Fast-Evolving Alignment Sites Are Highly Informative for Reconstructions of Deep Tree of Life Phylogenies
Abstract
:1. Introduction
2. Results
2.1. Slow-Evolving Sites Contain Inconsistent Phylogenetic Signals in Conserved Protein Datasets
2.2. Short-Branched Bipartitions Are Less Consistently Recovered from Slow-Evolving Sites
2.3. Slow-Evolving Sites in Simulated Alignments Are Less Likely to Recover True Tree Bipartitions
2.4. Slow-Evolving Sites in Simulated Sequence Datasets Are Biased against Reconstructing True Short-Branched Bipartitions
2.5. Substitution Saturation Does Not Explain the Loss of Phylogenetic Signal from Fast-Evolving Sites
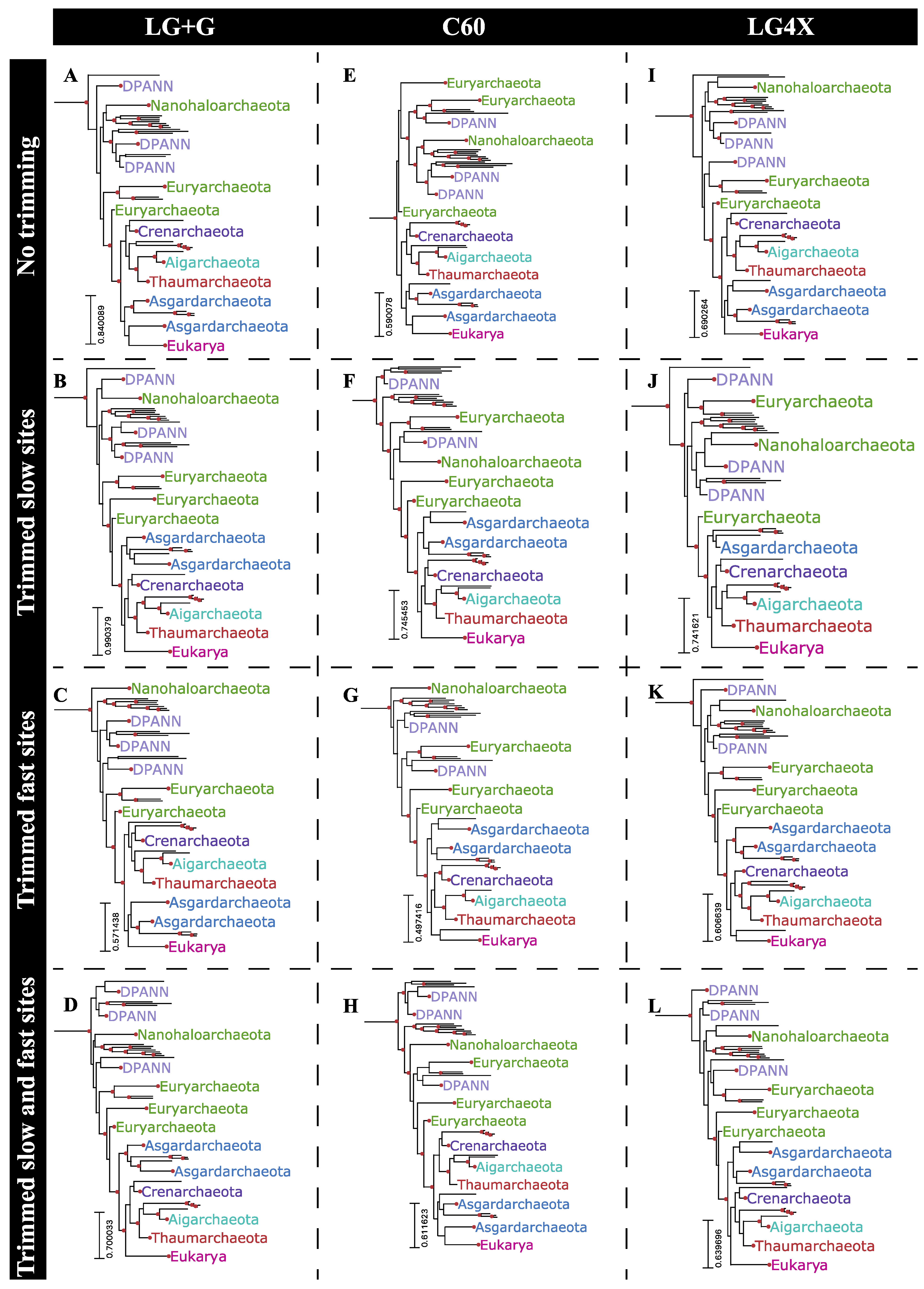
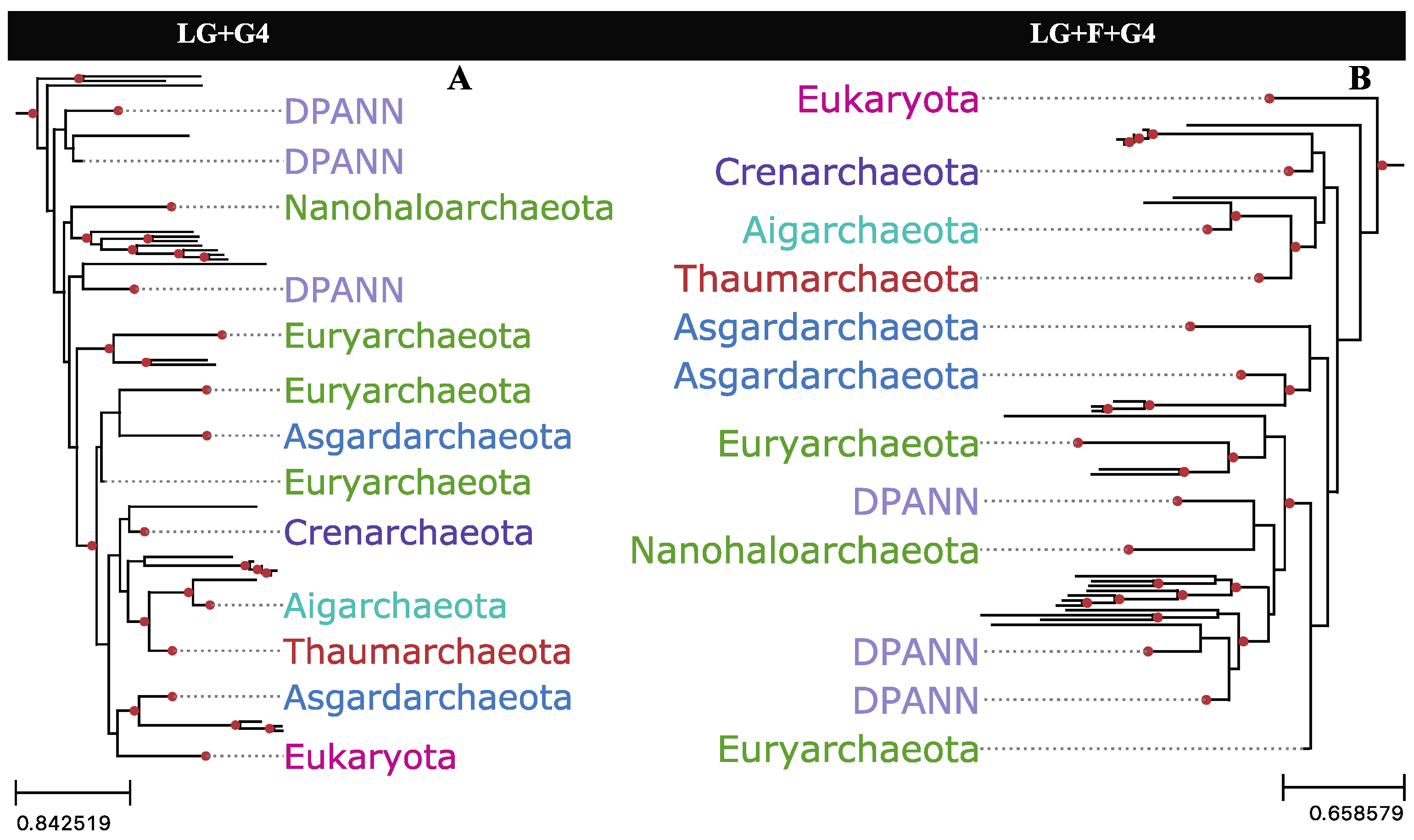
2.6. Phylogenetic Reconstructions Using Rate-Specific Subsets of Sequence Alignment Data
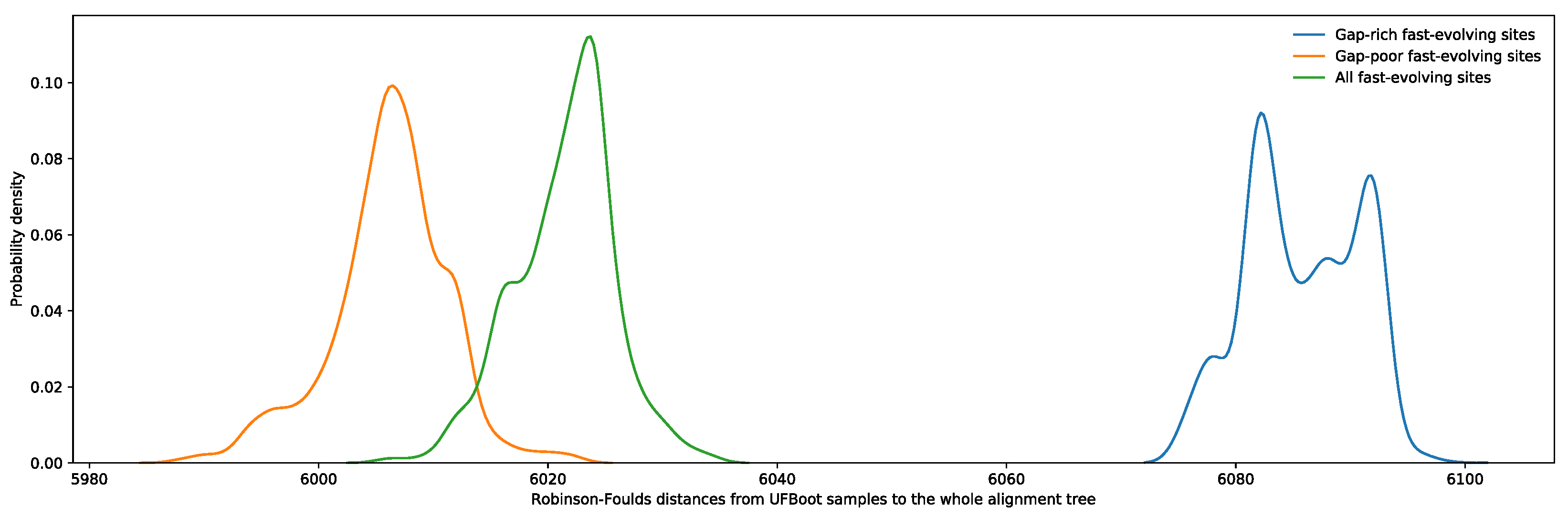
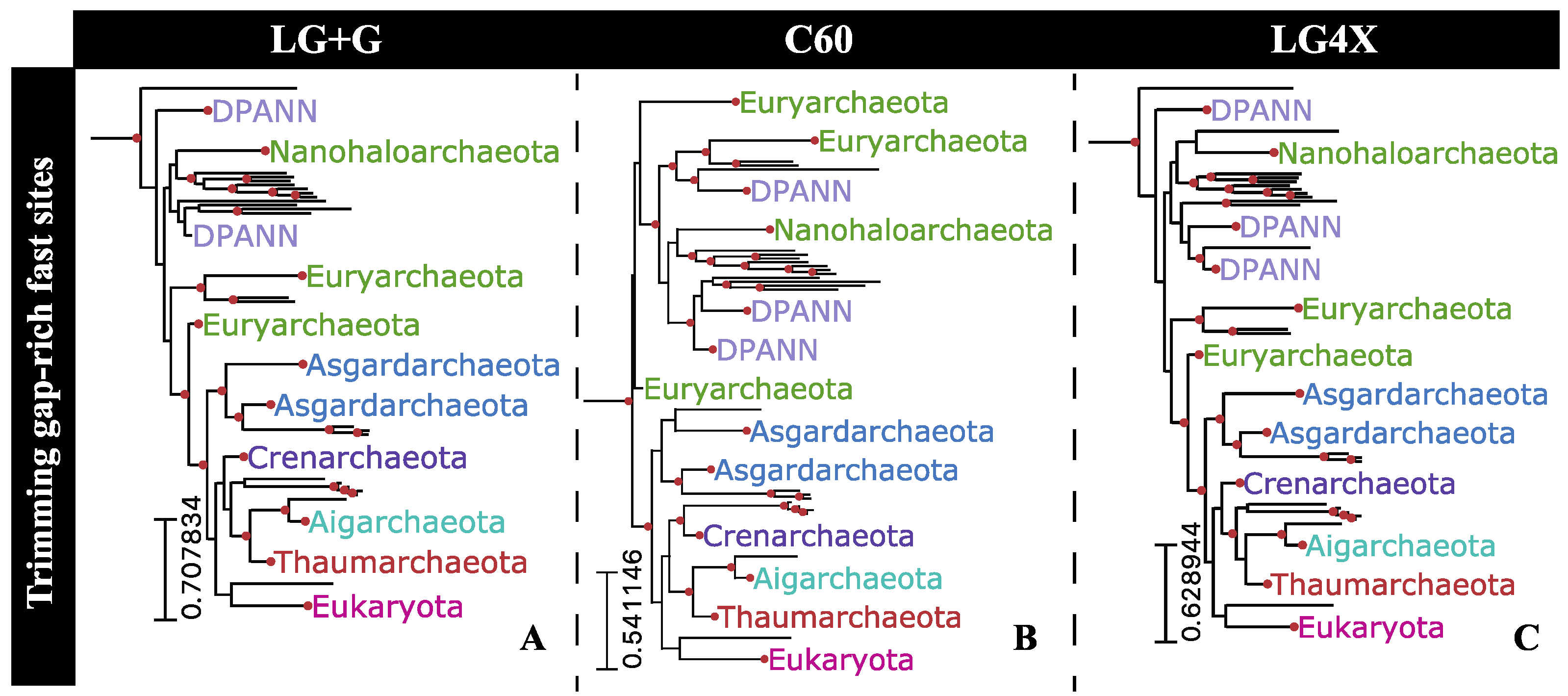
2.7. Phylogenetic Impact of Fast-Evolving Sites within Gap-Rich Regions
2.8. Composition Heterogeneity among Substitution Rate Categories
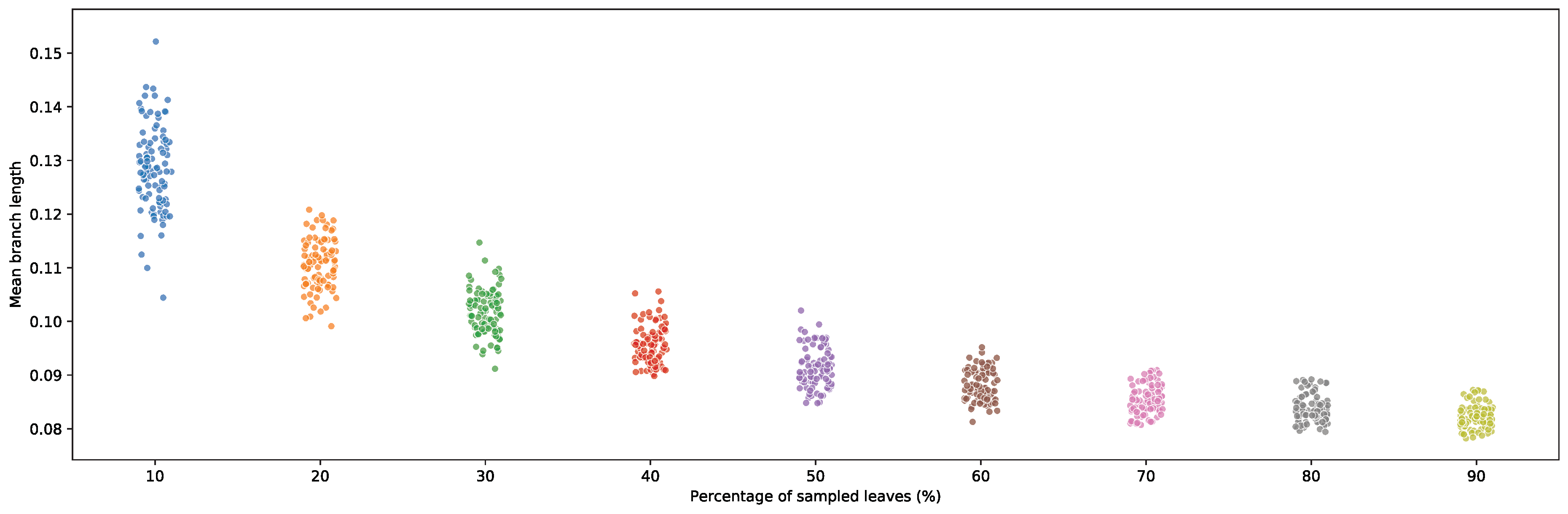
2.9. Short Deep Branches Increase in Frequency with Increased Taxon Sampling
3. Conclusions
4. Materials and Methods
