Cellular Stress Impact on Yeast Activity in Biotechnological Processes—A Short Overview
Abstract
:1. Introduction
2. Saccharomyces cerevisiae Species
3. Types of Cellular Stress
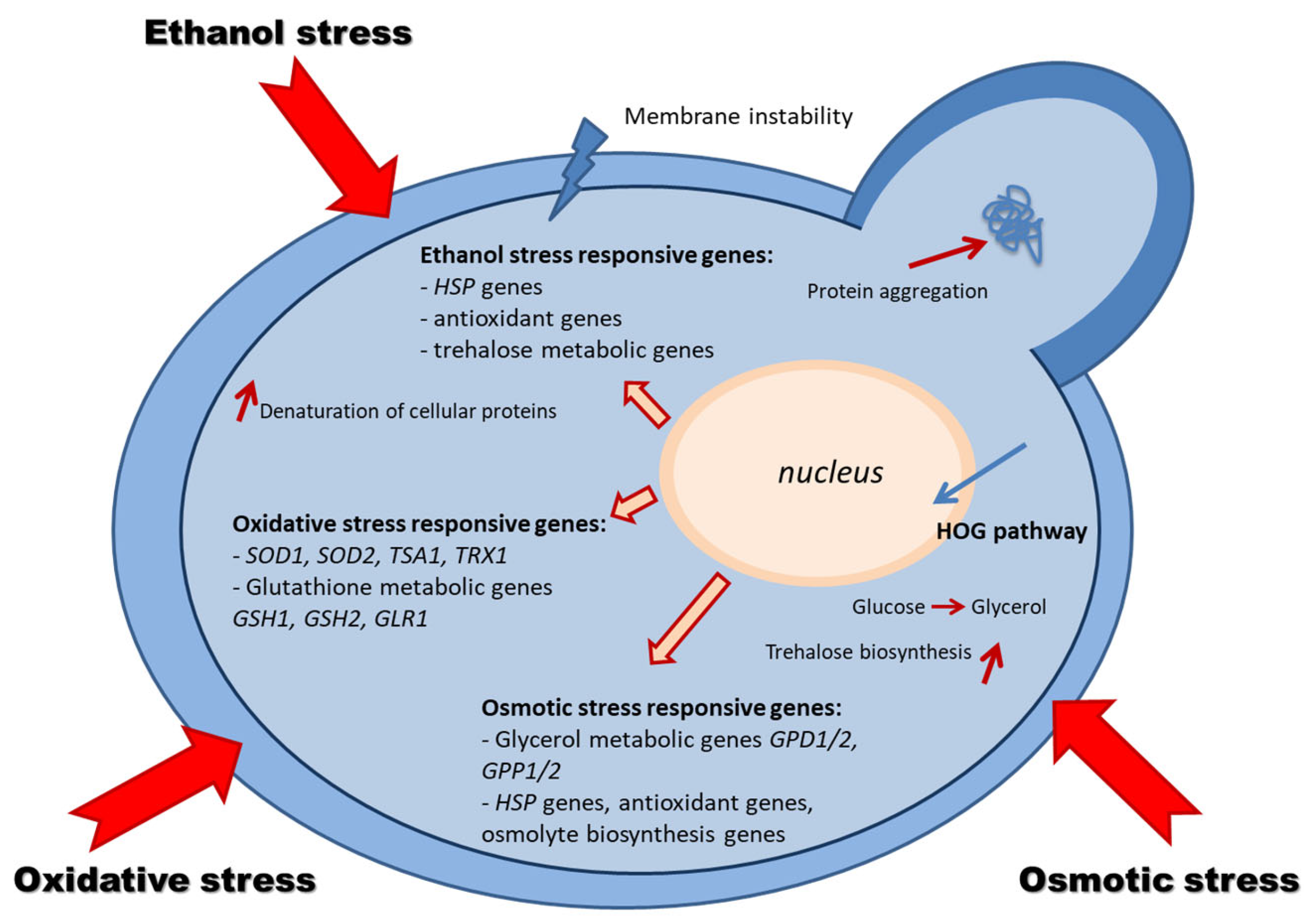
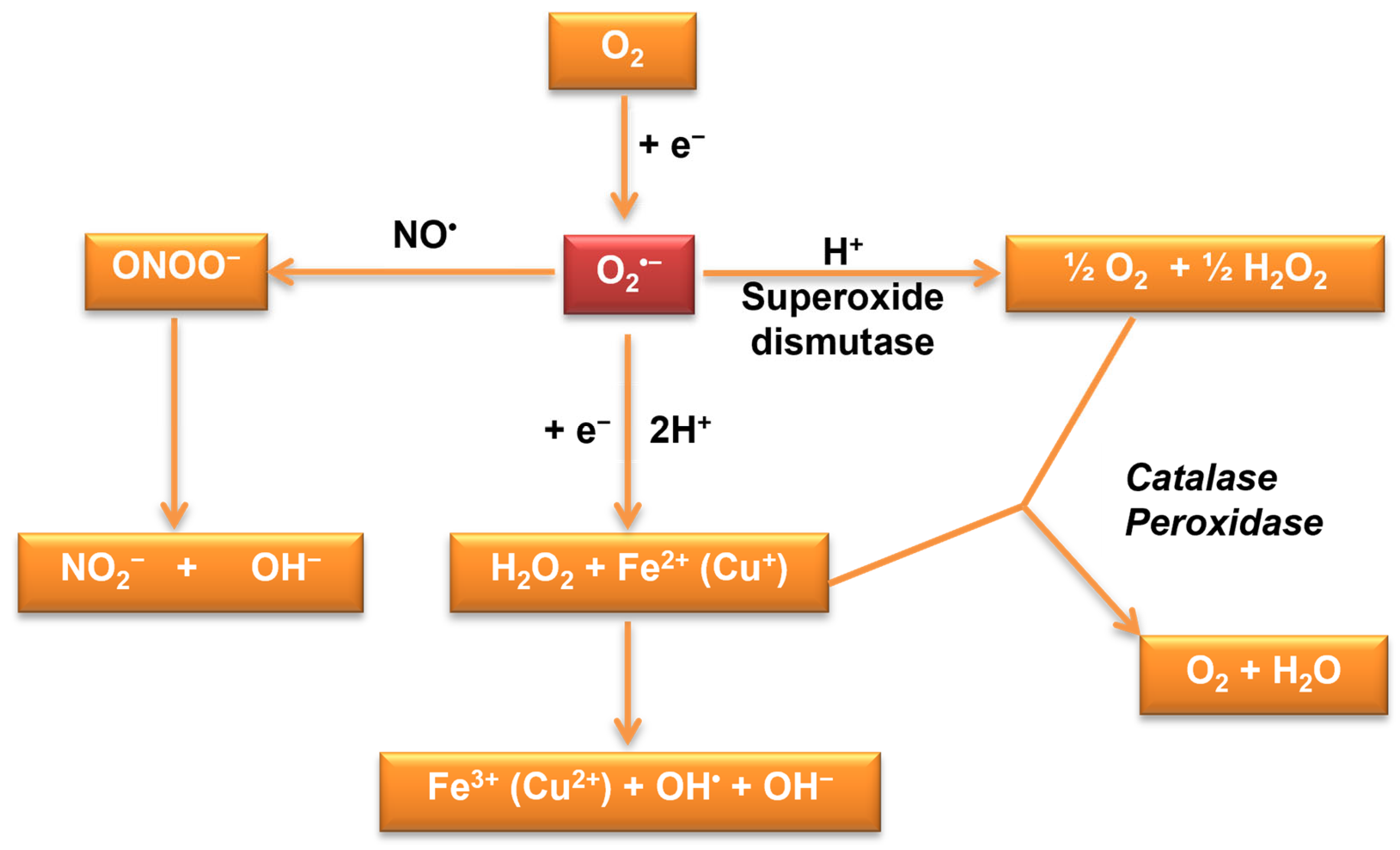
3.1. Oxidative Stress
3.2. Osmotic Stress—Stress Due to Environmental Salts
3.3. Stress Due to Ethanol Accumulation
4. The Behavior of Saccharomyces cerevisiae in Stressors Action
5. Conclusions
Funding
Conflicts of Interest
References
- Costa, A.C.T.; Russo, M.; Fernandes, A.A.R.; Broach, J.R.; Fernandes, P.M.B. Transcriptional Response of Multi-Stress-Tolerant Saccharomyces cerevisiae to Sequential Stresses. Fermentation 2023, 9, 195. [Google Scholar] [CrossRef]
- Walker, G.M.; Basso, T.O. Mitigating stress in industrial yeasts. Fungal Biol. 2020, 124, 387–397. [Google Scholar] [CrossRef]
- Kyriakou, M.; Christodoulou, M.; Ioannou, A.; Fotopoulos, V.; Koutinas, M. Improvement of stress multi-tolerance and bioethanol production by Saccharomyces cerevisiae immobilised on biochar: Monitoring transcription from defence-related genes. Biochem. Eng. J. 2023, 195, 108914. [Google Scholar] [CrossRef]
- Gibson, B.R.; Lawrence, S.J.; Leclaire, J.P.R.; Powell, C.D.; Smart, K.A. Yeast response to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [CrossRef]
- Cavalieri, D.; McGovern, P.E.; Hartl, D.L.; Mortimer, R.; Polsinelli, M. Evidence for S. cerevisiae Fermentation in Ancient Wine. J. Mol. Evol. 2003, 57, S226–S232. [Google Scholar] [CrossRef]
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nunez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.; et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef]
- De Llanos, R.; Llopis, S.; Molero, G.; Querol, A.; Gil, C.; Fernandez-Espinar, M.T. In vivo virulence of commercial Saccharomyces cerevisiae strains with pathogenicity-associated phenotypical traits. Int. J. Food Microbiol. 2011, 144, 393–399. [Google Scholar] [CrossRef]
- Pillai, U.; Devasahayam, J.; Kurup, A.N.; Lacasse, A. Invasive Saccharomyces cerevisiae Infection: A Friend Turning Foe? Saudi J. Kidney Dis. Transpl. 2014, 25, 1266–1269. [Google Scholar] [CrossRef]
- Cui, L.; Lucht, L.; Tipton, L.; Rogers, M.B.; Fitch, A.; Kessinger, C.; Camp, D.; Kingsley, L.; Leo, N.; Greenblatt, R.M.; et al. Topographic Diversity of the Respiratory Tract Mycobiome and Alteration in HIV and Lung Disease. Am. J. Respir. Crit. Care Med. 2015, 191, 932–942. [Google Scholar] [CrossRef]
- Zhu, Y.O.; Sherlock, G.; Petrov, D.A. Whole Genome Analysis of 132 Clinical Saccharomyces cerevisiae Strains Reveals Extensive Ploidy Variation. G3 2016, 6, 2421–2434. [Google Scholar] [CrossRef]
