Author Contributions
Conceptualization, A.E., H.A. and I.E.; formal analysis, A.E., H.A. and H.M.A.; methodology, A.E., H.A., I.E. and H.M.A.; writing—original draft preparation, A.E., H.A. and I.E.; writing—review and editing, H.A., H.M.A. and I.M.B. All authors have read and agreed to the published version of the manuscript.
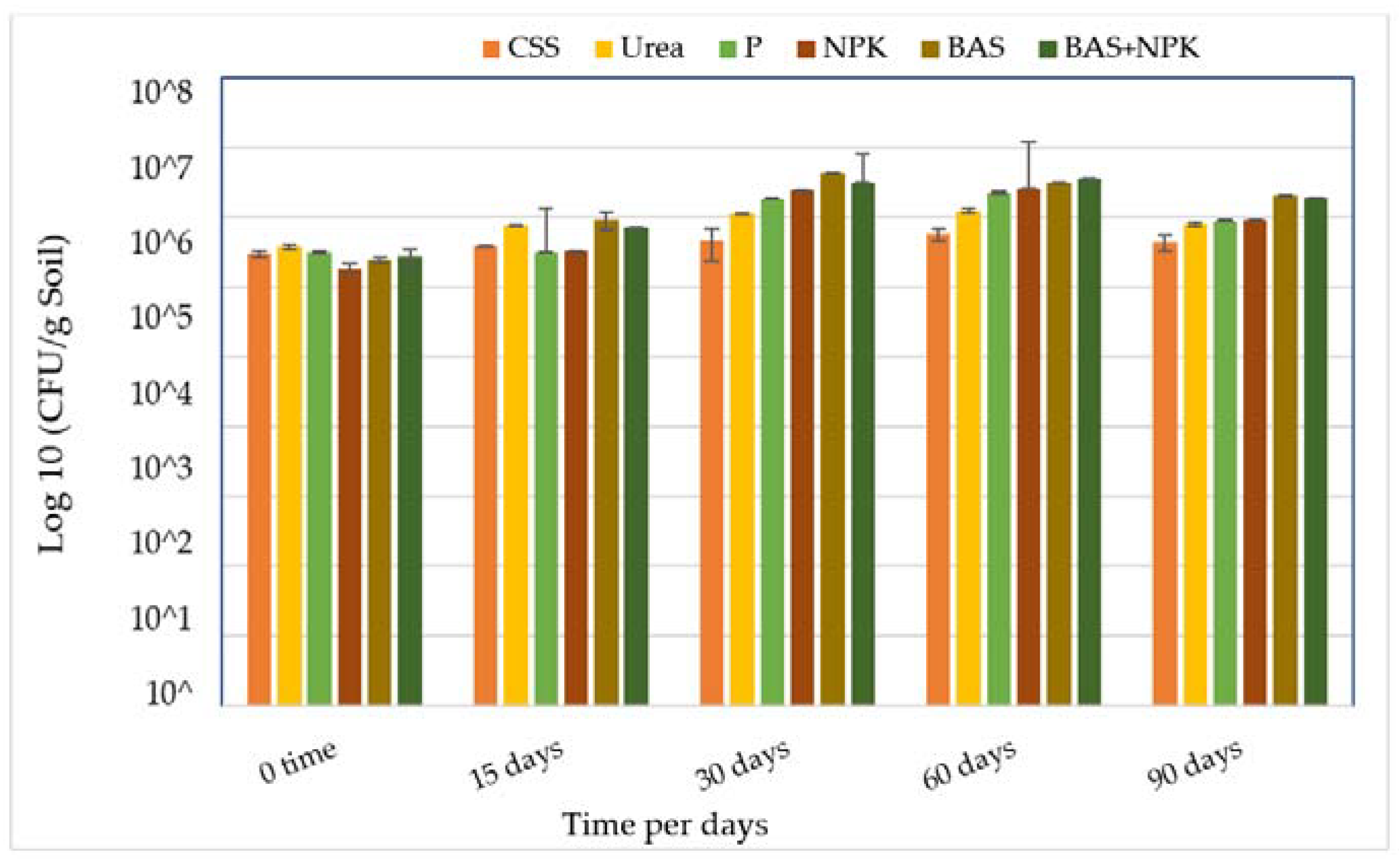
Figure 1.
