Transcriptional Profiling of Homologous Recombination Pathway Genes in Mycobacterium bovis BCG Moreau
Abstract
:1. Introduction
2. Materials and Methods
2.1. M. bovis BCG Culture Conditions
2.2. UV Irradiation Assay
2.3. RNA Extraction and RT-qPCR
2.4. Statistical Analysis
3. Results
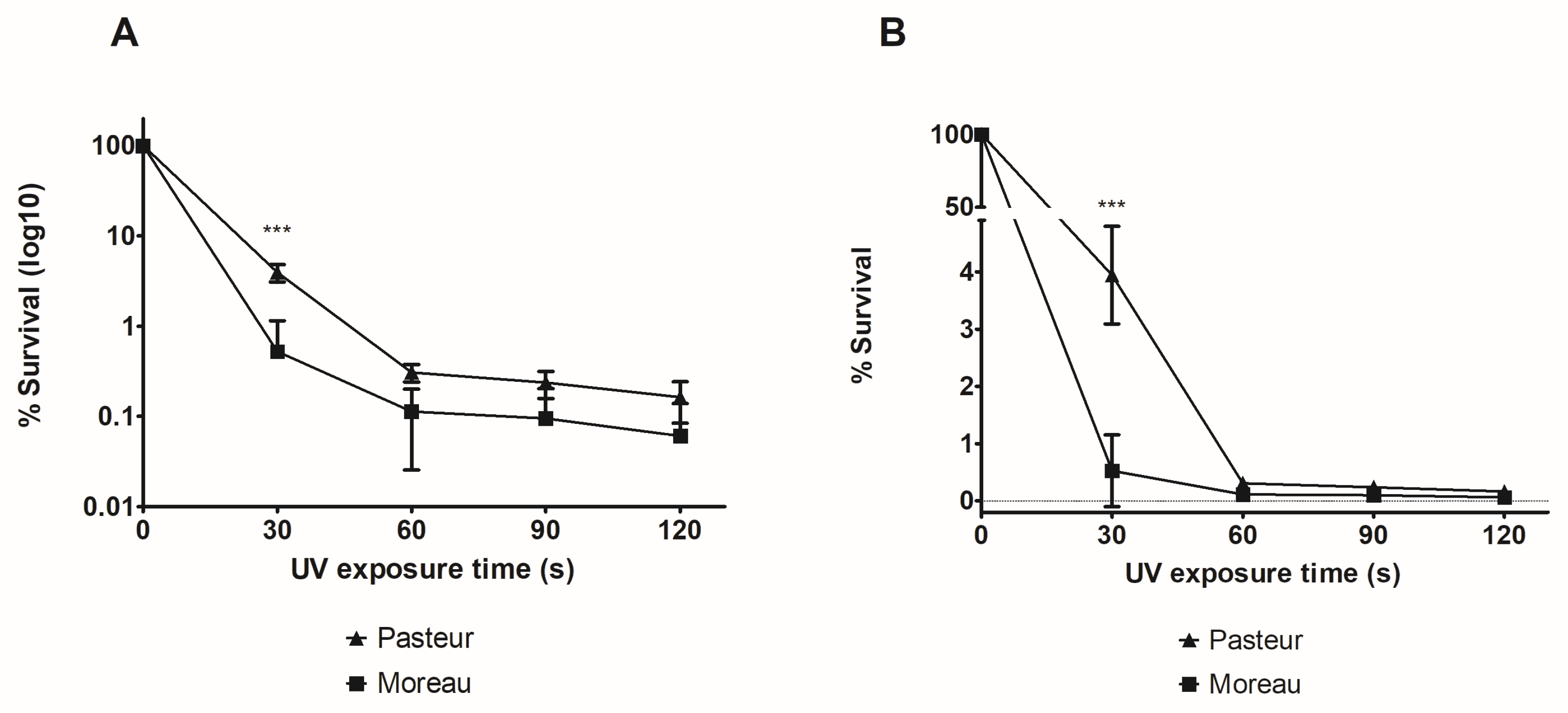
3.1. M. bovis BCG Moreau Is More Susceptible to UV Exposure Than BCG Pasteur