- Ackerman, A.L.; Underhill, D.M. The mycobiome of the human urinary tract: Potential roles for fungi in urology. Ann. Transl. Med. 2017, 5, 31. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Raghavan, V.; Aquadro, C.F.; Alani, E. Baker’s Yeast Clinical Isolates Provide a Model for How Pathogenic Yeasts Adapt to Stress. Trends Genet. 2019, 35, 804–817. [Google Scholar] [CrossRef]
- Kitagaki, H.; Takagi, H. Mitochondrial metabolism and stress response of yeast: Applications in fermentation technologies. J. Biosci. Bioeng. 2014, 117, 383–393. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Bai, F.W. Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J. Biotechnol. 2009, 144, 23–30. [Google Scholar] [CrossRef]
- Helalat, S.H.; Bidaj, S.; Samani, S.; Moradi, M. Producing alcohol and salt stress tolerant strain of Saccharomyces cerevisiae by heterologous expression of pprI gene. Enzyme Microb. Technol. 2019, 124, 17–22. [Google Scholar] [CrossRef]
- Steen, E.J.; Chan, R.; Prasad, N.; Myers, S.; Petzold, C.J.; Redding, A.; Ouellet, M.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 2008, 7, 36. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnson, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Barnett, J.A. Beginnings of microbiology and biochemistry: The contribution of yeast research. Microbiology 2003, 149, 557–567. [Google Scholar] [CrossRef]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef]
- Hagman, A.; Säll, T.; Compagno, C.; Piskur, J. Yeast “Make-Accumulate-Consume” Life Strategy Evolved as a Multi-Step Process That Predates the Whole Genome Duplication. PLoS ONE 2013, 8, e68734. [Google Scholar] [CrossRef]
- Engel, S.R.; Dietrich, F.S.; Fisk, D.G.; Binkley, G.; Balakrishnan, R.; Costanzo, M.C.; Dwight, S.S.; Hitz, B.C.; Karra, K.; Nash, R.S.; et al. The reference genome sequence of Saccharomyces cerevisiae: Then and now. G3 2014, 4, 389–398. [Google Scholar] [CrossRef]
- Williams, K.M.; Liu, P.; Fay, J.C. Evolution of ecological dominance of yeast species in high-sugar environments. Evolution 2015, 69, 2079–2093. [Google Scholar] [CrossRef]
- Basile, A.; De Pascale, F.; Bianca, F.; Rossi, A.; Frizzarin, M.; De Bernardini, N.; Bosaro, M.; Baldisseri, A.; Antoniali, P.; Lopreiato, R.; et al. Large-scale sequencing and comparative analysis of oenological Saccharomyces cerevisiae strains supported by nanopore refinement of key genomes. Food Microbiol. 2021, 97, 103753. [Google Scholar] [CrossRef]
