Taxonomic Re-Classification and Expansion of the Phylum Chloroflexota Based on over 5000 Genomes and Metagenome-Assembled Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Public Genome and MAG Collection
2.2. Metagenome Dataset Selection, Categorization, and Downloading
2.3. Metagenomes Obtained from Novel Environmental Samples
2.4. Assembling, Mapping, and Binning
2.5. Quality Processing, Preliminary Taxonomic Classification, and Clustering
2.6. Annotations
2.7. Phylogenomics Analyses
3. Results and Discussion
3.1. Novel Chloroflexota MAGs Assembled from Public Metagenome Datasets and Newly Sampled Habitats
3.2. Ca. Dormibacteria as Class of Chloroflexota According to Relative Evolutionary Divergence
3.3. Chloroflexota Classes According to Genome-Based Phylogenetic Analysis
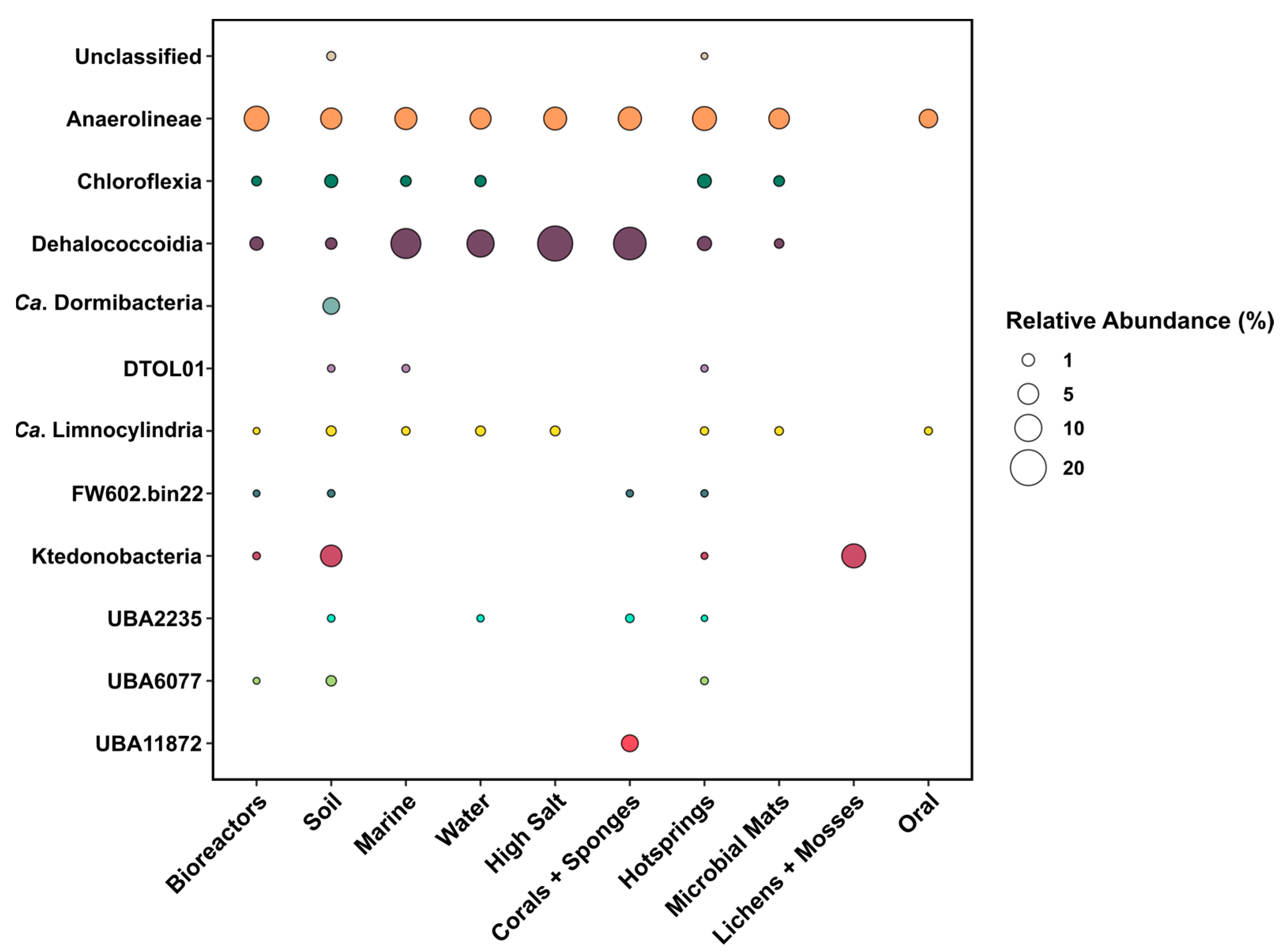
3.4. Features of Chloroflexota Classes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biddle, J.F.; Fitz-Gibbon, S.; Schuster, S.C.; Brenchley, J.E.; House, C.H. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. USA 2008, 105, 10583–10588. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Holt, J.G. The Archaea and the Deeply Branching and Phototrophic Bacteria. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Phylum BVI. Chloroflexi phy. nov.; Boone, D.R., Castenholz, R.W., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2001; Volume 1, pp. 427–446. [Google Scholar]
- Hug, L.A.; Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Frischkorn, K.R.; Williams, K.H.; Tringe, S.G.; Banfield, J.F. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 2013, 1, 22. [Google Scholar] [CrossRef] [PubMed]
- Kaster, A.K.; Mayer-Blackwell, K.; Pasarelli, B.; Spormann, A.M. Single cell genomic study of Dehalococcoidetes species from deep-sea sediments of the Peruvian Margin. ISME J. 2014, 8, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- Sewell, H.L.; Kaster, A.K.; Spormann, A.M. Homoacetogenesis in Deep-Sea Chloroflexi, as Inferred by Single-Cell Genomics, Provides a Link to Reductive Dehalogenation in Terrestrial Dehalococcoidetes. mbio 2017, 8, 10–1128. [Google Scholar] [CrossRef]
- Speirs, L.B.M.; Rice, D.T.F.; Petrovski, S.; Seviour, R.J. The Phylogeny, Biodiversity, and Ecology of the Chloroflexi in Activated Sludge. Front. Microbiol. 2019, 10, 2015. [Google Scholar] [CrossRef]
- Yamada, T.; Sekiguchi, Y.; Hanada, S.; Imachi, H.; Ohashi, A.; Harada, H.; Kamagata, Y. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2006, 56 Pt 6, 1331–1340. [Google Scholar] [CrossRef]
- Denef, V.J.; Mueller, R.S.; Chiang, E.; Liebig, J.R.; Vanderploeg, H.A. Chloroflexi CL500-11 Populations That Predominate Deep-Lake Hypolimnion Bacterioplankton Rely on Nitrogen-Rich Dissolved Organic Matter Metabolism and C1 Compound Oxidation. Appl. Environ. Microbiol. 2015, 82, 1423–1432. [Google Scholar] [CrossRef]
