, Can We Identify Potential Probiotics by Culturomics? Front. Microbiol. 2017, 8, 899. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.D.; Ye, Z.S.; Yang, L.Z.; Jin, L.X.; Wei, W.J.; Deng, Y.Y.; Chen, X.X.; Xiao, C.X.; Yu, X.F.; Xu, H.Z.; et al. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology 2017, 65, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Kumar, R.; Sharma, S.; Mahanta, M.; Vayuuru, S.K.; Nayak, B.; Kumar, S.; Shalimar. Fecal Microbiota Transplantation in Hepatitis B e Antigen-Positive Chronic Hepatitis B Patients: A Pilot Study. Dig. Dis. Sci. 2021, 66, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, J.; Lan, L.-L.; Li, X.-A. The effect of fecal microbiota transplantation on Hepatic myelopathy: A case report. Medicine 2019, 98, e16430. [Google Scholar] [CrossRef] [PubMed]
- Thilakarathna, W.P.D.W.; Rupasinghe, H.P.V.; Ridgway, N.D. Mechanisms by Which Probiotic Bacteria Attenuate the Risk of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 2606. [Google Scholar] [CrossRef] [PubMed]
- Bagga, D.; Reichert, J.L.; Koschutnig, K.; Aigner, C.S.; Holzer, P.; Koskinen, K.; Moissl-Eichinger, C.; Schöpf, V. Probiotics drive gut microbiome triggering emotional brain signatures. Gut Microbes 2018, 9, 486–496. [Google Scholar] [CrossRef]
- Ziada, D.H.; Soliman, H.H.; El Yamany, S.A.; Hamisa, M.F.; Hasan, A.M. Can Lactobacillus acidophilus improve minimal hepatic encephalopathy? A neurometabolite study using magnetic resonance spectroscopy. Arab. J. Gastroenterol. 2013, 14, 116–122. [Google Scholar] [CrossRef]
- Xia, X.; Chen, J.; Xia, J.; Wang, B.; Liu, H.; Yang, L.; Wang, Y.; Ling, Z. Role of probiotics in the treatment of minimal hepatic encephalopathy in patients with HBV-induced liver cirrhosis. J. Int. Med. Res. 2018, 46, 3596–3604. [Google Scholar] [CrossRef]
- Agrawal, A.; Sharma, B.C.; Sharma, P.; Sarin, S.K. Secondary Prophylaxis of Hepatic Encephalopathy in Cirrhosis: An Open-Label, Randomized Controlled Trial of Lactulose, Probiotics, and No Therapy. Off. J. Am. Coll. Gastroenterol.|ACG. 2012, 107, 1043–1050. [Google Scholar] [CrossRef]
- Bellali, S.; Lagier, J.C.; Million, M.; Anani, H.; Haddad, G.; Francis, R.; Kuete Yimagou, E.; Khelaifia, S.; Levasseur, A.; Raoult, D.; et al. Running after ghosts: Are dead bacteria the dark matter of the human gut microbiota? Gut Microbes 2021, 13, 1897208. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.N.; Blanchard, J.L. Reclassification of the Clostridium clostridioforme and Clostridium sphenoides clades as Enterocloster gen. nov. and Lacrimispora gen. nov.; including reclassification of 15 taxa. Int. J. Syst. Evol. Microbiol. 2020, 70, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Liu, C.; Molitoris, D.R.; Tomzynski, T.J.; Lawson, P.A.; Collins, M.D.; Finegold, S.M. Clostridium bolteae sp. nov.; Isolated from Human Sources. Syst. Appl. Microbiol. 2003, 26, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Pequegnat, B.; Sagermann, M.; Valliani, M.; Toh, M.; Chow, H.; Allen-Vercoe, E.; Monteiro, M.A. A vaccine and diagnostic target for Clostridium bolteae, an autism-associated bacterium. Vaccine 2013, 31, 2787–2790. [Google Scholar] [CrossRef] [PubMed]