- Saito, T.L.; Ohtani, M.; Sawai, H.; Sano, F.; Saka, A.; Watanabe, D.; Yukawa, M.; Ohya, Y.; Morishita, S. SCMD: Saccharomyces cerevisiae Morphological Database. Nucleic Acids Res. 2004, 32, D319–D322. [Google Scholar] [CrossRef]
- Stewart, G.G. SACCHAROMYCES|Saccharomyces cerevisiae. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 309–315. [Google Scholar] [CrossRef]
- Granek, J.A.; Magwene, P.M. Environmental and Genetic Determinants of Colony Morphology in Yeast. PLoS Genet. 2010, 6, e1000823. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Burtner, C.R.; Kennedy, B.K. Recent developments in yeast aging. PLoS Genet. 2007, 3, e84. [Google Scholar] [CrossRef]
- Inderpal, M. Oxidative Stress in Saccharomyces cerevisiae. Doctoral Dissertation, University of KwaZulu-Natal, Durban, South Africa, 2017. [Google Scholar]
- Cazzanelli, G.; Pereira, F.; Alves, S.; Francisco, R.; Azevedo, L.; Carvalho, P.D.; Almeida, A.; Corte-Real, M.; Oliveira, M.J.; Lucas, C.; et al. The Yeast Saccharomyces cerevisiae as a Model for Understanding RAS Proteins and Their Role in Human Tumorigenesis. Cells 2018, 7, 14. [Google Scholar] [CrossRef]
- Nielsen, J. Yeast Systems Biology: Model Organism and Cell Factory. Biotechnol. J. 2019, 14, 1800421. [Google Scholar] [CrossRef]
- Eleutherio, E.; Brasil, A.A.; França, M.B.; de Almeida, D.S.G.; Rona, G.B.; Magalhães, S.S. Oxidative stress and aging: Learning from yeast lessons. Fungal Biol. 2018, 122, 514–525. [Google Scholar] [CrossRef]
- Walker, G.M.; Van Dijck, P. Physiological and Molecular Responses of Yeasts to the Environment. In Yeasts in Food and Beverages; Querol, A., Fleet, G.H., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 111–152. [Google Scholar]
- Penninckx, M. A short review on the role of glutathione in the response of yeasts to nutritional, environmental, and oxidative stresses. Enzyme Microb. Technol. 2000, 26, 737–742. [Google Scholar] [CrossRef]
- Crawford, R.A.; Pavitt, G.D. Translational regulation in response to stress in Saccharomyces cerevisiae. Yeast 2019, 36, 5–21. [Google Scholar] [CrossRef]
- Eardley, J.; Timson, D.J. Yeast Cellular Stress: Impacts on Bioethanol Production. Fermentation 2020, 6, 109. [Google Scholar] [CrossRef]
- Sunyer-Figueres, M.; Mas, A.; Beltran, G.; Torija, M.J. Protective Effects of Melatonin on Saccharomyces cerevisiae under Ethanol Stress. Antioxidants 2021, 10, 1735. [Google Scholar] [CrossRef]
- Herrero, E.; Ros, J.; Belli, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta 2008, 1780, 1217–1235. [Google Scholar] [CrossRef]
- Moreno-Garcia, J.; Mauricio, J.C.; Moreno, J.; Garcia-Martinez, T. Stress responsive proteins of a flor yeast strain during the early stages of biofilm formation. Process Biochem. 2016, 51, 578–588. [Google Scholar] [CrossRef]