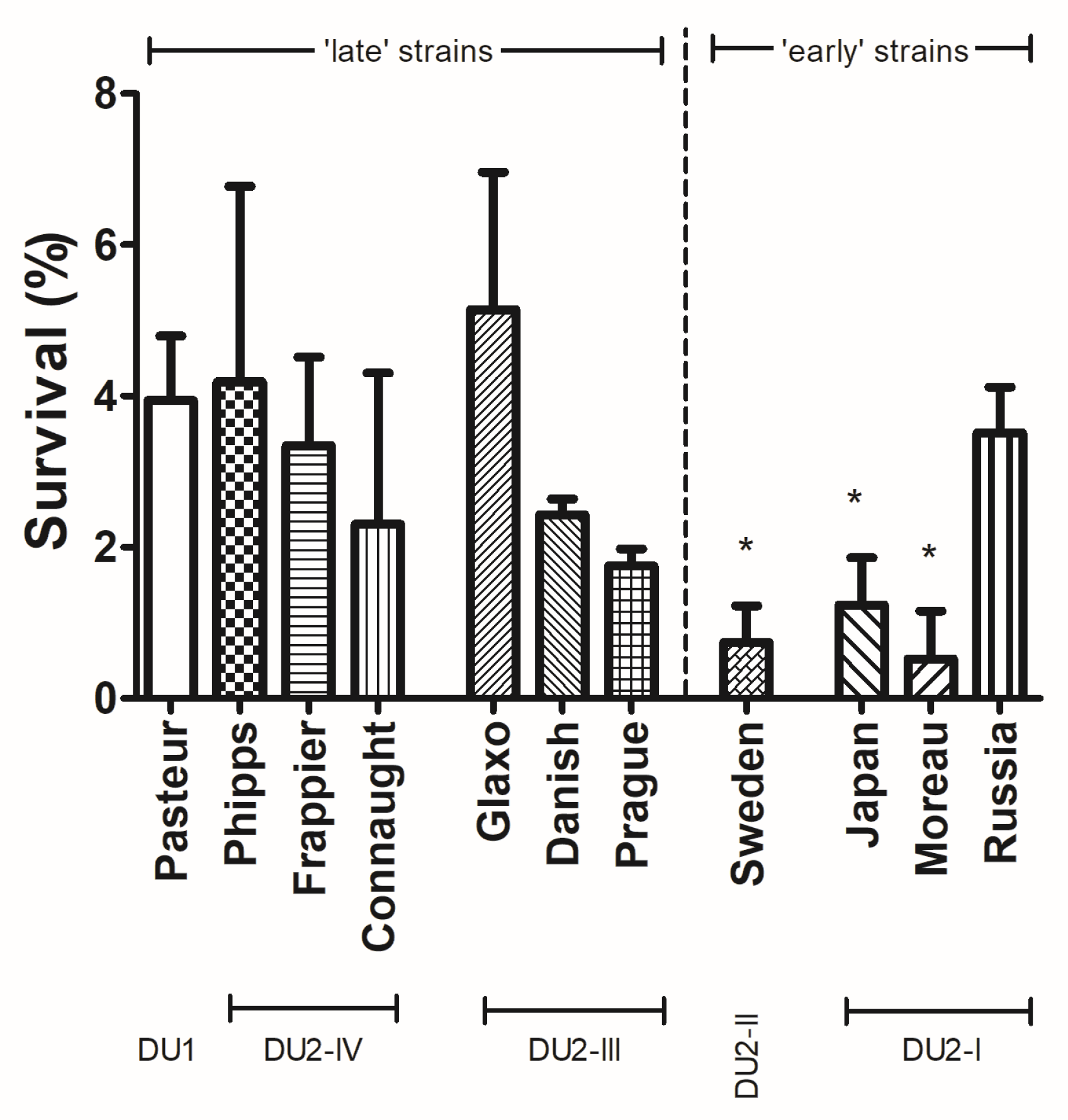
3.2. Early BCG Strains, except Russia, Are like Moreau, Having Lower UV Exposure Survival Rates
3.3. Transcriptional Profile Reveals Lower Expression of DNA Damage Repair Pathway Genes in BCG Moreau Compared to Pasteur
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- World Health Organization Global Tuberculosis Report 2022. Lancet Microbe 2023, 4, e20.