- Warren, Y.A.; Tyrrell, K.L.; Citron, D.M.; Goldstein, E.J.C. Clostridium aldenense sp. nov. and Clostridium citroniae sp. nov. isolated from human clinical infections. J. Clin. Microbiol. 2006, 44, 2416–2422. [Google Scholar] [CrossRef] [PubMed]
- Finegold, S.M.; Molitoris, D.; Song, Y.; Liu, C.; Vaisanen, M.L.; Bolte, E.; McTeague, M.; Sandler, R.; Wexler, H.; Marlowe, E.M.; et al. Gastrointestinal microflora studies in late-onset autism. Clin. Infect. Dis. 2002, 35, S6–S16. [Google Scholar] [CrossRef] [PubMed]
- Ruuskanen, M.O.; Åberg, F.; Männistö, V.; Havulinna, A.S.; Méric, G.; Liu, Y.; Loomba, R.; Vázquez-Baeza, Y.; Tripathi, A.; Valsta, L.M.; et al. Links between gut microbiome composition and fatty liver disease in a large population sample. Gut Microbes 2021, 13, 1888673. [Google Scholar] [CrossRef] [PubMed]
- Frame, N.W.; Allas, M.J.; Pequegnat, B.; Vinogradov, E.; Liao, V.C.; Al-Abdul-Wahid, S.; Arroyo, L.; Allen-Vercoe, E.; Lowary, T.L.; Monteiro, M.A. Structure and synthesis of a vaccine and diagnostic target for Enterocloster bolteae, an autism-associated gut pathogen—Part II. Carbohydr. Res. 2023, 526, 108805. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Namsolleck, P.; Lawson, P.A.; Osterhoff, M.; Collins, M.D.; Alpert, C.A.; Blaut, M. Clostridium asparagiforme sp. nov., isolated from a human faecal sample. Syst. Appl. Microbiol. 2006, 29, 292–299. [Google Scholar] [CrossRef]
- Mota, A.; Guedes, F.; Areias, J.; Pinho, L.; Cardoso, M.F. Alcohol consumption among patients with hepatitis B infection in northern Portugal considering gender and hepatitis B virus genotype differences. Alcohol 2010, 44, 149–156. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. Epidemiology 2007, 18, 800. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef] [PubMed]
- Handcock, M.S.; Gile, K.J. Comment: On the Concept of Snowball Sampling. Sociol. Methodol. 2011, 41, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Liver EA for the S of the. EASL Clinical Practice Guidelines: Management of chronic hepatitis B virus infection. J. Hepatol. 2012, 57, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The Effects of Vegetarian and Vegan Diets on Gut Microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and past strategies for bacterial culture in clinical microbiology. Clin. Microbiol. Rev. 2015, 28, 208–236. [Google Scholar] [CrossRef] [PubMed]
- Naud, S.; Khelaifia, S.; Mbogning Fonkou, M.D.; Dione, N.; Lagier, J.-C.; Raoult, D. Proof of Concept of Culturomics Use of Time of Care. Front. Cell. Infect. Microbiol. 2020, 10, 524769. [Google Scholar] [CrossRef]
- Lagier, J.C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef]
- Seng, P.; Abat, C.; Rolain, J.M.; Colson, P.; Lagier, J.C.; Gouriet, F.; Fournier, P.E.; Drancourt, M.; La Scola, B.; Raoult, D. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: Impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2013, 51, 2182–2194. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.-E.; Lagier, J.-C.; Dubourg, G.; Raoult, D. From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 2015, 36, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Diakite, A.; Dubourg, G.; Raoult, D. Updating the repertoire of cultured bacteria from the human being. Microb. Pathog. 2021, 150, 104698. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Bachar, D.; Henrissat, B.; Armougom, F.; Audoly, G.; Lagier, J.C.; Robert, C.; Raoult, D. Glycans affect DNA extraction and induce substantial differences in gut metagenomic studies. Sci. Rep. 2016, 6, 26276. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.M.; Eisen, J.A.; Zivkovic, A.M. The microbes we eat: Abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types. PeerJ 2014, 2, e659. [Google Scholar] [CrossRef] [PubMed]
