Abstract
Plants exposed to abiotic stress such as drought and salinity produce 1-aminocyclopropane-1-carboxylic acid (ACC) that is converted into the stress hormone ethylene. However, plant growth-promoting bacteria (PGPB), which synthesize the enzyme ACC deaminase, may lower the ACC concentration thereby reducing the concentration of ethylene and alleviating the abiotic stress. The PGPB Pseudomonas hormoni G20-18T (previously named P. fluorescens G20-18) harbors the genes acdR and acdS that encode regulation and synthesis of ACC deaminase, respectively. Regulation of the acdS gene has been investigated in several studies, but so far, it has been an open question whether plants can regulate microbial synthesis of ACC deaminase. In this study, small molecules in wheat root exudates were identified using untargeted metabolomics, and compounds belonging to amino acids, organic acids, and sugars were selected for evaluation of their influence on the expression of the acdS and acdR genes in P. hormoni G20-18T. acdS and acdR promoters were fused to the fluorescence reporter gene mCherry enabling the study of acdS and acdR promoter activity. In planta studies in wheat seedlings indicated an induced expression of acdS in association with the roots. Exudate molecules such as aspartate, alanine, arginine, and fumarate as well as glucose, fructose, and mannitol actively induced the acdS promoter, whereas the plant hormone indole-3-acetic acid (IAA) inhibited expression. Here, we present a model for how stimulatory and inhibitory root exudate molecules influence acdS promoter activity in P. hormoni G20-18T.
Keywords:
ACC deaminase; regulation; wheat root exudates; promoter fusion; Pseudomonas; metabolomics 1. Introduction
Ethylene acts as an important plant growth hormone that plays a vital role in breaking seed dormancy [1]. However, constant high levels of ethylene can also induce several detrimental effects with a predominant negative impact on root elongation and early senescence in plants [2,3]. Plant tissues use S-adenosyl methionine (SAM) to produce 1-aminocyclopropane-1-carboxylic acid (ACC) that serves as a precursor in the biosynthesis of ethylene. In addition to synthesis in developing seedlings, ACC is also increasingly formed in plants under stress. Plant-associated bacteria exhibit a unique capability to cleave this precursor molecule and thus to regulate the subsequent ethylene formation. This reaction is regulated by the enzyme ACC deaminase [4]. Therefore, being related to plant sustenance under stressful circumstances is considered one of the key determinants of microbial plant growth-promoting (PGP) characteristics and also a key factor regulating the microbe’s associative lifestyle, for instance in Rhizobia [5]. The presence of the ACC deaminase gene, acdS, has been documented in a variety of endo- and epiphytic bacteria including members of the Pseudomonas group [6,7]. ACC deamination appears to vary among microbial species. For example, in root nodules, acdS expression only accounted for 2–10% of the activity expressed by free-living microbes [8]. However, acdS function in symbionts is mainly focused on enhancing and maintaining nodulation [9] and may not necessarily participate in stress mitigation through lowering ethylene levels. Thus, although the levels of activity differ significantly with habitat and associative interactions with the host, the expression of ACC deamination among PGPR appears relatively evenly distributed.
In most organisms, acdS is located close to its regulator gene, acdR, that encodes a leucine-responsive regulator (Lrp), and this acdR–acdS gene cluster has been mapped to the chromosome in most Beta- and Gammaproteobacteria, whereas the cluster is located on plasmids in Alphaproteobacteria [10,11]. In silico analyses of the DNA region between the acdR and acdS genes in Pseudomonas putida UW4, Azospirillum lipoferum 4B, and Methylobacterium radiotolerans JCM 2831 showed the presence of putative binding sites for Lrp, cAMP receptor protein (CRP), and fumarate-nitrate reduction regulator (FNR) [8,12,13,14], and in vitro binding experiments showed that AcdR/Lrp indeed bound to the region between acdR and acdS [8,14]. In vivo experiments confirmed that the P. fluorescens UW4 acdS gene, when expressed recombinantly in Escherichia coli, was regulated by E. coli AcdR/Lrp, CRP, and FNR [12,13] and results obtained by Pringent-Combaret et al. [11] indicated that expression of A. lipoferum 4B acdS might be regulated by AcdR/Lrp and FNR but not CRP. The expression of acdS is highly dependent on oxygen availability, substate concentration, feedback from the reaction products, and also catabolite repression effects due to specific substrates [15]. The regulatory mechanisms of acdS are well-explained in the model organism P. putida UW4 by Li et al. [15].
However, from the plants’ perspective, both the expression as well as the regulatory mechanisms of acdS need further exploration in order to understand the highly intricate plant–microbial crosstalk and plant-regulatory influence on microbial gene expression, physiology, and metabolism. Since it is very likely that exudates from plant roots are involved in the regulation of the acdS and/or acdR genes, we analyzed root exudate molecules from wheat and investigated if some of the exudate molecules were involved in regulating acdS and/or acdR expression using promoter fusion technology with the fluorescence reporter mCherry.
2. Materials and Methods
2.1. Bacterial Strains and Growth Media
The P. hormoni G20-18T strain (previously named P. fluorescens G20-18; now described as a novel species, P. hormoni G20-18T, manuscript in press) used throughout this study was cultivated at 20–25 °C in Lysogenic Broth (LB) or minimal medium (M9). E. coli HST08 (StellarTM, Takara Bio Europe) was used in the construction of the acdS replacement mutant and the promoter fusion constructs. E. coli cells were cultivated in LB at 37 °C.
2.2. Construction of Promoter Fusions
Promoter fusion strains of P. hormoni G20-18T were constructed to investigate the characteristic influence of plant metabolites on acdS expression in detail. Mining the annotated genome sequence of strain G20-18 (GenBank accession no. CP075566) showed the presence of an acdS gene (locus tag KJF94_09105) and an acdR gene (locus tag KJF94_09100) (Supplementary Figure S1). The 200-base pair fragment comprising the acdS and acdR promoters was cloned into the reporter plasmid pSEVA237R [16] using In-Fusion cloning (Takara Bio Europe) with primers 5′-GCGGCCGCGCGAATTGTGGCTTCTGCACAATAAAAATATG-3′ and 5′-CGACTCTAGAGGATCGACTCTGCTCCTTGTTATTGG-3′ (Figure 1). Gene replacement in which the P. hormoni G20-18T acdS gene was replaced by the gentamycin resistance gene (ΔacsS) was carried out as described by Hennessy et al. [17] and Michelsen et al. [18] using the gene replacement vector pEX100T. The promoter fusion constructs and the acdS mutant, ΔacsS, were sequenced prior to conducting expression experiments. The activity of the acdR and acdS promoters in P. hormoni G20-18T was assayed by measuring fluorescence from the mCherry reporter gene using a fluorescence plate reader (CLARIOstar Plus–BMG Labtech, Ortenberg, Germany) as reported by Hennessy et al. [17].