- Behr, M.A.; Small, P.M. A Historical and Molecular Phylogeny of BCG Strains. Vaccine 1999, 17, 915–922. [Google Scholar] [CrossRef]
- Ritz, N.; Curtis, N. Mapping the Global Use of Different BCG Vaccine Strains. Tuberculosis 2009, 89, 248–251. [Google Scholar] [CrossRef]
- Abdallah, A.M.; Hill-Cawthorne, G.A.; Otto, T.D.; Coll, F.; Guerra-Assunção, J.A.; Gao, G.; Naeem, R.; Ansari, H.; Malas, T.B.; Adroub, S.A.; et al. Genomic Expression Catalogue of a Global Collection of BCG Vaccine Strains Show Evidence for Highly Diverged Metabolic and Cell-Wall Adaptations. Sci. Rep. 2015, 5, 15443. [Google Scholar] [CrossRef]
- Faustman, D.L.; Lee, A.; Hostetter, E.R.; Aristarkhova, A.; Ng, N.C.; Shpilsky, G.F.; Tran, L.; Wolfe, G.; Takahashi, H.; Dias, H.F.; et al. Multiple BCG Vaccinations for the Prevention of COVID-19 and Other Infectious Diseases in Type 1 Diabetes. Cell Rep. Med. 2022, 3, 100728. [Google Scholar] [CrossRef]
- Hart, B.E.; Asrican, R.; Lim, S.-Y.; Sixsmith, J.D.; Lukose, R.; Souther, S.J.R.; Rayasam, S.D.G.; Saelens, J.W.; Chen, C.; Seay, S.A.; et al. Stable Expression of Lentiviral Antigens by Quality-Controlled Recombinant Mycobacterium bovis BCG Vectors. Clin. Vaccine Immunol. 2015, 22, 726–741. [Google Scholar] [CrossRef]
- Soto, J.A.; Díaz, F.E.; Retamal-Díaz, A.; Gálvez, N.M.S.; Melo-González, F.; Piña-Iturbe, A.; Ramírez, M.A.; Bohmwald, K.; González, P.A.; Bueno, S.M.; et al. BCG-Based Vaccines Elicit Antigen-Specific Adaptive and Trained Immunity against SARS-CoV-2 and Andes Orthohantavirus. Vaccines 2022, 10, 721. [Google Scholar] [CrossRef]
- Bastos, R.G.; Borsuk, S.; Seixas, F.K.; Dellagostin, O.A. Recombinant Mycobacterium bovis BCG. Vaccine 2009, 27, 6495–6503. [Google Scholar] [CrossRef]
- Stover, C.K.; de la Cruz, V.F.; Fuerst, T.R.; Burlein, J.E.; Benson, L.A.; Bennett, L.T.; Bansal, G.P.; Young, J.F.; Lee, M.H.; Hatfull, G.F.; et al. New Use of BCG for Recombinant Vaccines. Nature 1991, 351, 456–460. [Google Scholar] [CrossRef]
- Van Kessel, J.C.; Hatfull, G.F. Recombineering in Mycobacterium tuberculosis. Nat. Methods 2007, 4, 147–152. [Google Scholar] [CrossRef]
- Van Kessel, J.C.; Marinelli, L.J.; Hatfull, G.F. Recombineering Mycobacteria and Their Phages. Nat. Rev. Microbiol. 2008, 6, 851–857. [Google Scholar] [CrossRef]
- Muttucumaru, N.; Parish, T. The Molecular Biology of Recombination in Mycobacteria: What Do We Know and How Can We Use It? Curr. Issues Mol. Biol. 2004, 6, 145–158. [Google Scholar] [CrossRef]
- Schwarz, M.G.A.; Corrêa, P.R.; Malaga, W.; Guilhot, C.; Mendonça-Lima, L. Mycobacterium bovis BCG Moreau Is Naturally Deficient in Homologous Recombination. Tuberculosis 2020, 123, 101956. [Google Scholar] [CrossRef]