- Vázquez, J.; Grillitsch, K.; Daum, G.; Mas, A.; Beltran, G.; Torija, M.J. The role of the membrane lipid composition in the oxidative stress tolerance of different wine yeasts. Food Microbiol. 2019, 78, 143–154. [Google Scholar] [CrossRef]
- Ayer, A.; Fazakerley, D.J.; James, D.E.; Stocker, R. The role of mitochondrial reactive oxygen species in insulin resistance. Free Radic. Biol. Med. 2022, 179, 339–362. [Google Scholar] [CrossRef]
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 175–190. [Google Scholar] [CrossRef]
- Itto-Nakama, K.; Watanabe, S.; Ohnuki, S.; Kondo, N.; Kikuchi, R.; Nakamura, T.; Ogasawara, W.; Kasahara, K.; Ohya, Y. Prediction of ethanol fermentation under stressed conditions using yeast morphological data. J. Biosci. Bioeng. 2023, 135, 210–216. [Google Scholar] [CrossRef]
- Lushchak, V.I. Oxidative stress in yeast. Biochemistry 2010, 75, 346–364. [Google Scholar] [CrossRef]
- Ji, Z.; Zhang, Y.; Tian, J.; Wang, F.; Song, M.; Li, H. Oxidative stress and cytotoxicity induced by tetrachlorobisphenol A in Saccharomyces cerevisiae cells. Ecotoxicol. Environ. Saf. 2018, 161, 1–7. [Google Scholar] [CrossRef]
- Yashimoto, N.; Kawai, T.; Yoshida, M.; Izawa, S. Xylene causes oxidative stress and pronounced translation repression in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2019, 128, 697–703. [Google Scholar] [CrossRef]
- Puddu, P.; Puddu, G.M.; Cravero, E.; Rosati, M.; Muscari, A. The molecular sources of reactive oxygen species in hypertension. Blood Press. 2008, 17, 70–77. [Google Scholar] [CrossRef]
- Ndukwe, J.K.; Aliyu, G.O.; Onwosi, C.; Chukwu, K.O.; Ezugworie, F.N. Mechanisms of weak acid-induced stress tolerance in yeasts: Prospects for improved bioethanol production form lignocellulosic biomass. Process Biochem. 2020, 90, 118–130. [Google Scholar] [CrossRef]
- Romero, A.; Ramos, E.; de Los Rios, C.; Egea, J.; del Pino, J.; Reiter, R.J. A review of metal-catalyzed molecular damage: Protection by melatonin. J. Pineal Res. 2014, 56, 343–370. [Google Scholar] [CrossRef]
- Bisquert, R.; Muniz-Calvo, S.; Guillamon, J.M. Protective Role of Intracellular Melatonin Against Oxidative Stress and UV Radiation in Saccharomyces cerevisiae. Front. Microbiol. 2018, 9, 318. [Google Scholar] [CrossRef]
- Morcillo-Parra, M.A.; Beltran, G.; Mas, A.; Torija, M.J. Effect of Several Nutrients and Environmental Conditions on Intracellular Melatonin Synthesis in Saccharomyces cerevisiae. Microorganims 2020, 8, 853. [Google Scholar] [CrossRef]