- Mehrshad, M.; Salcher, M.M.; Okazaki, Y.; Nakano, S.I.; Simek, K.; Andrei, A.S.; Ghai, R. Hidden in plain sight-highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome 2018, 6, 176. [Google Scholar] [CrossRef]
- Okazaki, Y.; Salcher, M.M.; Callieri, C.; Nakano, S.I. The Broad Habitat Spectrum of the CL500-11 Lineage (Phylum Chloroflexi), a Dominant Bacterioplankton in Oxygenated Hypolimnia of Deep Freshwater Lakes. Front. Microbiol. 2018, 9, 2891. [Google Scholar] [CrossRef]
- Suominen, S.; van Vliet, D.M.; Sanchez-Andrea, I.; van der Meer, M.T.J.; Sinninghe Damste, J.S.; Villanueva, L. Organic Matter Type Defines the Composition of Active Microbial Communities Originating From Anoxic Baltic Sea Sediments. Front. Microbiol. 2021, 12, 628301. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, E.R.; Payne, J.A.; Young, R.M.; Starr, M.G.; Perry, M.P.; Fahnestock, S.; Ellis, D.E.; Ebersole, R.C. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 2002, 68, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Müller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Cavaletti, L.; Monciardini, P.; Bamonte, R.; Schumann, P.; Rohde, M.; Sosio, M.; Donadio, S. New lineage of filamentous, spore-forming, gram-positive bacteria from soil. Appl. Environ. Microbiol. 2006, 72, 4360–4369. [Google Scholar] [CrossRef]
- Zheng, Y.; Saitou, A.; Wang, C.M.; Toyoda, A.; Minakuchi, Y.; Sekiguchi, Y.; Ueda, K.; Takano, H.; Sakai, Y.; Abe, K.; et al. Genome Features and Secondary Metabolites Biosynthetic Potential of the Class Ktedonobacteria. Front. Microbiol. 2019, 10, 893. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Chander, P.; George, S. Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexia class. nov. [corrected] into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. nov. Antonie Van Leeuwenhoek 2013, 103, 99–119. [Google Scholar] [CrossRef] [PubMed]
- Pierson, B.K.; Castenholz, R.W. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 1974, 100, 5–24. [Google Scholar] [CrossRef]
- Ludwig, W.; Viver, T.; Westram, R.; Francisco Gago, J.; Bustos-Caparros, E.; Knittel, K.; Amann, R.; Rossello-Mora, R. Release LTP_12_2020, featuring a new ARB alignment and improved 16S rRNA tree for prokaryotic type strains. Syst. Appl. Microbiol. 2021, 44, 126218. [Google Scholar] [CrossRef]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Coleman, G.A.; Davin, A.A.; Mahendrarajah, T.A.; Szantho, L.L.; Spang, A.; Hugenholtz, P.; Szollosi, G.J.; Williams, T.A. A rooted phylogeny resolves early bacterial evolution. Science 2021, 372, eabe0511. [Google Scholar] [CrossRef]
- Martinez-Gutierrez, C.A.; Aylward, F.O. Phylogenetic Signal, Congruence, and Uncertainty across Bacteria and Archaea. Mol. Biol. Evol. 2021, 38, 5514–5527. [Google Scholar] [CrossRef] [PubMed]