- Maukonen, J.; Saarela, M. Human gut microbiota: Does diet matter? Proc. Nutr. Soc. 2015, 74, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Dubourg, G.; Lagier, J.C.; Armougom, F.; Robert, C.; Hamad, I.; Brouqui, P.; Raoult, D. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 637–645. [Google Scholar] [CrossRef]
- Bellali, S.; Bou Khalil, J.; Fontanini, A.; Raoult, D.; Lagier, J.-C. A new protectant medium preserving bacterial viability after freeze drying. Microbiol Res. 2020, 236, 126454. [Google Scholar] [CrossRef]
- Dubourg, G.; Lagier, J.C.; Robert, C.; Armougom, F.; Hugon, P.; Metidji, S.; Dione, N.; Dangui, N.P.; Pfleiderer, A.; Abrahao, J.; et al. Culturomics and pyrosequencing evidence of the reduction in gut microbiota diversity in patients with broad-spectrum antibiotics. Int. J. Antimicrob. Agents 2014, 44, 117–124. [Google Scholar] [CrossRef]
- Lagier, J.C.; Hugon, P.; Khelaifia, S.; Fournier, P.E.; La Scola, B.; Raoult, D. The Rebirth of Culture in Microbiology through the Example of Culturomics to Study Human Gut Microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Pukall, R.; Abt, B.; Foesel, B.U.; Meier-Kolthoff, J.P.; Kumar, N.; Bresciani, A.; Martínez, I.; Just, S.; Ziegler, C.; et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 2016, 1, 16131. [Google Scholar] [CrossRef] [PubMed]
- Yeation, W.H.; Langenbrunner, J.C.; Smyth, J.M.; Wortman, P.M. Exploratory Research Synthesis: Methodological Considerations for Addressing Limitations in Data Quality. Eval. Health Prof. 1995, 18, 283–303. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.R.; Carley, S.; Harrison, M. An introduction to power and sample size estimation. Emerg. Med. J. 2003, 20, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Joo, E.-J.; Cheong, H.S.; Kwon, M.-J.; Sohn, W.; Kim, H.-N.; Cho, Y.K. Relationship between gut microbiome diversity and hepatitis B viral load in patients with chronic hepatitis B. Gut Pathogens 2021, 13, 65. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; Felis, G.E.; Dellaglio, F.; Castioni, A.; Torriani, S.; Lawson, P.A. Reclassification of Lactobacillus catenaformis (Eggerth 1935) Moore and Holdeman 1970 and Lactobacillus vitulinus Sharpe et al. 1973 as Eggerthia catenaformis gen. nov.; comb. nov. and Kandleria vitulina gen. nov.; comb. nov.; respectively. Int. J. Syst. Evol. Microbiol. 2011, 61, 2520–2524. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Nobu, M.K.; Kusada, H.; Meng, X.Y.; Hosogi, N.; Uematsu, K.; Yoshioka, H.; Kamagata, Y.; Tamaki, H. Isolation of a member of the candidate phylum ‘Atribacteria’ reveals a unique cell membrane structure. Nat. Commun. 2020, 11, 6381. [Google Scholar] [CrossRef]
- Burckhardt, J.C.; Chong, D.H.Y.; Pett, N.; Tropini, C. Gut commensal Enterocloster species host inoviruses that are secreted in vitro and in vivo. Microbiome 2023, 11, 65. [Google Scholar] [CrossRef]