- Covo, S.; Ma, W.; Westmoreland, J.W.; Gordenin, D.A.; Resnick, M.A. Understanding the Origins of UV-Induced Recombination through Manipulation of Sister Chromatid Cohesion. Cell Cycle 2012, 11, 3937–3944. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef]
- Smollett, K.L.; Smith, K.M.; Kahramanoglou, C.; Arnvig, K.B.; Buxton, R.S.; Davis, E.O. Global Analysis of the Regulon of the Transcriptional Repressor LexA, a Key Component of SOS Response in Mycobacterium tuberculosis. J. Biol. Chem. 2012, 287, 22004–22014. [Google Scholar] [CrossRef]
- Singh, A.; Bhagavat, R.; Vijayan, M.; Chandra, N. A Comparative Analysis of the DNA Recombination Repair Pathway in Mycobacterial Genomes. Tuberculosis 2016, 99, 109–119. [Google Scholar] [CrossRef]
- Davis, E.O.; Dullaghan, E.M.; Rand, L. Definition of the Mycobacterial SOS Box and Use To Identify LexA-Regulated Genes in Mycobacterium tuberculosis. J. Bacteriol. 2002, 184, 3287–3295. [Google Scholar] [CrossRef]
- Müller, A.U.; Imkamp, F.; Weber-Ban, E. The Mycobacterial LexA/RecA-Independent DNA Damage Response Is Controlled by PafBC and the Pup-Proteasome System. Cell Rep. 2018, 23, 3551–3564. [Google Scholar] [CrossRef]
- Fudrini Olivencia, B.; Müller, A.U.; Roschitzki, B.; Burger, S.; Weber-Ban, E.; Imkamp, F. Mycobacterium Smegmatis PafBC Is Involved in Regulation of DNA Damage Response. Sci. Rep. 2017, 7, 13987. [Google Scholar] [CrossRef]
- Rand, L.; Hinds, J.; Springer, B.; Sander, P.; Buxton, R.S.; Davis, E.O. The Majority of Inducible DNA Repair Genes in Mycobacterium tuberculosis Are Induced Independently of RecA. Mol. Microbiol. 2003, 50, 1031–1042. [Google Scholar] [CrossRef]
- Adefisayo, O.O.; Dupuy, P.; Nautiyal, A.; Bean, J.M.; Glickman, M.S. Division of Labor between SOS and PafBC in Mycobacterial DNA Repair and Mutagenesis. Nucleic Acids Res. 2021, 49, 12805–12819. [Google Scholar] [CrossRef]
- Prasad, D.; Arora, D.; Nandicoori, V.K.; Muniyappa, K. Elucidating the Functional Role of Mycobacterium Smegmatis RecX in Stress Response. Sci. Rep. 2019, 9, 10912. [Google Scholar] [CrossRef]
- Brosch, R.; Gordon, S.V.; Garnier, T.; Eiglmeier, K.; Frigui, W.; Valenti, P.; Dos Santos, S.; Duthoy, S.; Lacroix, C.; Garcia-Pelayo, C.; et al. Genome Plasticity of BCG and Impact on Vaccine Efficacy. Proc. Natl. Acad. Sci. USA 2007, 104, 5596–5601. [Google Scholar] [CrossRef]
- Orduña, P.; Cevallos, M.A.; de León, S.P.; Arvizu, A.; Hernández-González, I.L.; Mendoza-Hernández, G.; López-Vidal, Y. Genomic and Proteomic Analyses of Mycobacterium bovis BCG Mexico 1931 Reveal a Diverse Immunogenic Repertoire against Tuberculosis Infection. BMC Genom. 2011, 12, 493. [Google Scholar] [CrossRef]