- Taib, N.; Megrian, D.; Witwinowski, J.; Adam, P.; Poppleton, D.; Borrel, G.; Beloin, C.; Gribaldo, S. Genome-wide analysis of the Firmicutes illuminates the diderm/monoderm transition. Nat. Ecol. Evol. 2020, 4, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Battistuzzi, F.U.; Hedges, S.B. A major clade of prokaryotes with ancient adaptations to life on land. Mol. Biol. Evol. 2009, 26, 335–343. [Google Scholar] [CrossRef]
- Sutcliffe, I.C. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010, 18, 464–470. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Montgomery, K.; Williams, T.J.; Brettle, M.; Berengut, J.F.; Ray, A.E.; Zhang, E.; Zaugg, J.; Hugenholtz, P.; Ferrari, B.C. Persistence and resistance: Survival mechanisms of Candidatus Dormibacterota from nutrient-poor Antarctic soils. Environ. Microbiol. 2021, 23, 4276–4294. [Google Scholar] [CrossRef]
- Ji, M.; Greening, C.; Vanwonterghem, I.; Carere, C.R.; Bay, S.K.; Steen, J.A.; Montgomery, K.; Lines, T.; Beardall, J.; van Dorst, J.; et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 2017, 552, 400–403. [Google Scholar] [CrossRef]
- Zhou, J.; Xia, B.; Huang, H.; Treves, D.S.; Hauser, L.J.; Mural, R.J.; Palumbo, A.V.; Tiedje, J.M. Bacterial phylogenetic diversity and a novel candidate division of two humid region, sandy surface soils. Soil Biol. Biochem. 2003, 35, 915–924. [Google Scholar] [CrossRef]
- Costello, E.K.; Schmidt, S.K. Microbial diversity in alpine tundra wet meadow soil: Novel Chloroflexi from a cold, water-saturated environment. Environ. Microbiol. 2006, 8, 1471–1486. [Google Scholar] [CrossRef]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, D136–D143. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glockner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.A.; Hugenholtz, P. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef] [PubMed]
- Kawaichi, S.; Ito, N.; Kamikawa, R.; Sugawara, T.; Yoshida, T.; Sako, Y. Ardenticatena maritima gen. nov., sp. nov., a ferric iron- and nitrate-reducing bacterium of the phylum ‘Chloroflexi’ isolated from an iron-rich coastal hydrothermal field, and description of Ardenticatenia classis nov. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 8, 2992–3002. [Google Scholar] [CrossRef] [PubMed]
- Kochetkova, T.V.; Zayulina, K.S.; Zhigarkov, V.S.; Minaev, N.V.; Chichkov, B.N.; Novikov, A.A.; Toshchakov, S.V.; Elcheninov, A.G.; Kublanov, I.V. Tepidiforma bonchosmolovskayae gen. nov., sp. nov., a moderately thermophilic Chloroflexi bacterium from a Chukotka hot spring (Arctic, Russia), representing a novel class, Tepidiformia, which includes the previously uncultivated lineage OLB14. Int. J. Syst. Evol. Microbiol. 2020, 70, 1192–1202. [Google Scholar] [CrossRef]