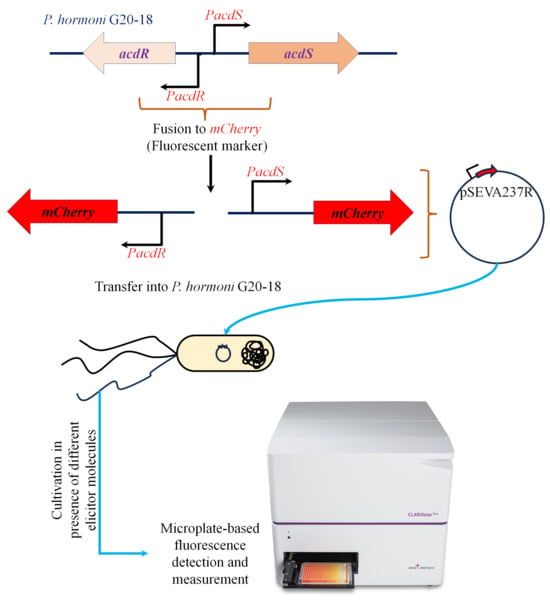
Figure 1.
Schematic representation of the workflow adapted to generate an mCherry-tagged acdS and acdR promoter fusion in P. hormoni G20-18T.
2.3. Production and Characterization of Wheat Root Exudates
Wheat root exudates were produced in hydroponic as well as in sterile soil conditions. The hydroponic growth was achieved by growing surface sterilized seeds (4% sodium hypochlorite for 8 min followed by 70% ethanol for 30 s) in sterile mass-spectrometry-grade water for 10 days. The water was changed every second day, and the final fraction at the 10th day was collected and enriched using solid-phase extraction as mentioned below and subjected to untargeted metabolomic analysis by high-resolution mass spectrometry.
The root exudate production in sterile soil was performed by growing surface sterilized seeds for 4 weeks under axenic conditions, followed by gently harvesting the roots. Excess soil on the root surface was removed by gentle shaking until a fine layer of ≤3 mm remained on the root surface. Although a soil cylinder with a maximum size of 4.0 mm around the roots is considered the rhizosphere [19], we considered a soil cylinder of ≤3 mm for the collection of the rhizosphere as the experimental plants were grown in pots in this study. The roots, along with the adhering rhizosphere soil layer were then immediately flash frozen in liquid nitrogen, transferred to −80 °C overnight, and freeze-dried at −100 °C under vacuum for 72 h. The rhizosphere soil from the dried roots was then collected by gentle tapping on the container. A one hundred-milligram fraction of the soil was then weighed and used for the extraction of root exudates. The solvent system for the extraction of root exudates was specifically standardized for polar to moderately polar compounds, to achieve optimal extraction of mobile root exudate moieties. Briefly, the solvent system for extraction contained 0.05% aqueous formic acid (A); 50% methanol in A (B); and 95% methanol in A (C). Each soil sample was sequentially extracted using each of the three extraction solvents at a proportion of 1:2 (w/v). The contents were vortex mixed at 3000 rpm for 15 min, followed by bath-ultrasonication for another 15 min, and the supernatants were collected. Finally, the solvent phase was isolated using high-speed centrifugation at 20,000× g for 20 min; the three aliquots were pooled and subjected to solid-phase extraction using 1cc HLB-30 mg extraction cartridges (Oasis, Waters–Ireland) and eluted with 1 mL methanol. The elutes were evaporated to dryness under nitrogen flow at room temperature, and the resultant pellet was dissolved in 1 mL 5% methanol. The extracts were then filtered using a Ø13 mm 0.22 μm PVDF syringe filter (Millex-Durapore, Merck, Kenilworth, NJ, USA). Ten microliters of each sample was injected into a UHPLC-Orbitrap-HRMS/MS platform (ThermoFisher Scientific, Waltham, MA, USA). The UHPLC (Ultimate3000, ThermoFisher Scientific) was fitted with a Kinetex biphenyl analytical column (2.1 × 100 mm, 2.6 µm, Phenomenex, Denmark) and operated with a biphasic acetonitrile/water gradient at a flow rate of 400 µL/minute. The mass spectrometer was operated in positive and negative electrospray ionization mode with data-dependent acquisition (Top3) mode with stepped collision (30 and 70 NCE). The data were processed using in-house Compound Discoverer and GNPS pipelines as described elsewhere [20]. The initial peak picking from the raw data into centroided form was performed using MSConvert (URL (1 August 2023), https://proteowizard.sourceforge.io/download.html). Features with annotated (confidence level 2) [21] molecular identities were pin picked for analyzing their influence on the expression of either acdR or acdS promoters. Similarly, the knowledge on root exudates from the available literature was also considered during the selection of test compounds that could influence the expression of acdR and acdS promoters.
2.4. Activity of acdR and acdS Promoters under the Influence of Root Exudate Analogues
The influence of synthetic root exudate analogues on the promoter expression was achieved using a growth system constructed in a 96-well microtiter plate design. The growth substrate contained 1x M9 medium with or without ammonium or glucose, (20% v/v stock mix of 0.25 M Na2HPO4, 0.11 M KH2PO4, and 0.0425 M NaCl; 2% v/v 1 M MgSO4; 0.01% v/v 1 M CaCl2; 10 or 25 mM test compounds; with or without 0.1% NH4Cl and 100 ppm of ACC). Furthermore, in order to determine the influence of the test compounds alone, phosphate-buffered saline (PBS) or aqueous systems were also used. Cells in the logarithmic growth phase were prepared by growing overnight in LB medium, pelleting and washing with sterile milliQ water, and resuspending in milliQ water in such a way that the final concentration of the inoculum in the test reaction medium was adjusted to approximately 0.05–0.07 OD600. Expression from the acdR and acdS promoters was then monitored in terms of rise in mCherry fluorescence (excitation at 570–15 nm; and emission at 620–20 nm), while cell growth was measured in terms of OD600. The measurements were recorded for 12–48 h at 3.75- or 30-min scan-cycles in a fluorescence plate reader (CLARIOstar Plus—BMG Labtech, Ortenberg, Germany) at 25 °C. The data were analyzed in terms of relative growth vs. fluorescence, and a ratio of fluorescence/growth dynamics.