- Sander, P.; Papavinasasundaram, K.G.; Dick, T.; Stavropoulos, E.; Ellrott, K.; Springer, B.; Colston, M.J.; Böttger, E.C. Mycobacterium bovis BCG RecA Deletion Mutant Shows Increased Susceptibility to DNA-Damaging Agents but Wild-Type Survival in a Mouse Infection Model. Infect. Immun. 2001, 69, 3562–3568. [Google Scholar] [CrossRef]
- Boshoff, H.I.M.; Reed, M.B.; Barry, C.E.; Mizrahi, V. DnaE2 Polymerase Contributes to In Vivo Survival and the Emergence of Drug Resistance in Mycobacterium tuberculosis. Cell 2003, 113, 183–193. [Google Scholar] [CrossRef]
- Schefe, J.H.; Lehmann, K.E.; Buschmann, I.R.; Unger, T.; Funke-Kaiser, H. Quantitative Real-Time RT-PCR Data Analysis: Current Concepts and the Novel “Gene Expression’s C T Difference” Formula. J. Mol. Med. 2006, 84, 901–910. [Google Scholar] [CrossRef]
- Manganelli, R.; Voskuil, M.I.; Schoolnik, G.K.; Dubnau, E.; Gomez, M.; Smith, I. Role of the Extracytoplasmic-Function Sigma Factor SigmaH in Mycobacterium tuberculosis Global Gene Expression. Mol. Microbiol. 2002, 45, 365–374. [Google Scholar] [CrossRef]
- Manganelli, R.; Dubnau, E.; Tyagi, S.; Kramer, F.R.; Smith, I. Differential Expression of 10 Sigma Factor Genes in Mycobacterium tuberculosis. Mol. Microbiol. 1999, 31, 715–724. [Google Scholar] [CrossRef]
- Norman, E.; Dellagostin, O.A.; McFadden, J.; Dale, J.W. Gene Replacement by Homologous Recombination in Mycobacterium bovis BCG. Mol. Microbiol. 1995, 16, 755–760. [Google Scholar] [CrossRef]
- Cirz, R.T.; Chin, J.K.; Andes, D.R.; de Crécy-Lagard, V.; Craig, W.A.; Romesberg, F.E. Inhibition of Mutation and Combating the Evolution of Antibiotic Resistance. PLoS Biol. 2005, 3, e176. [Google Scholar] [CrossRef]
- Darwin, K.H.; Nathan, C.F. Role for Nucleotide Excision Repair in Virulence of Mycobacterium tuberculosis. Infect. Immun. 2005, 73, 4581–4587. [Google Scholar] [CrossRef]
- Shuman, S.; Glickman, M.S. Bacterial DNA Repair by Non-Homologous End Joining. Nat. Rev. Microbiol. 2007, 5, 852–861. [Google Scholar] [CrossRef]
- Darmon, E.; Leach, D.R.F. Bacterial Genome Instability. Microbiol. Mol. Biol. Rev. 2014, 78, 1–39. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, X.; Lin, J.; Gao, X.; Xiong, J.; Liu, J.; Fei, Z.; Chen, C. A High Homologous Recombination Deficiency Score Is Associated with Poor Survival and a Non-Inflamed Tumor Microenvironment in Head and Neck Squamous Cell Carcinoma Patients. Oral. Oncol. 2022, 128, 105860. [Google Scholar] [CrossRef]
- Mi, Y.; Gurumurthy, R.K.; Zadora, P.K.; Meyer, T.F.; Chumduri, C. Chlamydia trachomatis Inhibits Homologous Recombination Repair of DNA Breaks by Interfering with PP2A Signaling. mBio 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Gupta, R.; Ryzhikov, M.; Koroleva, O.; Unciuleac, M.; Shuman, S.; Korolev, S.; Glickman, M.S. A Dual Role for Mycobacterial RecO in RecA-Dependent Homologous Recombination and RecA-Independent Single-Strand Annealing. Nucleic Acids Res. 2013, 41, 2284–2295. [Google Scholar] [CrossRef]