- Dodsworth, J.A.; Gevorkian, J.; Despujos, F.; Cole, J.K.; Murugapiran, S.K.; Ming, H.; Li, W.J.; Zhang, G.; Dohnalkova, A.; Hedlund, B.P. Thermoflexus hugenholtzii gen. nov., sp. nov., a thermophilic, microaerophilic, filamentous bacterium representing a novel class in the Chloroflexi, Thermoflexia classis nov., and description of Thermoflexaceae fam. nov. and Thermoflexales ord. nov. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 6, 2119–2127. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Stackebrandt, E. Reclassification of Sphaerobacter thermophilus from the subclass Sphaerobacteridae in the phylum Actinobacteria to the class Thermomicrobia (emended description) in the phylum Chloroflexi (emended description). Int. J. Syst. Evol. Microbiol. 2004, 54 Pt 6, 2049–2051. [Google Scholar] [CrossRef][Green Version]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Ortiz, M.; Leung, P.M.; Shelley, G.; Jirapanjawat, T.; Nauer, P.A.; Van Goethem, M.W.; Bay, S.K.; Islam, Z.F.; Jordaan, K.; Vikram, S.; et al. Multiple energy sources and metabolic strategies sustain microbial diversity in Antarctic desert soils. Proc. Natl. Acad. Sci. USA 2021, 118, e2025322118. [Google Scholar] [CrossRef]
- Anantharaman, K.; Hausmann, B.; Jungbluth, S.P.; Kantor, R.S.; Lavy, A.; Warren, L.A.; Rappe, M.S.; Pester, M.; Loy, A.; Thomas, B.C.; et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018, 12, 1715–1728. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Cai, R.; Wang, C.; Liu, R.; Sun, C. Characterization of the First Cultured Representative of “Candidatus Thermofonsia” Clade 2 within Chloroflexi Reveals Its Phototrophic Lifestyle. mBio 2022, 13, e0028722. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Imelfort, M.; Parks, D.; Woodcroft, B.J.; Dennis, P.; Hugenholtz, P.; Tyson, G.W. GroopM: An automated tool for the recovery of population genomes from related metagenomes. PeerJ 2014, 2, e603. [Google Scholar] [CrossRef]
- Yue, Y.; Huang, H.; Qi, Z.; Dou, H.M.; Liu, X.Y.; Han, T.F.; Chen, Y.; Song, X.J.; Zhang, Y.H.; Tu, J. Evaluating metagenomics tools for genome binning with real metagenomic datasets and CAMI datasets. BMC Bioinform. 2020, 21, 334. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.M.; Huntemann, M.; et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 2021, 39, 499–509. [Google Scholar] [CrossRef]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Katz, K.S.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. STAT: A fast, scalable, MinHash-based k-mer tool to assess Sequence Read Archive next-generation sequence submissions. Genome Biol. 2021, 22, 270. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Dam, H.T.; Vollmers, J.; Sobol, M.S.; Cabezas, A.; Kaster, A.K. Targeted Cell Sorting Combined with Single Cell Genomics Captures Low Abundant Microbial Dark Matter with Higher Sensitivity Than Metagenomics. Front. Microbiol. 2020, 11, 1377. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef]