4.1. Hug et al. [10] Dataset
4.2. Sequence Simulation
4.3. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Moreira, D.; Philippe, H. Molecular phylogeny: Pitfalls and progress. Int. Microbiol. 2000, 3, 9–16. [Google Scholar]
- Aouad, M.; Flandrois, J.-P.; Jauffrit, F.; Gouy, M.; Gribaldo, S.; Brochier-Armanet, C. A divide-and-conquer phylogenomic approach based on character supermatrices resolves early steps in the evolution of the Archaea. BMC Ecol. Evol. 2022, 22, 1. [Google Scholar] [CrossRef] [PubMed]
- Coleman, G.A.; Davín, A.A.; Mahendrarajah, T.A.; Szánthó, L.L.; Spang, A.; Hugenholtz, P.; Szöllősi, G.J.; Williams, T.A. A rooted phylogeny resolves early bacterial evolution. Science 2021, 372, eabe0511. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, H.; Philippe, H. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol. Biol. Evol. 1999, 16, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Brochier-Armanet, C.; Gribaldo, S. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl. Acad. Sci. USA 2015, 112, 6670–6675. [Google Scholar] [CrossRef]
- Philippe, H.; Sorhannus, U.; Baroin, A.; Perasso, R.; Gasse, F.; Adoutte, A. Comparison of molecular and paleontological data in diatoms suggests a major gap in the fossil record. J. Evol. Biol. 1994, 7, 247–265. [Google Scholar] [CrossRef]
- Kostka, M.; Uzlikova, M.; Cepicka, I.; Flegr, J. SlowFaster, a user-friendly program for slow-fast analysis and its application on phylogeny of Blastocystis. BMC Bioinform. 2008, 9, 4–9. [Google Scholar] [CrossRef]
- Yang, Z. On the best evolutionary rate for phylogenetic analysis. Syst. Biol. 1998, 47, 125–133. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 1–6. [Google Scholar] [CrossRef]
- Spang, A.; Saw, J.H.; Jørgensen, S.L.; Zaremba-Niedzwiedzka, K.; Martijn, J.; Lind, A.E.; van Eijk, R.; Schleper, C.; Guy, L.; Ettema, T.J.G. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015, 521, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Mayrose, I. Comparison of Site-Specific Rate-Inference Methods for Protein Sequences: Empirical Bayesian Methods Are Superior. Mol. Biol. Evol. 2004, 21, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.F.; Foulds, L.R. Comparison of phylogenetic trees. Math. Biosci. 1981, 53, 131–147. [Google Scholar] [CrossRef]