- Ilyina, T.S. Filamentous bacteriophages and their role in the virulence and evolution of pathogenic bacteria. Mol. Genet. Microbiol. Virol. 2015, 30, 1–9. [Google Scholar] [CrossRef]
- Hay, I.D.; Lithgow, T. Filamentous phages: Masters of a microbial sharing economy. EMBO Rep. 2019, 20, e47427. [Google Scholar] [CrossRef] [PubMed]
- Williams, O.M.; Brazier, J.; Peraino, V.; Goldstein, E.J.C. A review of three cases of Clostridium aldenense bacteremia. Anaerobe 2010, 16, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Leal, J.; Gregson, D.B.; Ross, T.; Church, D.L.; Laupland, K.B. Epidemiology of Clostridium species bacteremia in Calgary, Canada, 2000-2006. J. Infect. 2008, 57, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Tidjani Alou, M.; Lagier, J.-C.; Raoult, D. Diet influence on the gut microbiota and dysbiosis related to nutritional disorders. Hum. Microbiome J. 2016, 1, 3–11. [Google Scholar] [CrossRef]
- Song, Y.; Liu, C.; Finegold, S.M. Real-Time PCR Quantitation of Clostridia in Feces of Autistic Children. Appl. Environ. Microbiol. 2004, 70, 6459–6465. [Google Scholar] [CrossRef] [PubMed]
- Cox, L.M.; Maghzi, A.H.; Liu, S.; Tankou, S.K.; Dhang, F.H.; Willocq, V.; Song, A.; Wasén, C.; Tauhid, S.; Chu, R.; et al. The Gut Microbiome in Progressive Multiple Sclerosis. Ann. Neurol. 2021, 89, 1195–1211. [Google Scholar] [CrossRef] [PubMed]
- Pandit, L.; Cox, L.M.; Malli, C.; D’Cunha, A.; Rooney, T.; Lokhande, H.; Willocq, V.; Saxena, S.; Chitnis, T. Clostridium bolteae is elevated in neuromyelitis optica spectrum disorder in India and shares sequence similarity with AQP4. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e907. [Google Scholar] [CrossRef]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Takei, H.; Narushima, S.; Suzuki, M.; Kakiyama, G.; Sasaki, T.; Murai, T.; Yamashiro, Y.; Nittono, H. Characterization of long-chain fatty acid-linked bile acids: A major conjugation form of 3β-hydroxy bile acids in feces. J. Lipid Res. 2022, 63, 100275. [Google Scholar] [CrossRef]
- He, S.; Xiong, Q.; Tian, C.; Li, L.; Zhao, J.; Lin, X.; Guo, X.; He, Y.; Liang, W.; Zuo, X.; et al. Inulin-type prebiotics reduce serum uric acid levels via gut microbiota modulation: A randomized, controlled crossover trial in peritoneal dialysis patients. Eur. J. Nutr. 2022, 61, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, R.; Nugent, M.; Cai, W.; Nadkarni, G.N.; Chaves, L.D.; Abyad, S.; Honan, A.M.; Thomas, S.A.; Zheng, W.; Valiyaparambil, S.A.; et al. Advanced glycation end products dietary restriction effects on bacterial gut microbiota in peritoneal dialysis patients; a randomized open label controlled trial. PLoS ONE 2017, 12, e0184789. [Google Scholar] [CrossRef] [PubMed]
- Skerman, V.B.D.; Mcgowanvicki Sneath, P.H.A.Y. Approved Lists of Bacterial Names. Int. J. Syst. Evol. Microbiol. 1980, 30, 225–420. [Google Scholar] [CrossRef]
- Li, J.; Adams, V.; Bannam, T.L.; Miyamoto, K.; Garcia, J.P.; Uzal, F.A.; Rood, J.I.; McClane, B.A. Toxin plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 2013, 77, 208–233. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Lee, H.C.; Chang, C.; Chuang, Y.; Ko, W. Clostridium bacteremia: Emphasis on the poor prognosis in cirrhotic patients. J. Microbiol. Immunol. Infect. = Wei Mian Yu Gan Ran Za Zhi 2001, 34, 113–118. [Google Scholar] [PubMed]