- Vollmers, J.; Wiegand, S.; Lenk, F.; Kaster, A.K. How clear is our current view on microbial dark matter? (Re-)assessing public MAG & SAG datasets with MDMcleaner. Nucleic Acids Res. 2022, 50, e76. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, S.; Dam, H.T.; Riba, J.; Vollmers, J.; Kaster, A.K. Printing Microbial Dark Matter: Using Single Cell Dispensing and Genomics to Investigate the Patescibacteria/Candidate Phyla Radiation. Front. Microbiol. 2021, 12, 635506. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Shi, Z.J.; Seshadri, R.; Pollard, K.S.; Kyrpides, N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature 2019, 568, 505–510. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bayer, K.; Jahn, M.T.; Slaby, B.M.; Moitinho-Silva, L.; Hentschel, U. Marine Sponges as Chloroflexi Hot Spots: Genomic Insights and High-Resolution Visualization of an Abundant and Diverse Symbiotic Clade. mSystems 2018, 3, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Bowers, R.M.; Nayfach, S.; Schulz, F.; Jungbluth, S.P.; Ruhl, I.A.; Sheremet, A.; Lee, J.; Goudeau, D.; Eloe-Fadrosh, E.A.; Stepanauskas, R.; et al. Dissecting the dominant hot spring microbial populations based on community-wide sampling at single-cell genomic resolution. ISME J. 2022, 16, 1337–1347. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, S.; Xue, Q.; Chen, J.; Zhou, J.; Cheng, F.; Li, M.; Zhu, Y.; Yu, H.; Hu, S.; et al. Abundant Taxa and Favorable Pathways in the Microbiome of Soda-Saline Lakes in Inner Mongolia. Front. Microbiol. 2020, 11, 1740. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Shih, P.M.; Ward, L.M.; Fischer, W.W. Evolution of the 3-hydroxypropionate bicycle and recent transfer of anoxygenic photosynthesis into the Chloroflexi. Proc. Natl. Acad. Sci. USA 2017, 114, 10749–10754. [Google Scholar] [CrossRef]
- Reva, O.; Tümmler, B. Think big—Giant genes in bacteria. Environ. Microbiol. 2008, 10, 768–777. [Google Scholar] [CrossRef]
- Xia, Y.; Wang, Y.; Wang, Y.; Chin, F.Y.; Zhang, T. Cellular adhesiveness and cellulolytic capacity in Anaerolineae revealed by omics-based genome interpretation. Biotechnol. Biofuels 2016, 9, 111. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Behrens, S.F.; Müller, J.A.; Goke, J.; Ritalahti, K.M.; Wagner, R.; Goltsman, E.; Lapidus, A.; Holmes, S.; Loffler, F.E.; et al. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet 2009, 5, e1000714. [Google Scholar] [CrossRef]
- Wasmund, K.; Schreiber, L.; Lloyd, K.G.; Petersen, D.G.; Schramm, A.; Stepanauskas, R.; Jørgensen, B.B.; Adrian, L. Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J. 2014, 8, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Ahsanul Islam, M.; Edwards, E.A.; Mahadevan, R. Characterizing the metabolism of Dehalococcoides with a constraint-based model. PLoS Comput. Biol. 2010, 6, e1000887. [Google Scholar] [CrossRef] [PubMed]