- Li, C.; Zhang, N.; Li, B.; Xu, Q.; Song, J.; Wei, N.; Wang, W.; Zou, H. Increased torulene accumulation in red yeast Sporidiobolus pararoseus NGR as stress response to high salt conditions. Food Chem. 2017, 237, 1041–1047. [Google Scholar] [CrossRef]
- Illarionov, A.; Lahtvee, P.-J.; Kumar, R. Potassium and Sodium Salt Stress Characterization in the Yeasts Saccharomyces cerevisiae, Kluyveromyces marxianus, and Rhodotorula toruloides. Appl. Environ. Microbiol. 2021, 87, e0310020. [Google Scholar] [CrossRef]
- Dakal, T.C.; Solieri, L.; Giudici, P. Adaptive response and tolerance to sugar and salt stress in the food yeast Zygosaccharomyces rouxii. Int. J. Food Microbiol. 2014, 185, 140–157. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Klipp, E.; Nordlander, B.; Kruger, R.; Gennemark, P.; Hohmann, S. Integrative model of the response of yeast to osmotic shock. Nat. Biotechnol. 2005, 23, 975–982. [Google Scholar] [CrossRef]
- Shinto, H.; Kojima, M.; Shigaki, C.; Hirohashi, Y.; Seto, H. Effect of salt concentration and exposure temperature on adhesion and cytotoxicity of positively charged nanoparticles toward yeast cells. Adv. Powder Technol. 2022, 33, 103835. [Google Scholar] [CrossRef]
- Tokuoka, K. Sugar- and salt-tolerant yeasts. J. Appl. Bacteriol. 1993, 74, 101–110. [Google Scholar] [CrossRef]
- Dinu, L.D.; Craciun, T. Salt stress resistance in yeast is heterogeneous. Period. Mineral. 2022, 91, 18–29. [Google Scholar] [CrossRef]
- Wen, H.; Xiong, K.; Yang, H.; Zhang, P.; Wang, X. Dynamic mechanism of the microbiota of high-salinity organic wastewater with salt-tolerant yeast and its application. J. Environ. Chem. Eng. 2022, 10, 107377. [Google Scholar] [CrossRef]
- Nguyet, V.T.A.; Furutani, N.; Ando, R.; Izawa, S. Acquired resistance to severe ethanol stress-induced inhibition of proteasomal proteolysis in Saccharomyces cerevisiae. Biochim. Biophys. Acta Gen. Subj. 2022, 1866, 130241. [Google Scholar] [CrossRef]
- Saini, P.; Beniwal, A.; Kokkiligadda, A.; Vij, S. Response and tolerance of yeast to changing environmental stress during ethanol fermentation. Process Biochem. 2018, 72, 1–12. [Google Scholar] [CrossRef]
- Charoenbhakdi, S.; Dokpikur, T.; Burphan, T.; Techo, T.; Auesukaree, C. Vacuolar H+-ATPase protects Saccharomyces cerevisiae cells against ethanol-induced oxidative and cell wall stresses. Appl. Environ. Microbiol. 2016, 82, 3121–3130. [Google Scholar] [CrossRef]
- Francois, J.M.; Walther, T.; Parrou, J.L. Genetics and Regulation of Glycogen and Trehalose Metabolism in Saccharomyces cerevisiae. In Microbial Stress Tolerance for Biofuels. Microbiology Monographs; Liu, Z., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 22, pp. 29–54. [Google Scholar] [CrossRef]