- Cherny, K.E.; Muscat, E.B.; Reyna, M.E.; Kociolek, L.K. Clostridium innocuum: Microbiological and Clinical Characteristics of a Potential Emerging Pathogen. Anaerobe 2021, 71, 102418. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.; Anfora, A.; Liu, J. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.N.; Kang, N.L.; Jiang, J.J.; Zhu, Y.Y.; Liu, Y.R.; Zeng, D.W.; Wang, F. Gut microbiota of hepatitis B virus-infected patients in the immune-tolerant and immune-active phases and their implications in metabolite changes. World J. Gastroenterol. 2022, 28, 5188–5202. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Z.; Mo, Z.S.; Yang, X.H.; Lin, B.L.; Peng, L.; Xu, Y.; Lei, C.Y.; Zhuang, X.D.; Lu, L.; et al. Gut microbiota as prognosis markers for patients with HBV-related acute-on-chronic liver failure. Gut Microbes 2021, 13, 1921925. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Dehoux, P.; Marvaud, J.C.; Abouelleil, A.; Earl, A.M.; Lambert, T.; Dauga, C. Comparative genomics of Clostridium bolteae and Clostridium clostridioforme reveals species-specific genomic properties and numerous putative antibiotic resistance determinants. BMC Genom. 2016, 17, 819. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-E.; Zhang, Y.; Zhang, J.; Dong, P.-L.; Chen, M.; Duan, Z.-P. Probiotic yogurt effects on intestinal flora of patients with chronic liver disease. Nurs. Res. 2010, 59, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Shukla, A.; Mehboob, S.; Guha, S. Meta-analysis: The effects of gut flora modulation using prebiotics, probiotics and synbiotics on minimal hepatic encephalopathy. Aliment. Pharmacol. Ther. 2011, 33, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Paratore, M.; Santopaolo, F.; Cammarota, G.; Pompili, M.; Gasbarrini, A.; Ponziani, F.R. Fecal Microbiota Transplantation in Patients with HBV Infection or Other Chronic Liver Diseases: Update on Current Knowledge and Future Perspectives. J. Clin. Med. 2021, 10, 2605. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Kakiyama, G.; Savidge, T.; Takei, H.; Kassam, Z.A.; Fagan, A.; Gavis, E.A.; Pandak, W.M.; Nittono, H.; Hylemon, P.B.; et al. Antibiotic-Associated Disruption of Microbiota Composition and Function in Cirrhosis Is Restored by Fecal Transplant. Hepatology 2018, 68, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Khoruts, A.; Sadowsky, M.J.; Hamilton, M.J. Development of Fecal Microbiota Transplantation Suitable for Mainstream Medicine. Clin. Gastroenterol. Hepatol. 2015, 13, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Grosu-Tudor, S.-S.; Stancu, M.-M.; Pelinescu, D.; Zamfir, M. Characterization of some bacteriocins produced by lactic acid bacteria isolated from fermented foods. World J. Microbiol. Biotechnol. 2014, 30, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- El-Nezami, H.S.; Polychronaki, N.N.; Ma, J.; Zhu, H.; Ling, W.; Salminen, E.K.; Juvonen, R.O.; Salminen, S.J.; Poussa, T.; Mykkänen, H.M. Probiotic supplementation reduces a biomarker for increased risk of liver cancer in young men from Southern China. Am. J. Clin. Nutr. 2006, 83, 1199–1203. [Google Scholar] [CrossRef]
- Hillman, J.D.; McDonell, E.; Hillman, C.H.; Zahradnik, R.T.; Soni, M.G. Safety assessment of ProBiora3, a probiotic mouthwash: Subchronic toxicity study in rats. Int. J. Toxicol. 2009, 28, 357–367. [Google Scholar] [CrossRef]