Changes in hydrocarbon degrader counts (CFU g−1 soil) in contaminated soil samples with treatments. Bars represent the average of triplicate plate counts; error bars indicate the standard deviation of the triplicates. Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 1.
Changes in hydrocarbon degrader counts (CFU g−1 soil) in contaminated soil samples with treatments. Bars represent the average of triplicate plate counts; error bars indicate the standard deviation of the triplicates. Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 2.
Reduction in TPHs in contaminated soil samples with treatments. Bars represent the average of triplicate microcosms readings; error bars indicate the standard deviation of the triplicates. Abbreviations: Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 2.
Reduction in TPHs in contaminated soil samples with treatments. Bars represent the average of triplicate microcosms readings; error bars indicate the standard deviation of the triplicates. Abbreviations: Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 3.
Representative GC chromatograms of TPHs extracted from petroleum-contaminated soil after 15 days of incubation, shown as percentages remaining compared to those at the initiation of the experiment (zero time). Bars represent the average of triplicate samples. Error bars indicate the standard deviation of the triplicates. Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 3.
Representative GC chromatograms of TPHs extracted from petroleum-contaminated soil after 15 days of incubation, shown as percentages remaining compared to those at the initiation of the experiment (zero time). Bars represent the average of triplicate samples. Error bars indicate the standard deviation of the triplicates. Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 4.
Representative GC chromatograms of TPHs extracted from petroleum-contaminated soil after 90 days of incubation shown as percentages remaining compared to those at the initiation of the experiment (zero time). Bars represent the means of triplicate samples. Error bars indicate the standard deviation of the triplicates. Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 4.
Representative GC chromatograms of TPHs extracted from petroleum-contaminated soil after 90 days of incubation shown as percentages remaining compared to those at the initiation of the experiment (zero time). Bars represent the means of triplicate samples. Error bars indicate the standard deviation of the triplicates. Abbreviations: CSS, control soil sample; P, phosphorus; NPK, nitrogen, phosphorous, and potassium; BAS: bio-augmentation soil.
Figure 5.
Biotoxicity values (acute toxicity assay) during bioremediation treatment determined by V. fisheri bacteria.
Figure 5.
Biotoxicity values (acute toxicity assay) during bioremediation treatment determined by V. fisheri bacteria.
Table 1.
Supplements added to the different treatments at the beginning of the experiments.
Table 1.
Supplements added to the different treatments at the beginning of the experiments.
| Treatments | Supplements Added |
|---|
| Treatment 1 | None (control) |
| Treatment 2 | Urea |
| Treatment 3 | Phosphorous |
| Treatment 4 | Agricultural fertilizers (N.P.K) |
| Treatment 5 | Bacterial inoculum (consortium 1) |
| Treatment 6 | Equal mixtures of N.P.K, bacterial inoculum (consortium 1) |
Table 2.
General physical and chemical properties of dry contaminated soil.
Table 2.
General physical and chemical properties of dry contaminated soil.
| Contaminated Soil Properties |
|---|
| Sampling location | Southeastern region of Tripoli |
| Texture | Sandy |
| pH | 7.64 |
| Water holding capacity (%) | 4.39 |
| Available C (ppm) | 1.58 |
| Available N (ppm) | 0.051 |
| Available P (ppm) | 44.25 |
| Available K (ppm) | 66.24 |
| TPH (mg/kg) | 6571.08 |
| CFU/g soil bacteria | 2.40 × 106 |
| CFU/g soil fungi | 6.9 × 105 |
| As (ppm) | 19.72 |
| Ba (ppm) | 48.01 |
| Cd (ppm) | <0.002 |
| Total Cr (ppm) | 105.06 |
| Total Hg (ppm) | 166.21 |
| Pb (ppm) | 205.32 |
| Ni (ppm) | 121.77 |
| Zn (ppm) | 234.14 |
| Hg (ppb) | 5.83 |
Table 3.
Minerology composition of soil sample using XRD analysis.
Table 3.
Minerology composition of soil sample using XRD analysis.
| Ref. Code | Compound Name | Chemical Name | Chemical Formula | Approximate % |
|---|
| 44-1415 | Lepidocrocite | Iron Oxide Hydroxide | FeO(OH) | 58.04 |
| 19-0932 | Microcline | Potassium Aluminum Silicate | KAISi3O8 | 41.46 |
| 06-0615 | Wustite | Iron Oxide | FeO | 0.50 |
Table 4.