- Gupta, R.; Barkan, D.; Redelman-Sidi, G.; Shuman, S.; Glickman, M.S. Mycobacteria Exploit Three Genetically Distinct DNA Double-Strand Break Repair Pathways. Mol. Microbiol. 2011, 79, 316–330. [Google Scholar] [CrossRef]
- Khanduja, J.S.; Tripathi, P.; Muniyappa, K. Mycobacterium tuberculosis RuvA Induces Two Distinct Types of Structural Distortions between the Homologous and Heterologous Holliday Junctions. Biochemistry 2009, 48, 27–40. [Google Scholar] [CrossRef]
- Keller, P.M.; Böttger, E.C.; Sander, P. Tuberculosis Vaccine Strain Mycobacterium bovis BCG Russia Is a Natural RecA Mutant. BMC Microbiol. 2008, 8, 120. [Google Scholar] [CrossRef]
- Lvovna, V.O.; Ivanovna, A.E.; Sergeevna, K.M.; Nikolaevna, R.N.; Nikolaevich, S.A.; Eugenievna, S.N.; Leonidovich, G.A. Mosaic Structure as the Main Feature of Mycobacterium bovis BCG Genomes. In Mycobacterium—Research and Development; InTech: London, UK, 2018. [Google Scholar]
- Patané, J.S.L.; Martins, J.; Castelão, A.B.; Nishibe, C.; Montera, L.; Bigi, F.; Zumárraga, M.J.; Cataldi, A.A.; Junior, A.F.; Roxo, E.; et al. Patterns and Processes of Mycobacterium bovis Evolution Revealed by Phylogenomic Analyses. Genome Biol. Evol. 2017, 9, 521–535. [Google Scholar] [CrossRef]
- Forse, L.N.; Houghton, J.; Davis, E.O. Enhanced Expression of RecX in Mycobacterium tuberculosis Owing to a Promoter Internal to RecA. Tuberculosis 2011, 91, 127–135. [Google Scholar] [CrossRef]
- Gopaul, K.K.; Brooks, P.C.; Prost, J.-F.; Davis, E.O. Characterization of the Two Mycobacterium tuberculosis RecA Promoters. J. Bacteriol. 2003, 185, 6005–6015. [Google Scholar] [CrossRef]
- Ganief, N.; Sjouerman, J.; Albeldas, C.; Nakedi, K.C.; Hermann, C.; Calder, B.; Blackburn, J.M.; Soares, N.C. Associating H2O2− and NO-Related Changes in the Proteome of Mycobacterium smegmatis with Enhanced Survival in Macrophage. Emerg. Microbes Infect. 2018, 7, 1–17. [Google Scholar] [CrossRef]
- Müller, A.U.; Kummer, E.; Schilling, C.M.; Ban, N.; Weber-Ban, E. Transcriptional Control of Mycobacterial DNA Damage Response by Sigma Adaptation. Sci. Adv. 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Finlay, B.B. Pathogenicity Islands: A Molecular Toolbox for Bacterial Virulence. Cell Microbiol. 2006, 8, 1707–1719. [Google Scholar] [CrossRef]
- Schmidt, H.; Hensel, M. Pathogenicity Islands in Bacterial Pathogenesis. Clin. Microbiol. Rev. 2004, 17, 14–56. [Google Scholar] [CrossRef]
- Desvaux, M.; Dalmasso, G.; Beyrouthy, R.; Barnich, N.; Delmas, J.; Bonnet, R. Pathogenicity Factors of Genomic Islands in Intestinal and Extraintestinal Escherichia coli. Front. Microbiol. 2020, 11, 1–35. [Google Scholar] [CrossRef]