- Ciobanu, C.; Tucaliuc, A.; Cascaval, D.; Turnea, M.; Galaction, A.I. Correlation between aeration and ergosterol production by yeasts. Environ. Eng. Manag. J. 2019, 18, 2747–2756. [Google Scholar] [CrossRef]
- Pupyshev, A.B.; Klyushnik, T.P.; Akopyan, A.A.; Singh, S.K.; Tikhonova, M.A. Disaccharide trehalose in experimental therapies for neurodegenerative disorders: Molecular targets and translational potential. Pharmacol. Res. 2022, 183, 106373. [Google Scholar] [CrossRef]
- Singh, L.; Sharma, S.C.; Rai, J. Role of Hal5p protein kinase under ethanol stress in Saccharomyces cerevisiae. Appl. Biol. Chem. J. (TABCJ) 2023, 4, 44–53. [Google Scholar] [CrossRef]
- Lei, J.; Zhao, X.; Ge, X.; Bai, F. Ethanol tolerance and the variation of plasma membrane composition of yeast floc populations with different size distribution. J. Biotechnol. 2007, 131, 270–275. [Google Scholar] [CrossRef]
- Martinez-Munoz, G.A.; Kane, P. Vacuolar and Plasma Membrane Proton Pumps Collaborate to Achieve Cytosolic pH Homeostasis in Yeast. J. Biol. Chem. 2008, 283, 20309–20319. [Google Scholar] [CrossRef]
- Auesukaree, C. Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J. Biosci. Bioeng. 2017, 124, 133–142. [Google Scholar] [CrossRef]
- Westergard, L.; True, H.L. Wild yeast harbour a variety of distinct amyloid structures with strong prion-inducing capabilities. Mol. Microbiol. 2014, 92, 183–193. [Google Scholar] [CrossRef]
- Yona, A.H.; Frumkin, I.; Pilpel, Y. A relay race on the evolutionary adaptation spectrum. Cell 2015, 163, 549–559. [Google Scholar] [CrossRef]
- Fabrizio, P.; Garvis, S.; Palladino, F. Histone Methylation and Memory of Environmental Stress. Cells 2019, 8, 339. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Bai, F.W. Zinc and yeast stress tolerance: Micronutrient plays a big role. J. Biotechnol. 2012, 158, 176–183. [Google Scholar] [CrossRef]
- Ramos, J.; Ariño, J.; Sychrová, H. Alkali-metal-cation influx and efflux systems in nonconventional yeast species. FEMS Microbiol. Lett. 2011, 317, 1–8. [Google Scholar] [CrossRef]
- De Nadal, E.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Tanino, T.; Sato, S.; Oshige, M.; Ohshima, T. Analysis of the stress response of yeast Saccharomyces cerevisiae toward pulsed electric field. J. Electrost. 2012, 70, 212–216. [Google Scholar] [CrossRef]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 2001, 12, 323–337. [Google Scholar] [CrossRef]
- Saccharomyces Genome Database. Available online: https://www.yeastgenome.org/ (accessed on 26 September 2023).