2.5. Wheat Seedling in Planta Studies
In planta studies in wheat seedlings were carried out to determine the associative behavior of the strain P. hormoni G20-18T, and to study the dynamic expression of the acdS promoter during the associative lifestyle. Wheat seeds were surface sterilized as described above and subsequently treated with bacterial suspensions (~106 CFU − OD600 = 0.1) for 60 min. A Tn7 mCherry-tagged wild type P. hormoni G20-18T was provided by Prof. Thomas Roitsch (Dept. of Plant and Environmental Sciences, University of Copenhagen, Copenhagen, Denmark). The seeds were air-dried on sterile filter paper and cultivated in Petri dishes (n = 10 × 3) containing sterile filter paper beds. Seeds were allowed to germinate at 20 °C in the dark for 5 days, followed by light/dark exposure for 16/8 h for the next 5 days. The early roots were harvested from the cotyledon and 100 mg of fresh roots was washed with 5 mL of PBS containing 0.01% Tween-20 at 180 rpm for 30 min. The solution containing epiphytic cells was preserved and the roots were further washed with sterile milliQ water 5 times. The roots were then crushed in 5 mL of sterile milliQ water, the debris was allowed to settle, and the cleared solutions were read in a fluorescence reader as mentioned above.
2.6. Statistical Analysis
All the experiments were conducted in triplicate unless specified. ANOVA was applied using SPSS 16.0 (Windows 8.0, URL www.spss.com, (accessed on 1 August 2023)). The differences at the 95% confidence levels were considered significant.
3. Results and Discussion
The untargeted metabolomic data analysis of root exudates showed the presence of a variety of organic compounds belonging to different metabolic groups, viz., primary and secondary metabolites. The chemistry of the identified biomolecules predominantly highlighted the presence of important classes including low-molecular-weight organic acids, phenolics, amino acids, benzoxazinoides, plant hormones, etc. (cf. Table 1 and Supplementary Table S2). Many of the observed metabolites, e.g., amino acids and plant hormones, are already known for their diversified functioning within the rhizosphere microzone.

Table 1.
An overview of the biomolecules that could influence acdS expression.
In addition to the identified root exudate molecules in this study, we also included the root-secreted biomolecules already reported in the literature or the parent molecules (e.g., sugars) that were observed in modified forms in the root exudate composition [25,26,27] as well as their role in influencing acdS expression [28]. These molecules included amino acids and low-molecular-weight organic acids that mainly comprise primary metabolites and thus may also have a significant influence on the acdS and acdR promoters. Furthermore, known acdS regulatory compounds, such as ACC itself, α-ketobutyrate, leucine, and , were also included.
The analysis of the genome sequence of P. hormoni G20-18T revealed that the ACC deaminase gene, acdS, was co-localized with the Lrp-like regulator gene, acdR (Supplementary Figure S1). This gene organization has been reported before for a number of Proteobacteria, including Pseudomonas spp. [11]. The region between the acdR and acdS genes contains the promoters for the acdR and acdS genes including putative binding sites for regulatory molecules. This region was amplified by PCR and inserted into the reporter plasmid pSEVA237R [16]. The fragment was inserted in both orientations; in one orientation, the acdR promoter transcribed the mCherry gene, and in the opposite orientation, the mCherry gene was transcribed from the acdS promoter. The mCherry promoter fusion plasmids were transformed into wild-type P. hormoni G20-18T (acdS+) cells and in the replacement mutant, ΔacdS. However, since wild-type cells with active ACC deaminase reduce the concentration of ACC with the concomitant production of and α-ketobutyrate, experiments with wild-type cells and ACC would complicate the picture when long incubations were conducted (data not shown); thus, we used ΔacdS for the subsequent experiments. Both acdR and acdS promoter fusions were included in the study. However, as the acdR promoter was shown to be expressed constitutively, whereas the acdS promoter was regulated by a number of molecules, only the results from the acdS promoter fusion are shown below.
3.1. Influence of Nitrogenous Compounds on acdS Promoter Activity
Addition of ACC alone in M9 medium (no C or N supplements) confirmed earlier reports, which showed that ACC induced the acdS promoter (Supplementary Figure S2) [4,13,29,30]. The end products of the ACC deaminase reaction, α-ketobutyrate and ammonium, also influenced acdS promoter activity. Figure 2A shows that the induction of the acdS promoter by 100 ppm ACC was similar in experiments with or without the addition of 0.1% or . When 25 mM α-ketobutyrate and 100 ppm ACC were added, the promoter activity increased by a factor of 3–4, but this stimulation was lowered to approximately half if (or ) was added to the reaction (Figure 2B). This interaction between ammonium and α-ketobutyrate was also observed when glucose was added or if nitrate was substituted for ammonium (Figure 2C,D). This phenomenon underscores the interaction of ammonium nitrogen in regulating acdS activity, and also links the presence of acdS sequences to the Nif regulatory regions in symbiotic bacteria [2]. However, increased expression due to amino acids and ACC over that with the ACC alone without any nitrogen source (Figure 3) points towards the probable influence of the nitrogen component from the amino acids.
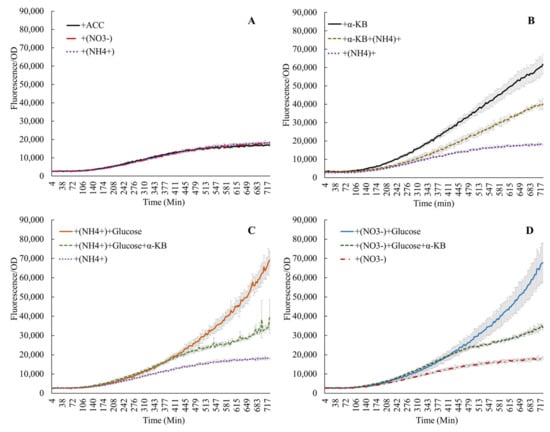
Figure 2.