- Carpenter, M.R.; Rozovsky, S.; Boyd, E.F. Pathogenicity Island Cross Talk Mediated by Recombination Directionality Factors Facilitates Excision from the Chromosome. J. Bacteriol. 2016, 198, 766–776. [Google Scholar] [CrossRef]
- Gorna, A.E.; Bowater, R.P.; Dziadek, J. DNA Repair Systems and the Pathogenesis of Mycobacterium tuberculosis: Varying Activities at Different Stages of Infection. Clin. Sci. 2010, 119, 187–202. [Google Scholar] [CrossRef]
- Zaczek, A.; Brzostek, A.; Augustynowicz-Kopec, E.; Zwolska, Z.; Dziadek, J. Genetic Evaluation of Relationship between Mutations in RpoB and Resistance of Mycobacterium tuberculosis to Rifampin. BMC Microbiol. 2009, 9, 10. [Google Scholar] [CrossRef]
- Bostanabad, S.; Titov, L.; Bahrmand, A.; Nojoumi, S. Detection of Mutation in Isoniazid-Resistant. Mycobacterium tuberculosis Isolates from Tuberculosis Patients in Belarus. Indian J. Med. Microbiol. 2008, 26, 143. [Google Scholar] [CrossRef]

| Primer Name | Sequence (5′-3′) | |
|---|---|---|
| Locus | Direction | |
| lexA | For | CTTACCGGAACCCACCTTTG |
| Rev | GCTTCGACCATCGAGTCAC | |
| recA | For | CTGAATAATTCGGGCACCAC |
| Rev | GCGTTGGTACCGTCCTTGA | |
| recX | For | CTGGTGGATGACACCGACT |
| Rev | GTACCAGCTTTTCCGCCCG | |
| pafB | For | ACAAGAACGAGCTGCGTGAC |
| Rev | ATCCGGGGTCAGCTCGACA | |
| pafC | For | GAACAGGCACCCACAGAAAG |
| Rev | ATCAACAGCACCCGGATGG | |
| adnA | For | GACCGCCTTGTTCGACATC |
| Rev | GGCATGTGCGCTAAGGAC | |
| adnB | For | CTGCTGGCTTATTTGGACGT |
| Rev | CACCACCTGCCATTCCAAG | |
| ruvC | For | GAGGTGGTGGCTATCGAAC |
| Rev | TGAGCCTTGTCTGCGGAAC | |
| ruvA | For | GAACGCATGGTGTTGGAACT |
| Rev | TGTTTGGCCGCAAAGCCCA | |
| ruvB | For | ATGGAAGACTTCCGCGTCGA |
| Rev | GTAGAAATCCATGTGCGCGG | |
| sigA | For | CTCGACGCTGAACCAGACT |
| Rev | AGGTCTTCGTGGTCTTCGC | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarz, M.G.A.; Corrêa, P.R.; Mendonça-Lima, L. Transcriptional Profiling of Homologous Recombination Pathway Genes in Mycobacterium bovis BCG Moreau. Microorganisms 2023, 11, 2534. https://doi.org/10.3390/microorganisms11102534
Schwarz MGA, Corrêa PR, Mendonça-Lima L. Transcriptional Profiling of Homologous Recombination Pathway Genes in Mycobacterium bovis BCG Moreau. Microorganisms. 2023; 11(10):2534. https://doi.org/10.3390/microorganisms11102534
Chicago/Turabian StyleSchwarz, Marcos Gustavo Araujo, Paloma Rezende Corrêa, and Leila Mendonça-Lima. 2023. "Transcriptional Profiling of Homologous Recombination Pathway Genes in Mycobacterium bovis BCG Moreau" Microorganisms 11, no. 10: 2534. https://doi.org/10.3390/microorganisms11102534
APA StyleSchwarz, M. G. A., Corrêa, P. R., & Mendonça-Lima, L. (2023). Transcriptional Profiling of Homologous Recombination Pathway Genes in Mycobacterium bovis BCG Moreau. Microorganisms, 11(10), 2534. https://doi.org/10.3390/microorganisms11102534