- Horak, C.E.; Snyder, M. Global analysis of protein expression in yeast. Funct. Integr. Genomicc. 2002, 2, 171–180. [Google Scholar] [CrossRef]
- Zahrl, R.J.; Prielhofer, R.; Burgard, J.; Mattanovich, D.; Gasser, B. Synthetic activation of yeast stress response improves secretion of recombinant proteins. N. Biotechnol. 2023, 73, 19–28. [Google Scholar] [CrossRef]
- Engelberg, D.; Perlman, R.; Levitzki, A. Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: State of the art after 25 years. Cell Signal. 2014, 26, 2865–2878. [Google Scholar] [CrossRef]
- Rødkaer, S.V.; Faergeman, N.J. Glucose- and nitrogen sensing and regulatory mechanism in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 683–696. [Google Scholar] [CrossRef]
- Kang, X.X.; Wang, Q.Q.; Chi, Z.; Liu, G.L.; Hu, Z.; Chi, Z.M. The GATA type transcriptional factors regulate pullulan biosynthesis in Aureobasidium melanogenum P16. Int. J. Biol. Macromol. 2021, 192, 161–168. [Google Scholar] [CrossRef]

| Category | Characteristic | Species Name | NaCl: M; % (w/v) |
|---|---|---|---|
| Halotolerant | Growth at NaCl > 2.0 M | C. parapsilosis | 3.0; 17.5 |
| P. membranifaciens | 3.0; 17.5 | ||
| I. orientalis | 2.0; 11.7 | ||
| P. sorbitophila | 3.0–4.0; 17.5–23.4 | ||
| Z. rouxii | 3.0; 17.5 | ||
| H. werneckii | 5.20; 30.8 | ||
| D. hansenii | 3.0–4.0; 17.5–23.4 | ||
| C. halophila | 4.0–5.0; 23.4–29.1 | ||
| Moderate halotolerant | Growth inhibition at NaCl > 2.0 M | S. cerevisiae | <1.70; 10 |
| S. pombe | 1.00; 5.8 | ||
| Z. florentinus | 1.00; 5.8 | ||
| C. albicans | 1.70; 10 | ||
| C. tropicalis | 1.7–2.0; 10–11.7 | ||
| Z. sapae | 2.0; 11.7 | ||
| Z. bailii | 1.0–2.0; 5.8–11.7 | ||
| Z. bisporus | 1.0–2.0; 5.8–11.7 |
| Stress Response | Details |
|---|---|
| Apoptosis | Near the end of life, senescent cells enter apoptosis |
| Genotypic changes | Mutations are common in yeasts subjected to stress during the brewing process |
| Changes in intracellular metal ion homeostasis | Several stressors lead to the loss of key metal ions such as Mg and Zn |
| Blocking the cell division cycle | Stress leads to the blocking of the yeast cell cycle, thus inhibiting growth |
| Induction of stress enzymes | For example, the induction of antioxidant enzymes during the action of oxidative stress |
| Activation of glycerol biosynthesis | Glycerol is produced in excess during osmotic stress |
| Induction of trehalose biosynthesis | Cells produce large amounts of trehalose in response to heat shock and other stressors |
| Structural changes in the cell membrane | Disorders of membrane integrity caused by stressors |
| Induction of heat shock protein synthesis (hot/cold) | Heat shock proteins are induced in response to high temperatures and other factors |
| Gene Type | Function |
|---|---|
| Ace2 | Transcription factor that early activates G1-specific gene expression; may be involved in acetic acid tolerance |
| Crz1 | Transcription factor that activates the copying of genes involved in the stress response |
| Nrg1 | Intervenes in glucose repression and negatively regulates many processes, including filamentous growth. Involved in tolerance to salt, oxidative stress, and acetic acid |
| Rim101 | Repressive transcription factor involved in cell wall structuring and pH response. It also plays a role in acetic acid tolerance |
| Stb5 | Transcription factor involved in multidrug resistance, oxidative stress response, and acetic acid tolerance |
| Usv1 | Transcriptional regulator of genes involved in growth of non-fermentable carbon sources and response to osmotic stress |
| Cmr3 | YPR015C-related transcription factor affects the copying of genes involved in cell defense and rescue, but also in DNA processing and the cell cycle |
| Msn2/4 | Activators of transcription, regulate the general stress response. Overwriting Msn2 improves ethanol tolerance |
| Prd1/Prd3 | Involved in tolerance to hydroxymethylfurfural (HMF) and ethanol, and in response to salt stress |
| Ume6 | Key transcriptional regulator of early meiotic genes, involved in acetic acid tolerance and salt stress response |
| System for | Gene | Sc 1 | Sp 2 | Zr 3 | Dh 4 | Ca 5 |
|---|---|---|---|---|---|---|
| K+ influx | TRK1 | + | + | + | + | + |
| TRK2 | + | + | − | − | − | |
| HAK1 | − | − | − | + | + | |
| ACU1 | − | − | − | − | pseudogene | |
| K+ efflux | TOK1 | + | − | + | − | + |
| K+ and Na+ efflux | NHA1 | + | + | + | + | + |
| SOD2 | − | + | + | undefined | − | |
| ENA1-4 | + | + | + | + | + |
| Method | Details |
|---|---|
| Microbial contamination control | Sanitization operations of fermentation processes, use of antimicrobial products |
| Ensuring optimal physiological conditions | Correct cold storage temperature, optimal pre-propagation conditions |