Activity of the acdS promoter in the presence of nitrate, ammonium, or ACC (A); α-ketobutyrate (α-KB) lowers the expression only in the presence of nitrogen (B), while the induced expression due to glucose is lowered in the presence of both the reaction products of ACC deaminase (C,D).
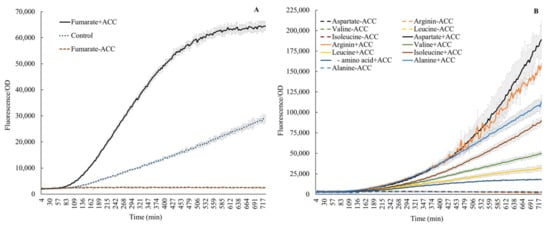
Figure 3.
Effect on acdS promoter activity by fumarate (A) and amino acids (B), with or without ACC. Control (A): without fumarate, but with ACC.
3.2. Influence of Amino Acids on acdS Promoter Activity
A significant amount of carbon is involved in amino acid fluxes in wheat. For instance, Phillips et al. [22] described fluxes of 16 amino acids from wheat and other plants under axenic and microbially associated conditions. The authors reported methionine as the lowest (4 nmol g−1 root h−1) and alanine as the highest efflux molecule (60 nmol g−1 root h−1). We also noticed the presence of amino acids in root exudates in a qualitative form (Table 1). Thus, the abundance of amino acids in root exudates generated an interest in evaluating their influence on acdS promoter activity. The expression of acdS dramatically changed following the addition of amino acids (Figure 3). As anticipated, no expression was observed in the absence of ACC. Previous publications have pointed to leucine as an inhibitor [7,11,12,13]. However, here, we show that the addition of 25 mM leucine and 100 ppm ACC to growing cells increased the acdS promoter activity by 78%, when compared to cells only induced with ACC. A similar, small stimulation was also observed by Honma [28], who reported that ACC deaminase activity in cells without leucine was 0.22 × 10−3 units/mg but if leucine was supplemented, the activity increased to 0.26 × 10−3 units/mg. Furthermore, Figure 3 shows that the addition of either fumarate, alanine, valine, arginine, or isoleucine further increased the acdS promoter activity. Honma [28] also reported that the specific activity of ACC deaminase in cells increased upon addition of valine. The conflicting results here, when compared to the literature, could be due to different concentrations of leucine used [11,13] and the fact that some of the previous results were based on in vitro gel retardation experiments [7]. However, the action of Lrp regulators may be very complex. Recent reports [31,32,33] describe how leucine may act as an inducer or inhibitor. In a recent review on Lrp, Ziegler and Freddolino describe that leucine acts on E. coli Lrp by causing a shift from a hexadecamer (16 mer) to an octamer (8 mer) if leucine is added [31]. Furthermore, they conclude that with respect to DNA binding, the general consensus in the field is that leucine increases the cooperative binding of Lrp to DNA, but overall, it reduces the affinity of Lrp to DNA. Finally, they point out that all in vitro assays on Lrp required anywhere from 1 to 10 mM leucine to elicit an effect on Lrp, whereas the typical concentration of leucine in cells is on the order of 0.1 mM. Furthermore, other amino acids, such as aspartate and arginine as demonstrated earlier in wheat root exudates by Phillips et al. [22], also stimulate acdS promoter activity (Figure 3). Prigent-Combaret et al. also reported that the addition of arginine increased the acdS mRNA levels in A. lipoferum 4B when measured by semiquantitative RT-PCR [11].
l-amino acid isomers without ACC did not induce the acdS promoter, aligning with the available knowledge [34,35]. However, the characteristic response of acdS observed in this study in the presence of ACC plus l-amino acids indicates a yet-unknown mechanism, especially synergistic, or antagonistic (probably dependent on interactions of structural confirmations of amino acids –, e.g., branched chain, hydrophobic groups, etc.) that could either up or down regulate expression (Figure 3B). Further, fumarate, alanine, and aspartic acid induced expression here (Figure 3A,B); both alanine and aspartic acid are intermediates in the same biosynthetic branch, where fumarate acts as a precursor [36]. Thus, fumarate, aspartate, and alanine together indicate a clear positive influence on the cellular biochemistry of acdS. Additionally, the response in the presence of fumarate also underscores the involvement of FNR activity in acdS regulation. The phenomenon also aligns with the higher acdS expression seen in endophytic environments (Figures 6 and 7). As FNR proteins are major contributors to the oxygen response, particularly for switching from an aerobic to anaerobic mode of metabolism [37], their involvement in adaptation to the endophytic lifestyle and acdS regulation highlights that the acdS response in an endophytic habitat is tightly regulated and may not be solely dependent on the ACC abundance in cellular environments.
3.3. Influence of Sugars on acdS Promoter Activity
Following the evaluation of amino acids, optimization of the system to investigate the influence of individual sugar moieties was carried out. Although we could not detect sugars in native form within the wheat root exudates, evidence from the available literature indicates a dominance of sugars in root-secreted carbon in plants such as maize [38]. Therefore, to generate a wider context for microbial acdS interactions in general, we also included representative sugars in this study. A combination of glucose, ammonium, nitrate, and α-ketobutyrate was tested in presence of 100 ppm ACC. Glucose without N sources but with ACC induced the activity of the acdS promoter, while inclusion of either of the N sources lowered the expression (Figure 2C,D). The addition of α-ketobutyrate indeed slightly enhanced the expression, indicating that the presence of nitrogen has an influence on the acdS promoter. Sugars other than glucose influenced acdS promoter activity. Figure 4 shows that the other sugars could also positively induce the acdS promoter. Within the first 11–12 h (Figure 4A), glucose induced the promoter the most, but after 24 h of incubation, fructose was the dominant inducer. The observation that fructose is a better inducer when compared to glucose has also been made by [11] in A. lipoferum 4B. Mannitol was almost as good an inducer as glucose, whereas l-rhamnose only displayed a small stimulation compared to the no sugar addition. Sucrose, however, in the present study, showed no induction although it was shown earlier to be present in the root exudates [38].