- Thorne, J.L.; Kishino, H. Freeing phylogenies from artifacts of alignment. Mol. Biol. Evol. 1992, 9, 1148–1162. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Selecting the Best-Fit Model of Nucleotide Substitution. Syst. Biol. 2001, 50, 580–601. Available online: http://www.ncbi.nlm.nih.gov/pubmed/12116655 (accessed on 1 August 2023). [CrossRef]
- Buckley, T.R.; Cunningham, C.W. The effects of nucleotide substitution model assumptions on estimates of nonparametric bootstrap support. Mol. Biol. Evol. 2002, 19, 394–405. [Google Scholar] [CrossRef]
- Hoff, M.; Orf, S.; Riehm, B.; Darriba, D.; Stamatakis, A. Does the choice of nucleotide substitution models matter topologically? BMC Bioinform. 2016, 17, 143. [Google Scholar] [CrossRef]
- Dayhoff, M.O.; Schwartz, R.M. Chapter 22: A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Silver Spring, MD, USA, 1978; pp. 345–352. [Google Scholar]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Da Cunha, V.; Gaia, M.; Gadelle, D.; Nasir, A.; Forterre, P. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 2017, 13, e1006810. [Google Scholar] [CrossRef]
- Gouy, R.; Baurain, D.; Philippe, H. Rooting the tree of life: The phylogenetic jury is still out. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140329. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Cases in which Parsimony or Compatibility Methods Will be Positively Misleading. Syst. Zool. 1978, 27, 401. [Google Scholar] [CrossRef]