- Kube, M.; Beck, A.; Zinder, S.H.; Kuhl, H.; Reinhardt, R.; Adrian, L. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 2005, 23, 1269–1273. [Google Scholar] [CrossRef] [PubMed]
- Pöritz, M.; Schiffmann, C.L.; Hause, G.; Heinemann, U.; Seifert, J.; Jehmlich, N.; von Bergen, M.; Nijenhuis, I.; Lechner, U. Dehalococcoides mccartyi strain DCMB5 respires a broad spectrum of chlorinated aromatic compounds. Appl. Environ. Microbiol. 2015, 81, 587–596. [Google Scholar] [CrossRef] [PubMed]
- West-Roberts, J.A.; Matheus-Carnevali, P.B.; Schoelmerich, M.C.; Al-Shayeb, B.; Thomas, A.D.; Sharrar, A.; He, C.; Chen, L.X.; Lavy, A.; Keren, R.; et al. The Chloroflexi supergroup is metabolically diverse and representatives have novel genes for non-photosynthesis based CO2 fixation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Maymo-Gatell, X.; Chien, Y.T.; Gossett, J.M.; Zinder, S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 1997, 276, 1568–1571. [Google Scholar] [CrossRef]
- Martinez-Torro, C.; Torres-Puig, S.; Marcos-Silva, M.; Huguet-Ramon, M.; Munoz-Navarro, C.; Lluch-Senar, M.; Serrano, L.; Querol, E.; Pinol, J.; Pich, O.Q. Functional Characterization of the Cell Division Gene Cluster of the Wall-less Bacterium Mycoplasma genitalium. Front. Microbiol. 2021, 12, 695572. [Google Scholar] [CrossRef]
- Orsi, W.D.; Schink, B.; Buckel, W.; Martin, W.F. Physiological limits to life in anoxic subseafloor sediment. FEMS Microbiol. Rev. 2020, 44, 219–231. [Google Scholar] [CrossRef]
- Daniel, R.A.; Errington, J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell 2003, 113, 767–776. [Google Scholar] [CrossRef]
- Zemskaya, T.I.; Cabello-Yeves, P.J.; Pavlova, O.N.; Rodriguez-Valera, F. Microorganisms of Lake Baikal-the deepest and most ancient lake on Earth. Appl. Microbiol. Biotechnol. 2020, 104, 6079–6090. [Google Scholar] [CrossRef]
- Colatriano, D.; Tran, P.Q.; Gueguen, C.; Williams, W.J.; Lovejoy, C.; Walsh, D.A. Genomic evidence for the degradation of terrestrial organic matter by pelagic Arctic Ocean Chloroflexi bacteria. Commun. Biol. 2018, 1, 90. [Google Scholar] [CrossRef]
- Landry, Z.; Swan, B.K.; Herndl, G.J.; Stepanauskas, R.; Giovannoni, S.J. SAR202 Genomes from the Dark Ocean Predict Pathways for the Oxidation of Recalcitrant Dissolved Organic Matter. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wei, X.; Song, W.; Wang, L.; Cao, J.; Wu, J.; Thomas, T.; Jin, T.; Wang, Z.; Wei, W.; et al. Novel Chloroflexi genomes from the deepest ocean reveal metabolic strategies for the adaptation to deep-sea habitats. Microbiome 2022, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Krzmarzick, M.J.; Crary, B.B.; Harding, J.J.; Oyerinde, O.O.; Leri, A.C.; Myneni, S.C.; Novak, P.J. Natural niche for organohalide-respiring Chloroflexi. Appl. Environ. Microbiol. 2012, 78, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Schipp, C.J.; Marco-Urrea, E.; Kublik, A.; Seifert, J.; Adrian, L. Organic cofactors in the metabolism of Dehalococcoides mccartyi strains. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120321. [Google Scholar] [CrossRef]