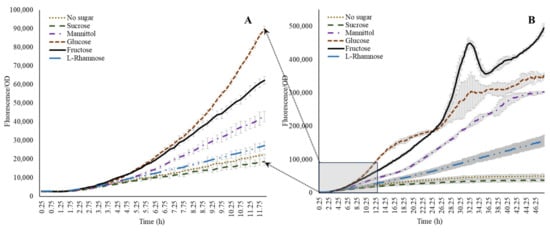
Figure 4.
Effect of sugars on acdS promoter expression. Note that glucose dominates during initial growth (A), while the induction is taken over by fructose during the later growth stages (B).
3.4. Influence of Indole-3-Acetic Acid (IAA) on acdS Promoter Activity
The major goal of this study is to study acdS expression from the plants’ perspective and thus, the measurements typically focus on biomolecules originating from the plant root exudations. Therefore, considering the rhizosphere microzone as a general habitat of the microbe, we used very low concentrations of the test molecules (10–25 mM) over the reported concentrations (e.g., 1% as reported by Honma [28]. Other molecules that microorganisms may encounter in the rhizosphere are plant hormones such as IAA. Here, we show that 1500 ppm IAA inhibited acdS promoter activity (Figure 5).
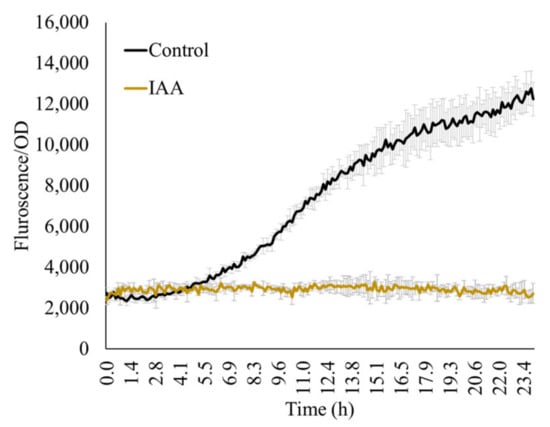
Figure 5.
IAA (1500 ppm) in the presence of ACC (100 ppm) inhibits expression from the acdS promoter. Control: 100 ppm ACC without IAA.
3.5. acdS Promoter Activity in Epiphytic vs. Endophytic Cells
Another important aspect to be considered in the acdS expression dynamics is linking the current results to the lifestyle of P. hormoni G20-18T. This bacterium was isolated from an arctic grass [39], and although there is significant knowledge available regarding beneficial interactions of P. hormoni G20-18T in crop plants [40,41], its associative behavior in wheat is not known. Here, we show that P. hormoni G20-18T can adopt both epiphytic and endophytic lifestyles. Figure 6A shows that P. hormoni G20-18T tagged with Tn7 mCherry could be retrieved both from the root surface and from the endophytic compartment. Similarly, fluorescent mCherry reporter proteins were observed in the epiphytic and endophytic compartments, respectively, when acdS promoter fusion cells were inoculated on wheat roots. The mCherry fluorescence from the Tn7-tagged constructs and from acdS promoter fusions was higher in epiphytic extracts compared to endophytic extracts, indicating that more cells were attached to the root surface than present in the endophytic compartment. However, as the ratio of acdS promoter fluorescence/Tn7-tagged fluorescence was higher in endophytic extracts than in epiphytic extracts, this could indicate that the acdS promoter was more active inside the plant root cells than on the outside (Figure 6B). This observation is in line with previous reports that showed higher acdS activity in cells cultivated in oxygen-limited conditions [11,12]. Furthermore, the results also align with the observations reported by [5] during Sinorhizobium meliloti colonization in the root zone of Medicago sativa. However, unlike Rhizobia, the range of activity of acdS in the case of the P. hormoni G20-18T might not be limited to establishment and maintenance of association, whereas the induced expression indicates active channelizing of ACC even from the epiphytic zone that could balance the internal ACC levels in the roots [8]. This phenomenon provides important links to the previous report mentioning that P. hormoni G20-18T mediated drought tolerance in tomato [41]. The results therefore align with the model of microbe-mediated deamination of ACC in plants that was proposed by Glick et al. [42], who mentioned that seeds and/or roots leak significant quantities of ACC that is utilized by rhizosphere microbes.
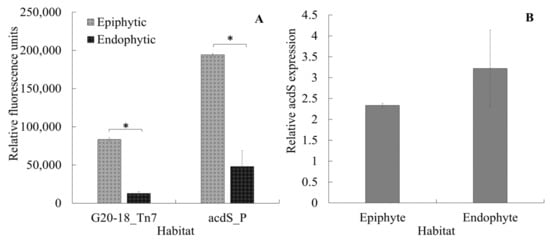
Figure 6.
Colonization of the mCherry-tagged acdS+ P. hormoni G20-18T strain on and in the wheat seedling roots (A-G20-18_Tn7). acdS promoter expression in promoter fusion strain (A) and relative expression of the acdS promoter (acdS promotor fluorescence/Tn7 tagged fluorescence) in epi- and endophytic environments (B). * Indicates that the differences are statistically significant at p ≤ 0.05.