- Sharp, P.M.; Emery, L.R.; Zeng, K. Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010, 365, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Hendy, M.D.; Penny, D. A Framework for the Quantitative Study of Evolutionary Trees. Syst. Zool. 1989, 38, 297. [Google Scholar] [CrossRef]
- Lax, G.; Eglit, Y.; Eme, L.; Bertrand, E.M.; Roger, A.J.; Simpson, A.G.B. Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes. Nature 2018, 564, 410–414. [Google Scholar] [CrossRef]
- Williams, T.A.; Szöllősi, G.J.; Spang, A.; Foster, P.G.; Heaps, S.E.; Boussau, B.; Ettema, T.J.G.; Embley, T.M. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl. Acad. Sci. USA 2017, 114, E4602–E4611. [Google Scholar] [CrossRef]
- Philippe, H.; Forterre, P. The rooting of the universal tree of life is not reliable. J. Mol. Evol. 1999, 49, 509–523. [Google Scholar] [CrossRef]
- Fletcher, W.; Yang, Z. INDELible: A flexible simulator of biological sequence evolution. Mol. Biol. Evol. 2009, 26, 1879–1888. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]



| RSAP | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average site-specific substitution rate | 0.023 | 0.089 | 0.175 | 0.278 | 0.4 | 0.545 | 0.721 | 0.937 | 1.214 | 1.595 | 2.194 | 3.824 |
| Sum of squared errors of aa compositional bias | 0.14 | 0.012 | 0.003 | 0.002 | 0.003 | 0.003 | 0.003 | 0.006 | 0.005 | 0.009 | 0.01 | 0.01 |
| Best-fit substitution model | WAG | LG | LG | LG+F | LG+F | LG+F | LG | LG | LG | WAG | WAG | WAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangel, L.T.; Fournier, G.P. Fast-Evolving Alignment Sites Are Highly Informative for Reconstructions of Deep Tree of Life Phylogenies. Microorganisms 2023, 11, 2499. https://doi.org/10.3390/microorganisms11102499
Rangel LT, Fournier GP. Fast-Evolving Alignment Sites Are Highly Informative for Reconstructions of Deep Tree of Life Phylogenies. Microorganisms. 2023; 11(10):2499. https://doi.org/10.3390/microorganisms11102499
Chicago/Turabian StyleRangel, L. Thibério, and Gregory P. Fournier. 2023. "Fast-Evolving Alignment Sites Are Highly Informative for Reconstructions of Deep Tree of Life Phylogenies" Microorganisms 11, no. 10: 2499. https://doi.org/10.3390/microorganisms11102499