- Zhuang, W.Q.; Yi, S.; Bill, M.; Brisson, V.L.; Feng, X.; Men, Y.; Conrad, M.E.; Tang, Y.J.; Alvarez-Cohen, L. Incomplete Wood-Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc. Natl. Acad. Sci. USA 2014, 111, 6419–6424. [Google Scholar] [CrossRef]
- Davis, K.E.; Sangwan, P.; Janssen, P.H. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 2011, 13, 798–805. [Google Scholar] [CrossRef]


| Class | Public 1 (All) | Public (h.q.) 4 | Binning 2 (All) | Binning (h.q.) | Environ. 3 (All) | Environ. (h.q.) | Sum (All) | Sum (h.q.) |
|---|---|---|---|---|---|---|---|---|
| Anaerolineae | 1522 | 563 | 968 | 367 | 28 | 11 | 2518 | 941 |
| Chloroflexia | 153 | 82 | 119 | 51 | 5 | 3 | 277 | 136 |
| Dehalococcoidia | 1100 | 331 | 605 | 207 | 23 | 10 | 1738 | 548 |
| Ca. Dormibacteria | 149 | 51 | 13 | 2 | 0 | 0 | 162 | 53 |
| Ktedonobacteria | 87 | 20 | 35 | 11 | 6 | 5 | 128 | 36 |
| Ca. Limnocylindria 5 | 254 | 50 | 23 | 8 | 5 | 4 | 282 | 62 |
| Thermomicrobia 6 | 112 | 39 | 22 | 12 | 7 | 4 | 141 | 55 |
| UBA11872 | 13 | 7 | 35 | 11 | 0 | 0 | 48 | 18 |
| UBA2235 | 43 | 15 | 5 | 4 | 2 | 1 | 50 | 20 |
| UBA4733 | 3 | 2 | 0 | 0 | 0 | 0 | 3 | 2 |
| UBA5177 | 9 | 2 | 0 | 0 | 0 | 0 | 9 | 2 |
| IMG ID 3300005529_81 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Sum | 3456 | 1162 | 1825 | 673 | 76 | 38 | 5357 | 1873 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiegand, S.; Sobol, M.; Schnepp-Pesch, L.K.; Yan, G.; Iqbal, S.; Vollmers, J.; Müller, J.A.; Kaster, A.-K. Taxonomic Re-Classification and Expansion of the Phylum Chloroflexota Based on over 5000 Genomes and Metagenome-Assembled Genomes. Microorganisms 2023, 11, 2612. https://doi.org/10.3390/microorganisms11102612
Wiegand S, Sobol M, Schnepp-Pesch LK, Yan G, Iqbal S, Vollmers J, Müller JA, Kaster A-K. Taxonomic Re-Classification and Expansion of the Phylum Chloroflexota Based on over 5000 Genomes and Metagenome-Assembled Genomes. Microorganisms. 2023; 11(10):2612. https://doi.org/10.3390/microorganisms11102612
Chicago/Turabian StyleWiegand, Sandra, Morgan Sobol, Luca Kristina Schnepp-Pesch, Geng Yan, Sajid Iqbal, John Vollmers, Jochen A. Müller, and Anne-Kristin Kaster. 2023. "Taxonomic Re-Classification and Expansion of the Phylum Chloroflexota Based on over 5000 Genomes and Metagenome-Assembled Genomes" Microorganisms 11, no. 10: 2612. https://doi.org/10.3390/microorganisms11102612
APA StyleWiegand, S., Sobol, M., Schnepp-Pesch, L. K., Yan, G., Iqbal, S., Vollmers, J., Müller, J. A., & Kaster, A.-K. (2023). Taxonomic Re-Classification and Expansion of the Phylum Chloroflexota Based on over 5000 Genomes and Metagenome-Assembled Genomes. Microorganisms, 11(10), 2612. https://doi.org/10.3390/microorganisms11102612