| Use of improved yeast strains | Application of genetic and physiological techniques to optimize stress resistance |
| Ensuring proper yeast nutrition | Micronutrient balance, bioavailability for metal ions |
| Control of fermentation parameters | pH, temperature, agitation, specific density of the medium, dissolved oxygen |
| Stress Type | Cell Response | Industrial Application |
|---|---|---|
| Osmotic shock | Glycerol and trehalose levels increase by lowering the osmotic potential of the growth medium | Usage of tolerant yeasts in fermentation processes |
| UV irradiation | Irradiated yeasts convert membrane ergosterol to vitamin D2 | Vitamin-D2-enriched yeasts for baking and nutraceuticals |
| Oxidative stress | Induction of catalase and superoxide dismutase, stimulation of membrane sterol, and unsaturated fatty acid synthesis in aerobic environment | Oxygenated yeasts will have a higher stress tolerance during the fermentation process due to the high amounts of ergosterol and oleic acid in the membrane |
| Heat shock | Yeast cells with high levels of trehalose have a higher tolerance against stress after heat shock | Usage of frozen dough for baking |
| Heat and salts | Self-digestion | Obtaining yeast extracts and yeast beta-glucan |
| Cell Stress Genes | Biological Stress Response |
|---|---|
| BSD2 | involved in metal ion transport (IMP), protein targeting to vacuole (IMP), ubiquitin-dependent protein catabolic process (IMP) |
| CTT1 | involved in response to reactive oxygen species (IMP), protection from oxidative damage by hydrogen peroxide |
| DAK1 | involved in glycerol to glycerone phosphate metabolic process (IGI), involved in stress adaptation |
| DDR48 | involved in DNA repair (IDA, IMP), expression is increased in response to heat-shock stress |
| GTT1 | involved in glutathione metabolic process (IDA) and protein glutathionylation (IMP) |
| HSP104 | involved in cellular heat acclimation (IMP), chaperone cofactor-dependent protein refolding (IDA), protein folding in endoplasmic reticulum (IMP), protein unfolding (IMP), trehalose metabolism in response to heat stress (IMP), responds to stresses including heat, ethanol, and sodium arsenite |
| HSP12 | involved in cell adhesion (IDA), in plasma membrane organization (IMP), cellular response to heat (IMP), osmotic stress (IMP), and oxidative stress (IMP) |
| HSP26 | involved in cellular response to heat (IDA) and in protein folding (IDA) |
| HSP42 | involved in cytoskeleton organization after heat shock (IMP) |
| HSP78 | involved in cellular response to heat (IMP, IGI), mitochondrial genome maintenance (IGI), protein refolding (IDA, IMP), stabilization (IMP, IGI), and unfolding (IMP) |
| NCE103 | involved in cellular response to carbon dioxide (IMP) and oxidative stress (IMP) |
| PPZ2 | involved in intracellular sodium ion homeostasis (IMP) and regulation of potassium transport, which affects osmotic stability |
| SSA4 | involved in protein folding (IGI), cellular response to heat, and role in SRP-dependent cotranslational protein-membrane targeting to membrane and translocation (IMP) |
| TRR2 | involved in protection against oxidative stress (IMP) |
| TTR1 | involved in cellular response to oxidative stress (IMP) and in glutathione metabolic process (IGI) |
| XBP1 | involved in negative regulation of transcription by RNA polymerase II (IMP), induced by stress or starvation during mitosis, and late in meiosis |
| TSA2 | involved in cell redox homeostasis (IDA, IMP), in cellular response to oxidative stress (IGI, IMP), and protein folding (IDA) |
| NQM1 | involved in cellular response to oxidative stress (IMP) |
| RCN1 | involved in response to osmotic and ionic changes concentration |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Postaru, M.; Tucaliuc, A.; Cascaval, D.; Galaction, A.-I. Cellular Stress Impact on Yeast Activity in Biotechnological Processes—A Short Overview. Microorganisms 2023, 11, 2522. https://doi.org/10.3390/microorganisms11102522
Postaru M, Tucaliuc A, Cascaval D, Galaction A-I. Cellular Stress Impact on Yeast Activity in Biotechnological Processes—A Short Overview. Microorganisms. 2023; 11(10):2522. https://doi.org/10.3390/microorganisms11102522
Chicago/Turabian StylePostaru, Madalina, Alexandra Tucaliuc, Dan Cascaval, and Anca-Irina Galaction. 2023. "Cellular Stress Impact on Yeast Activity in Biotechnological Processes—A Short Overview" Microorganisms 11, no. 10: 2522. https://doi.org/10.3390/microorganisms11102522
APA StylePostaru, M., Tucaliuc, A., Cascaval, D., & Galaction, A.-I. (2023). Cellular Stress Impact on Yeast Activity in Biotechnological Processes—A Short Overview. Microorganisms, 11(10), 2522. https://doi.org/10.3390/microorganisms11102522