3.6. Model for Regulation of acdS Promoter Activity in P. hormoni G20-18T
The results presented here led us to propose the following hypothesis for the interactions between P. hormoni G20-18T and wheat roots (Figure 7). In the root under normal growth conditions without abiotic stress, Figure 7A, there exists an intricate balance between ethylene and IAA biosynthesis and subsequent localization of IAA to the elongation zone. During stress (Figure 7B), induction of ethylene leads to ethylene-responsive cascades that induce auxin transport proteins and induce localization of IAA within the root elongation zone, causing ethylene-induced inhibition of root elongation. However, in stressed plant roots growing in the presence of P. hormoni G20-18T (Figure 7C), ACC produced by the plant will be channeled into the ACC deaminase-producing bacterial cells where ACC is converted to ammonia and α-ketobutyrate. Furthermore, root exudate molecules such as sugars (e.g., fructose, glucose, and mannitol) and amino acids (e.g., aspartate, alanine, and arginine) may further stimulate this conversion of ACC, and in the endophytic compartment, fumarate may further induce synthesis of ACC deaminase. Thus, the lower ACC concentration will result in lower ethylene-induced inhibition of root elongation. However, this will only work if ACC deaminase-producing bacteria are present on or in the root before the onset of the abiotic stress. If the bacteria associate with stressed plant roots, it is likely that the high IAA concentrations might induce an inhibitory influence on the production of ACC deaminase and no bacterium-induced lowering of the ethylene concentration will occur (Figure 7D). Overall, the plant IAA could exhibit a higher influence due to its ethylene-dependent specific localization in the roots that determines root growth and development. Although a general concentration of IAA ranges on the order of 250 pM g−1 of wheat roots on a fresh weight basis [43], the localization could be highly site-specific along the roots and might lead to the development of IAA-rich zones. Moreover, under a natural scenario, an association of high-IAA-producing microbes could further augment the local IAA concentration at the site of root colonization; thus, the phenomenon depicted in Figure 7C may occur along the root regions with IAA hyperactivity. Further, within the rhizosphere and endophytic habitat, acdS expression occurs in the presence of a complex mixture of biomolecules that could influence acdS activity in either a positive or negative manner. Therefore, net acdS activity under such a highly complex chemical scenario still remains unknown.
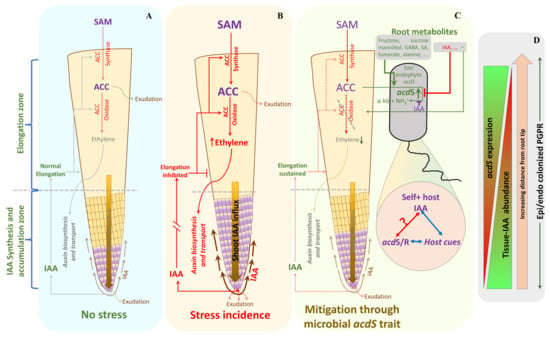
Figure 7.
A model of microbial acdS interactions with the host plant under abiotic stress conditions. Although IAA has an inhibitory effect on the acdS promoter, the regulation through self-IAA originating from the microbial cells still needs to be investigated. (SAM: S’-adenosyl methionine).
The influence of IAA in the strains with both the IAA+ and acdS+ traits remains an open question. In the case of IAA+, acdS+ microbial strains, the interactions might involve even more intricate molecular cascades. However, microbial IAA, being a secondary metabolite [44], could exhibit higher concentrations during the late growth phases extending from the late-log to stationary stage. Meanwhile, acdS expression seems more dependent on the induction by ACC, and could continue to be expressed even during the log phase of growth. Furthermore, as evident from the fumarate responses (Figure 3A), during the adaptation to an endophytic lifestyle, the process could probably trigger low oxygen-induced metabolic rearrangements through FNR proteins that also seem involved in acdS regulation. Therefore, the results strongly endorse the need to thoroughly investigate the acdS trait in P. hormoni G20-18T, and microbial acdS trait in general in the context of rhizosphere and/or endophytic microhabitats where both the inducers and inhibitors are found together in a cocktail of metabolites.
4. Conclusions
The results show that the acdS promoter activity in P. hormoni G20-18T is dynamic and can be regulated through a range of commonly occurring biomolecules, particularly those belonging to the sugars, amino acids, and phenolics including plant hormones. However, in all cases, ACC is required as a primary inducer of acdS; thereafter, the expression is further modulated in the presence of different biomolecules. Principally, the associative behavior and metabolite chemistry exert a significant influence on the acdS promoter, and hence, can be regarded as key determinants of microbial acdS traits under natural conditions. Furthermore, a decline in expression in the presence of the reaction product—particularly ammonia—indicates the need for detailed investigations in an agricultural scenario where N inputs are frequent and can cause a higher abundance of ammonia nitrogen. The aligning trends of acdS in the presence of ammonia and nitrate also highlight the possible involvement of cellular nitrogen fluxes in acdS regulation. The results also underscore the need to investigate the interactions in planta under drought conditions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11102504/s1. Supplementary Figure S1: acdR and acdS regions in Pseudomonas hormoni G20-18T with flanking genes as aligned with acdR and acdS regions in other PGPR strains. Supplementary Figure S2: Effect of ACC alone in M9 medium without glucose on the induction of acdS expression in Pseudomonas hormoni G20-18T with deleted acdS activity. Supplementary Table S1: Wheat root exudate diversity as identified using LC-MS/MS.
Author Contributions
Conceptualization, methodology, software, validation, formal analysis, and investigation, A.M.S., F.N. and P.S.; LC-MS/MS experiments: M.H.; writing—original draft preparation, project administration, and funding acquisition, A.M.S. and P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Novo Nordisk Foundation (NNF19SA0059360) and the European Commission H2020 MSCA-IF program (RhizoEng project, GAN 101028448). M.H. acknowledges the funding support from the Carlsberg Foundation (CF20-0422) and the Aarhus University Research Foundation (AUFF-T-2017-FLS-7-4).
Data Availability Statement
The genome sequence of P. hormoni G20-18T (P. fluorescens G20-18) had been determined before GenBank accession no. CP075566. The acdS gene (locus tag KJF94_09105) and acdR gene (locus tag KJF94_09100) sequences are derived from the genome sequence.