- Mbaye, B.; Borentain, P.; Magdy Wasfy, R.; Alou, M.T.; Armstrong, N.; Mottola, G.; Meddeb, L.; Ranque, S.; Gérolami, R.; Million, M.; et al. Endogenous Ethanol and Triglyceride Production by Gut Pichia kudriavzevii, Candida albicans and Candida glabrata Yeasts in Non-Alcoholic Steatohepatitis. Cells 2022, 11, 3390. [Google Scholar] [CrossRef]
- Million, M.; Armstrong, N.; Khelaifia, S.; Guilhot, E.; Richez, M.; Lagier, J.C.; Dubourg, G.; Chabriere, E.; Raoult, D. The Antioxidants Glutathione, Ascorbic Acid and Uric Acid Maintain Butyrate Production by Human Gut Clostridia in The Presence of Oxygen In Vitro. Sci. Rep. 2020, 10, 7705. [Google Scholar] [CrossRef] [PubMed]
- Borges, S.; Silva, J.; Teixeira, P. The Role of Lactobacilli and Probiotics in Maintaining Vaginal Health. Arch. Gynecol. Obstet. 2014, 289, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Mogami, O.; Matsueda, K. Characteristic of Milk Whey Culture with Propionibacterium Freudenreichii ET-3 and Its Application to the Inflammatory Bowel Disease Therapy. Inflammopharmacology 2007, 15, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Campaniello, D.; Bevilacqua, A.; Sinigaglia, M.; Altieri, C. Screening of Propionibacterium Spp. for Potential Probiotic Properties. Anaerobe 2015, 34, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Chanto, A.; Schmid-Meyer, A.C.; Schroeder, A.G.; Carvalho-Jonas, M.F.; Blanco, I.; Jonas, R. Effect of Oxygen Supply on Biomass, Organic Acids and Vitamin B12 Production by Propionibacterium Shermanii. World J. Microbiol. Biotechnol. 1998, 14, 843–846. [Google Scholar] [CrossRef]
- Chan, P.L.; Lauw, S.; Ma, K.L.; Kei, N.; Ma, K.L.; Wong, Y.O.; Lam, H.Y.; Ting, Y.Y.; Yau, T.K.; Nong, W.; et al. ProBioQuest: A Database and Semantic Analysis Engine for Literature, Clinical Trials and Patents Related to Probiotics. Database 2022, 2022, baac059. [Google Scholar] [CrossRef] [PubMed]
- Reents, R.; Dekkers, J.C.; Schaeffer, L.R. Genetic Evaluation for Somatic Cell Score with a Test Day Model for Multiple Lactations. J. Dairy Sci. 1995, 78, 2858–2870. [Google Scholar] [CrossRef]
- Kajander, K.; Hatakka, K.; Poussa, T.; Färkkilä, M.; Korpela, R. A Probiotic Mixture Alleviates Symptoms in Irritable Bowel Syndrome Patients: A Controlled 6-Month Intervention. Aliment. Pharmacol. Ther. 2005, 22, 387–394. [Google Scholar] [CrossRef]
- Myllyluoma, E.; Veijola, L.; Ahlroos, T.; Tynkkynen, S.; Kankuri, E.; Vapaatalo, H.; Rautelin, H.; Korpela, R. Probiotic Supplementation Improves Tolerance to Helicobacter Pylori Eradication Therapy--a Placebo-Controlled, Double-Blind Randomized Pilot Study. Aliment. Pharmacol. Ther. 2005, 21, 1263–1272. [Google Scholar] [CrossRef]
- Hatakka, K.; Holma, R.; El-Nezami, H.; Suomalainen, T.; Kuisma, M.; Saxelin, M.; Poussa, T.; Mykkänen, H.; Korpela, R. The Influence of Lactobacillus rhamnosus LC705 Together with Propionibacterium freudenreichii ssp. shermanii JS on Potentially Carcinogenic Bacterial Activity in Human Colon. Int. J. Food Microbiol. 2008, 128, 406–410. [Google Scholar] [CrossRef]