Identification of selected hydrocarbon-degrading bacterial isolates via 16S rDNA gene sequencing.
Table 4.
Identification of selected hydrocarbon-degrading bacterial isolates via 16S rDNA gene sequencing.
| Isolates | Species as Close
Relatives | Accession No. | Similarity (%) |
|---|
| HKI | Rhodococcus ruber | CP038030.2 | 100.0 |
| HKII | Beta proteobacterium | EF599311.1 | 99.1 |
Table 5.
Changes in soil pH during the experiment.
Table 5.
Changes in soil pH during the experiment.
| * Treatments | pH (In Double Distilled H2O) |
|---|
| 0 Day | 15 Days | 30 Days | 60 Days | 90 Days |
|---|
| CSS | 7.5 ± 0.1 | 7.1 ± 0.1 | 6.6 ± 0.2 | 7.0 ± 0.2 | 7.1 ± 0.2 |
| UREA | 7.8 ± 0.0 | 7.9 ± 0.3 | 7.7 ± 0.2 | 7.2 ± 0.1 | 6.8 ± 0.2 |
| P | 7.7 ± 0.1 | 7.9 ± 0.2 | 7.5 ± 0.1 | 6.7 ± 0.2 | 7.6 ± 0.1 |
| NPK | 7.6 ± 0.2 | 8.2 ± 0.1 | 7.3 ± 0.2 | 7.1 ± 0.1 | 7.1 ± 0.1 |
| BAS | 7.4 ± 0.2 | 8.1 ± 0.2 | 8.3 ± 0.3 | 7.1 ± 0.1 | 6.8 ± 0.2 |
| BAS + NPK | 7.7 ± 0.1 | 8.4 ± 0.3 | 7.9 ± 0.1 | 6.4 ± 0.1 | 7.2 ± 0.1 |
Table 6.
TPH reduction in contaminated soil samples with treatments.
Table 6.
TPH reduction in contaminated soil samples with treatments.
| Treatments | 0 Time | 15 Days | 30 Days | 60 Days | 90 Days |
|---|
| CSS | 0% | 12% | 24% | 24% | 31% |
| Urea | 0% | 15% | 27% | 30% | 43% |
| P | 0% | 12% | 24% | 31% | 33% |
| NPK | 0% | 22% | 27% | 40% | 45% |
| BAS | 0% | 27% | 33% | 52% | 61% |
| BAS + NPK | 0% | 30% | 43% | 58% | 73% |
Table 7.
Heavy metal quantity of five selected soil treatments (urea, P, NPK, BAS, BAS + NPK) measured in μg mL−1 at the start of the experiment.
Table 7.
Heavy metal quantity of five selected soil treatments (urea, P, NPK, BAS, BAS + NPK) measured in μg mL−1 at the start of the experiment.
| Treatments | As | Ba | Cd | Cr | Cu | Pb | Ni | Zn | Hg |
|---|
| Contaminated soil samples (PPM) | 56.95 | 48.01 | <0.002 | 105.06 | 166.21 | 205.32 | 121.77 | 234.14 | 5.83 ppb |
Table 8.
Heavy metal quantity of five selected soil treatments (urea, P, NPK, BAS, BAS + NPK) measured in μg mL−1 at 90 days.
Table 8.
Heavy metal quantity of five selected soil treatments (urea, P, NPK, BAS, BAS + NPK) measured in μg mL−1 at 90 days.
| Treatments | As | Ba | Cd | Cr | Cu | Pb | Ni | Zn | Hg |
|---|
| CSS | 37.29 | 45.59 | <0.002 | 91.18 | 144.99 | 178.27 | 87.78 | 150.86 | 5.37 |
| Urea | 38.57 | 41.94 | <0.002 | 67.85 | 94.74 | 147.52 | 62.23 | 145.55 | 3.94 |
| P | 31.2 | 38.01 | <0.002 | 85.6 | 108.27 | 166.21 | 73.72 | 143.36 | 3.94 |
| NPK | 19.72 | 33.59 | <0.002 | 69.47 | 128.2 | 142.27 | 65.45 | 145 | 4.33 |
| BAS | 20.59 | 32.35 | <0.002 | 72.91 | 133.19 | 142.34 | 96.77 | 139.25 | 4.13 |
| BAS + NPK | 13.35 | 26.51 | <0.002 | 66.87 | 103.9 | 142.34 | 74.61 | 125.79 | 2.86 |