Acknowledgments
Thomas Roitsch, Department of Plant and Environmental Sciences, University of Copenhagen, Denmark, is thanked for the Tn7 mCherry-tagged P. hormoni G20-18T strain.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Esashi, Y. Ethylene and seed germination. In The Plant Hormone Ethylene; Matoo, A.K., Suttle, J.C., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 133–157. [Google Scholar]
- Ma, W.; Guinel, F.C.; Glick, B.R. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 2003, 69, 4396–4402. [Google Scholar] [CrossRef]
- Jackson, M.B. Ethylene in root growth and development. In The Plant Hormone Ethylene; Matoo, A.K., Suttle, J.C., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 159–181. [Google Scholar]
- Grichko, V.P.; Glick, B.R. Amelioration of flooding stress by ACC deaminase-containing plant growth-promoting bacteria. Plant Physiol. Biochem. 2001, 39, 11–17. [Google Scholar] [CrossRef]
- Checcucci, A.; Azzarello, E.; Bazzicalupo, M.; De Carlo, A.; Emiliani, G.; Mancuso, S.; Spini, G.; Viti, C.; Mengoni, A. Role and Regulation of ACC Deaminase Gene in Sinorhizobium meliloti: Is It a Symbiotic, Rhizospheric or Endophytic Gene? Front. Genet. 2017, 8, 6. [Google Scholar] [CrossRef]
- Glick, B.R.; Jacobson, C.B.; Schwarze, M.M.K.; Pasternak, J.J. 1-Aminocyclopropane-1-carboxylic acid deaminase mutants of the plant growth promoting rhizobacterium Pseudomonas putida GR12-2 do not stimulate canola root elongation. Can. J. Microbiol. 1994, 40, 911–915. [Google Scholar] [CrossRef]
- Cheng, Z.; Duncker, B.P.; McConkey, B.J.; Glick, B.R. Transcriptional regulation of ACC deaminase gene expression in Pseudomonas putida UW4. Can. J. Microbiol. 2008, 54, 128–136. [Google Scholar] [CrossRef]
- Singh, R.P.; Shelke, G.M.; Kumar, A.; Jha, P.N. Biochemistry and genetics of ACC deaminase: A weapon to “stress ethylene” produced in plants. Front. Microbiol. 2015, 6, 937. [Google Scholar] [CrossRef]
- Uchiumi, T.; Ohwada, T.; Itakura, M.; Mitsui, H.; Nukui, N.; Dawadi, P.; Kaneko, T.; Tabata, S.; Yokoyama, T.; Tejima, K.; et al. Expression islands clustered on symbiosis island of Mesorhizobium loti genome. J. Bacteriol. 2004, 186, 2439–2448. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Rossi, M.J.; Soares, C.R.; McConkey, B.J.; Glick, B.R. New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE 2014, 9, e99168. [Google Scholar] [CrossRef] [PubMed]
- Prigent-Combaret, C.; Blaha, D.; Pothier, J.F.; Vial, L.; Poirier, M.A.; Wisniewski-Dyé, F.; Moënne-Loccoz, Y. Physical organization and phylogenetic analysis of acdR as leucine-responsive regulator of the 1-aminocyclopropane-1-carboxylate deaminase gene acdS in phytobeneficial Azospirillum lipoferum 4B and other Proteobacteria. FEMS Microbiol. Ecol. 2008, 65, 202–219. [Google Scholar] [CrossRef]
- Grichko, V.P.; Glick, B.R. Identification of DNA sequences that regulate the expression of the Enterobacter cloacae UW4 1-aminocyclopropane-1-carboxylic acid deaminase gene. Can. J. Microbiol. 2000, 46, 1159–1165. [Google Scholar] [CrossRef]
- Li, J.; Glick, B.R. Transcriptional regulation of the Enterobacter cloaceae UW41-aminocyclopropane-1-carboxylate (ACC) deaminase gene (AcdS). Can. J. Microbiol. 2001, 47, 259–267. [Google Scholar] [CrossRef]
- Ekimova, G.A.; Fedorov, D.N.; Doronina, N.V.; Khmelenina, V.N.; Mustakhimov, I.I. AcdR protein is an activator of transcription of 1-aminocyclopropane-1-carboxylate deaminase in Methylobacterium radiotolerans JCM 2831. Antonie Van Leeuwenhoek 2022, 115, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ovakim, D.H.; Charles, T.C.; Glick, B.R. An ACC deaminase minus mutant of Enterobacter cloacae UW4 no longer promotes root elongation. Curr. Microbiol. 2000, 41, 101–105. [Google Scholar] [CrossRef]
- Silva-Rocha, R.; Martínez-García, E.; Calles, B.; Chavarría, M.; Arce-Rodríguez, A.; de Las Heras, A.; Páez-Espino, A.D.; Durante-Rodríguez, G.; Kim, J. The Standard European Vector Architecture (SEVA): A coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, R.C.; Phippen, C.B.W.; Nielsen, K.F.; Olsson, S.; Stougaard, P. Biosynthesis of the antimicrobial cyclic lipopeptides nunamycin and nunapeptin by Pseudomonas fluorescens strain In5 is regulated by the LuxR-type transcriptional regulator NunF. Microbiol. Open 2017, 6, e00516. [Google Scholar] [CrossRef]
- Michelsen, C.F.; Watrous, J.; Glaring, M.A.; Kersten, R.; Koyama, N.; Dorrestein, P.C.; Stougaard, P. Nonribosomal peptides, key biocontrol components for Pseudomonas fluorescens In5, isolated from a Greenlandic suppressive soil. mBio 2015, 6, e00079. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Razavi, B.S. Rhizosphere Size and Shape: Temporal Dynamics and Spatial Stationarity. Soil Biol. Biochem. 2019, 135, 343–360. [Google Scholar] [CrossRef]
- Jensen, M.; Poulsen, R.; Langebæk, R.; Jenssen, B.M.; Moe, J.; Ciesielski, T.M.; Dietz, R.; Sonne, C.; Madsen, J.; Hansen, M. The metabolome of pink-footed goose: Heavy metals and lipid metabolism. Environ. Res. 2023, 231 Pt 1, 116043. [Google Scholar] [CrossRef]
- Viant, M.R.; Ebbels, T.M.D.; Beger, R.D.; Ekman, D.R.; Epps, D.J.T.; Kamp, H.; Leonards, P.E.G.; Loizou, G.D.; MacRae, J.I.; van Ravenzwaay, B.; et al. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat. Commun. 2019, 10, 3041. [Google Scholar] [CrossRef]
- Phillips, D.A.; Fox, T.C.; King, M.D.; Bhuvaneswari, T.V.; Teuber, L.R. Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 2004, 136, 2887–2894. [Google Scholar] [CrossRef]