- Kukkonen, K.; Savilahti, E.; Haahtela, T.; Juntunen-Backman, K.; Korpela, R.; Poussa, T.; Tuure, T.; Kuitunen, M. Probiotics and Prebiotic Galacto-Oligosaccharides in the Prevention of Allergic Diseases: A Randomized, Double-Blind, Placebo-Controlled Trial. J. Allergy Clin. Immunol. 2007, 119, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wu, Z.; Xu, W.; Yang, J.; Chen, Y.; Li, L. Intestinal Microbiota Was Assessed in Cirrhotic Patients with Hepatitis B Virus Infection. Intestinal Microbiota of HBV Cirrhotic Patients. Microb. Ecol. 2011, 61, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.-D.; Peng, X.-B.; Zhao, R.-R.; Ma, C.-Q.; Li, J.-N.; Yao, L.-Q. The Intestinal Microbial Community Dissimilarity in Hepatitis B Virus-Related Liver Cirrhosis Patients with and without at Alcohol Consumption. Gut Pathog. 2019, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.; Chang, Y.; Kim, H.-N.; Ryu, S.; Kwon, M.-J.; Cho, Y.K.; Kim, H.-L.; Cheong, H.S.; Joo, E.-J. Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level. J. Clin. Med. 2019, 8, E173. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, B.; Fu, Y.; Chen, Y.; Yang, F.; Lu, H.; Chen, Y.; Xu, J.; Li, L. Changes of Fecal Bifidobacterium Species in Adult Patients with Hepatitis B Virus-Induced Chronic Liver Disease. Microb. Ecol. 2012, 63, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Zhang, X.; Liu, J.; Zhang, Q.; Zhao, Y.; Peng, J.; Feng, Q.; Dai, J.; Sun, S.; et al. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front. Microbiol. 2017, 8, 2222. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-W.; Lu, H.-F.; Wu, J.; Zuo, J.; Chen, P.; Sheng, J.-F.; Zheng, S.-S.; Li, L.-J. Assessment of the Fecal Lactobacilli Population in Patients with Hepatitis B Virus-Related Decompensated Cirrhosis and Hepatitis B Cirrhosis Treated with Liver Transplant. Microb. Ecol. 2012, 63, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, F.; Zhuang, Y.; Xu, J.; Wang, J.; Mao, X.; Zhang, Y.; Liu, X. Alteration in Gut Microbiota Associated with Hepatitis B and Non-Hepatitis Virus Related Hepatocellular Carcinoma. Gut Pathog. 2019, 11, 1. [Google Scholar] [CrossRef]
- Huang, H.; Ren, Z.; Gao, X.; Hu, X.; Zhou, Y.; Jiang, J.; Lu, H.; Yin, S.; Ji, J.; Zhou, L.; et al. Integrated Analysis of Microbiome and Host Transcriptome Reveals Correlations between Gut Microbiota and Clinical Outcomes in HBV-Related Hepatocellular Carcinoma. Genome Med. 2020, 12, 102. [Google Scholar] [CrossRef]
- Shen, Y.; Wu, S.-D.; Chen, Y.; Li, X.-Y.; Zhu, Q.; Nakayama, K.; Zhang, W.-Q.; Weng, C.-Z.; Zhang, J.; Wang, H.-K.; et al. Alterations in Gut Microbiome and Metabolomics in Chronic Hepatitis B Infection-Associated Liver Disease and Their Impact on Peripheral Immune Response. Gut Microbes 2023, 15, 2155018. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, J.; Liu, Y.; Zeng, Y.; Jiang, Z.; Yan, H.; Lin, J.; Zhou, W.; Ou, Q.; Ao, L. Identification Reproducible Microbiota Biomarkers for the Diagnosis of Cirrhosis and Hepatocellular Carcinoma. AMB Express 2023, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Shanjian, C.; Jinpiao, L.; Qishui, O. Gut Microbiota Dysbiosis in Patients with Hepatitis B Virus-Related Cirrhosis. Ann. Hepatol. 2022, 27, 100676. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhang, Q.; Shi, K.; Zhang, Y.; Zhu, B.; Bi, Y.; Wang, X. Gut Microbiota Dysbiosis with Hepatitis B Virus Liver Disease and Association with Immune Response. Front. Cell. Infect. Microbiol. 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Yu, H.; Fan, G.; Xiang, H.-P.; Long, L.; Xu, H.; Wu, Z.; Chen, M.; Xi, W.; Gao, Z.; et al. Impact of the Gut Microbiome on the Progression of Hepatitis B Virus Related Acute-on-Chronic Liver Failure. Front. Cell. Infect. Microbiol. 2021, 11, 573923. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yi, X.; Yang, J.; Zhu, Z.; Wang, Y.; Liu, X.; Huang, X.; Wan, Y.; Fu, X.; Shu, W.; et al. Gut Microbiome Signatures in the Progression of Hepatitis B Virus-Induced Liver Disease. Front. Microbiol. 2022, 13, 916061. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-J.; Su, T.-H.; Chen, C.-C.; Wu, W.-K.; Hsu, S.-J.; Tseng, T.-C.; Liao, S.-H.; Hong, C.-M.; Yang, H.-C.; Liu, C.-J.; et al. Diversity and Composition of Gut Microbiota in Healthy Individuals and Patients at Different Stages of Hepatitis B Virus-Related Liver Disease. Gut Pathog. 2023, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Thorat, V.; Kirdat, K.; Tiwarekar, B.; Dhanavade, P.; Karodi, P.; Shouche, Y.; Sathe, S.; Lodha, T.; Yadav, A. Paenibacillus albicereus sp. nov. and Niallia alba sp. nov., Isolated from Digestive Syrup. Arch. Microbiol. 2022, 204, 127. [Google Scholar] [CrossRef]
- Magdy Wasfy, R.; Zoaiter, M.; Bilen, M.; Tidjani Alou, M.; Lo, C.I.; Bellali, S.; Caputo, A.; Alibar, S.; Andrieu, C.; Raoult, D.; et al. Description of Agathobaculum massiliense sp. nov., a New Bacterial Species Prevalent in the Human Gut and Predicted to Produce Indole and Tryptophan Based on Genomic Analysis. Antonie Van Leeuwenhoek 2023, 116, 541–555. [Google Scholar] [CrossRef]
- Liu, C.; Du, M.-X.; Abuduaini, R.; Yu, H.-Y.; Li, D.-H.; Wang, Y.-J.; Zhou, N.; Jiang, M.-Z.; Niu, P.-X.; Han, S.-S.; et al. Enlightening the Taxonomy Darkness of Human Gut Microbiomes with a Cultured Biobank. Microbiome 2021, 9, 119. [Google Scholar] [CrossRef]
- Molinero, N.; Conti, E.; Sánchez, B.; Walker, A.W.; Margolles, A.; Duncan, S.H.; Delgado, S. Ruminococcoides bili gen. nov., sp. nov., a Bile-Resistant Bacterium from Human Bile with Autolytic Behavior. Int. J. Syst. Evol. Microbiol. 2021, 71, 004960. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magdy Wasfy, R.; Mbaye, B.; Borentain, P.; Tidjani Alou, M.; Murillo Ruiz, M.L.; Caputo, A.; Andrieu, C.; Armstrong, N.; Million, M.; Gerolami, R. Ethanol-Producing Enterocloster bolteae Is Enriched in Chronic Hepatitis B-Associated Gut Dysbiosis: A Case–Control Culturomics Study. Microorganisms 2023, 11, 2437. https://doi.org/10.3390/microorganisms11102437
Magdy Wasfy R, Mbaye B, Borentain P, Tidjani Alou M, Murillo Ruiz ML, Caputo A, Andrieu C, Armstrong N, Million M, Gerolami R. Ethanol-Producing Enterocloster bolteae Is Enriched in Chronic Hepatitis B-Associated Gut Dysbiosis: A Case–Control Culturomics Study. Microorganisms. 2023; 11(10):2437. https://doi.org/10.3390/microorganisms11102437
Chicago/Turabian StyleMagdy Wasfy, Reham, Babacar Mbaye, Patrick Borentain, Maryam Tidjani Alou, Maria Leticia Murillo Ruiz, Aurelia Caputo, Claudia Andrieu, Nicholas Armstrong, Matthieu Million, and Rene Gerolami. 2023. "Ethanol-Producing Enterocloster bolteae Is Enriched in Chronic Hepatitis B-Associated Gut Dysbiosis: A Case–Control Culturomics Study" Microorganisms 11, no. 10: 2437. https://doi.org/10.3390/microorganisms11102437