- Jalali, B.L.; Suryanarayana, D. Shift in the carbohydrate spectrum of root exudates of wheat in relation to its root-rot disease. Plant Soil. 1971, 34, 261–267. [Google Scholar] [CrossRef]
- Xiao, J.X.; Zheng, Y.; Tang, L. Effect of wheat and faba bean intercropping on root exudation of low molecular weight organic acids. J. Appl. Ecol. 2014, 25, 1739–1744. [Google Scholar]
- Zhang, A.-H.; Ma, W.-L.; Lei, F.-J.; An, N.-B.; Zhang, X.-X.; Liu, Z.-Q.; Zhang, L.-X. Research on chemotaxis response of Alternaria panax to amino acid of ginseng root exudates. China J. Chin. Mater. Medica 2017, 42, 2052–2057. [Google Scholar] [CrossRef]
- Li, J.; Lin, S.; Zhang, Q.; Zhang, Q.; Hu, W.; He, H. Fine-Root Traits of Allelopathic Rice at the Seedling Stage and Their Relationship with Allelopathic Potential. PeerJ 2019, 7, e7006. [Google Scholar] [CrossRef] [PubMed]
- Sebastiana, M.; Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.; Monteiro, F.; Figueiredo, A.; Maia, M.; Nascimento, R.; Silva, M.S.; Ferreira, A.N.; et al. Metabolomics and Transcriptomics to Decipher Molecular Mechanisms Underlying Ectomycorrhizal Root Colonization of an Oak Tree. Sci. Rep. 2021, 11, 8576. [Google Scholar] [CrossRef]
- Honma, M. Enzymatic determination of 1-aminocyclopropane-1-carboxylate deaminase. Agric. Biol. Chem. 1983, 47, 617–618. [Google Scholar]
- Viterbo, A.; Landau, U.; Kim, S.; Chernin, L.; Chet, I. Characterization of ACC deaminase from the biocontrol and plant growth-promoting agent Trichoderma asperellum T203. FEMS Microbiol. Lett. 2010, 305, 42–48. [Google Scholar] [CrossRef]
- Jacobson, C.B.; Pasternak, J.J.; Glick, B.R. Partial purification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can. J. Microbiol. 2011, 40, 1019–1025. [Google Scholar] [CrossRef]
- Ziegler, C.A.; Freddolino, P.L. The leucine-responsive regulatory proteins/feast-famine regulatory proteins: An ancient and complex class of transcriptional regulators in bacteria and archaea. Critic. Rev. Biochem. Mol. Biol. 2021, 56, 373–400. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; He, H.; Dong, S.; Tang, L.; Yang, E.; Wang, W.; Zhang, B. The leucine-responsive regulatory protein SCAB_Lrp modulates thaxtomin biosynthesis, pathogenicity, and morphological development in Streptomyces scabies. Mol. Plant Pathol. 2023, 24, 167–178. [Google Scholar] [CrossRef]
- Ziegler, C.A.; Freddolino, P.L. Escherichia coli Leucine-Responsive Regulatory Protein Bridges DNA In Vivo and Tunably Dissociates in the Presence of Exogenous Leucine. mBio 2023, 14, e0269022. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.; Pascal, R.A.; Johnston, M.; Raines, R.; Dikshit, D.; Krantz, A.; Honma, M. Mechanistic studies on the pyridoxal phosphate enzyme 1-aminocyclopropane-1-carboxylate from Pseudomonas sp. Biochemistry 1981, 20, 7509–7519. [Google Scholar] [CrossRef] [PubMed]
- Honma, M. Chemically reactive sulfhydryl groups of 1-aminocyclopropane-1-carboxylate deaminase. Agric. Biol. Chem. 1985, 49, 567–571. [Google Scholar] [CrossRef]
- Bulusu, V.; Jayaraman, V.; Balaram, H. Metabolic fate of fumarate, a side product of the purine salvage pathway in the intraerythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 2011, 286, 9236–9245. [Google Scholar] [CrossRef]
- Guest, J.R.; Green, J.; Irvine, A.S.; Spiro, S. The FNR modulon and FNR-regulated gene expression. In Regulation of Gene Expression in Escherichia coli; Lin, E.C.C., Lynch, A.S., Eds.; Champman and Hall: New York, NY, USA, 1996; pp. 317–342. [Google Scholar] [CrossRef]
- Lopes, L.D.; Wang, P.; Futrell, S.L.; Schachtman, D.P. Sugars and Jasmonic Acid Concentration in Root Exudates Affect Maize Rhizosphere Bacterial Communities. Appl. Environ. Microbiol. 2022, 88, e0097122. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Scher, F.M.; Laliberte, M.; Tipping, B. Emergence-promoting rhizobacteria: Description and implications for agricuture. In Iron, Siderophores, and Plant Diseases; Swinburne, T.R., Ed.; Plenum Press: New York, NY, USA, 1986; pp. 155–164. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Tafner, R.; Moreno, M.V.; Stenglein, S.A.; García de Salamone, I.E.; Nelson, L.M.; Novák, O.; Strnad, M.; van der Graaff, E.; Roitsch, T. Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci. Rep. 2016, 6, 23310. [Google Scholar] [CrossRef] [PubMed]
- Mekureyaw, M.F.; Pandey, C.; Hennessy, R.C.; Nicolaisen, M.H.; Liu, F.; Nybroe, O.; Roitsch, T. The cytokinin-producing plant beneficial bacterium Pseudomonas fluorescens G20-18 primes tomato (Solanum lycopersicum) for enhanced drought stress responses. J. Plant Physiol. 2022, 270, 153629. [Google Scholar] [CrossRef]
- Glick, B.R.; Penrose, D.M.; Li, J. A model for lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theoret. Biol. 1998, 190, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Yemelyanov, V.V.; Lastochkin, V.V.; Chirkova, T.V.; Lindberg, S.M.; Shishova, M.F. Indoleacetic Acid Levels in Wheat and Rice Seedlings under Oxygen Deficiency and Subsequent Reoxygenation. Biomolecules 2020, 10, 276. [Google Scholar] [CrossRef]
- Liu, W.H.; Chen, F.F.; Wang, C.E.; Fu, H.H.; Fang, X.Q.; Ye, J.R.; Shi, J.Y. Indole-3-Acetic Acid in Burkholderia pyrrocinia JK-SH007: Enzymatic Identification of the Indole-3-Acetamide Synthesis Pathway. Front. Microbiol. 2019, 10, 2559. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).