Cellulose Synthase in Atacama Cyanobacteria and Bioethanol Production from Their Exopolysaccharides
Abstract
1. Introduction
2. Materials and Methods
2.1. Cyanobacteria Culture Conditions
2.2. Extraction and Quantification of Genomic DNA
2.3. Molecular Study of Cellulose Synthase Gene
2.3.1. PCR Amplification, Sequencing, and Similarity Searches for Cellulose Synthase Gene
2.3.2. Analysis of the Conserved Motif D,D,D,35QXXRW
2.3.3. Phylogenetic Analyses
2.4. EPS Purification
2.5. Enzymatic EPS Digestion and Fermentation
2.6. Glucose and Ethanol Determination
2.7. EPS Surface Morphology and Chemical Composition
3. Results and Discussion
3.1. Molecular Analysis
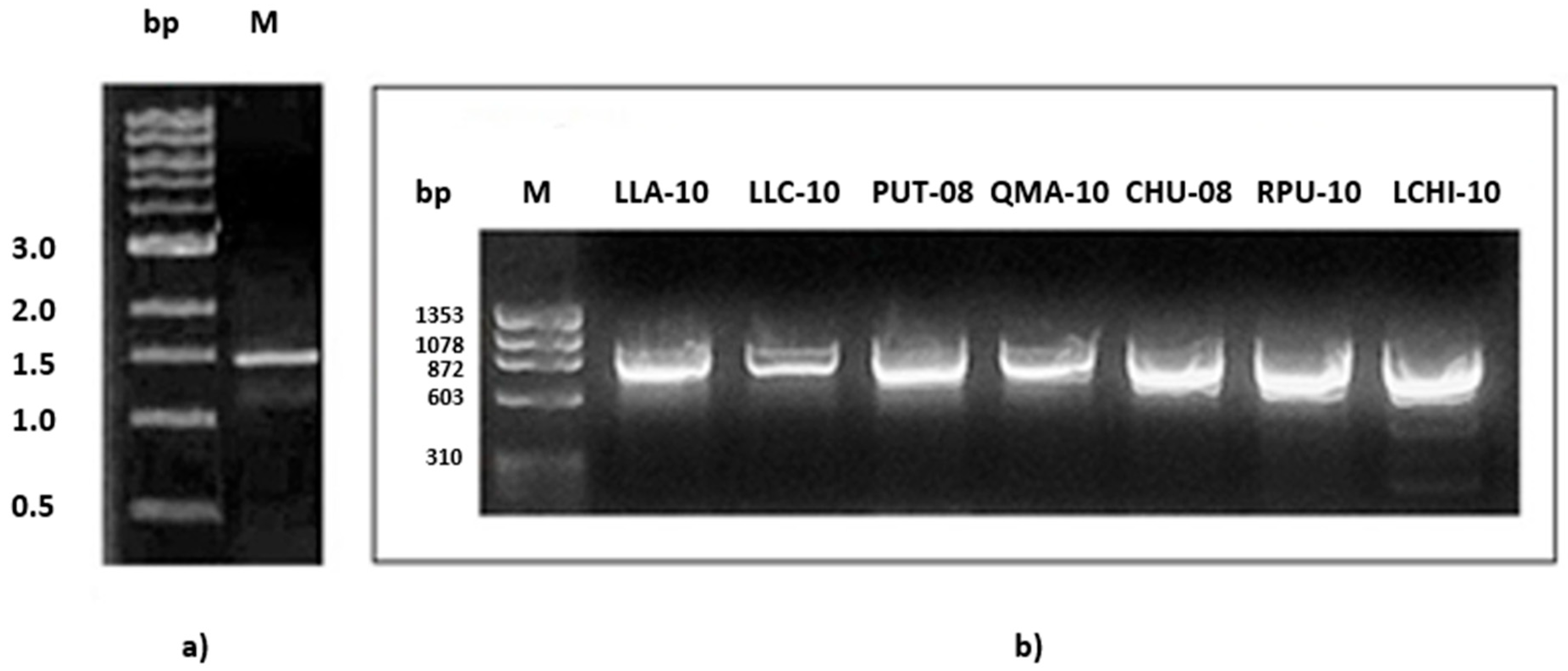
3.1.1. Cellulose Synthase Gene Amplification
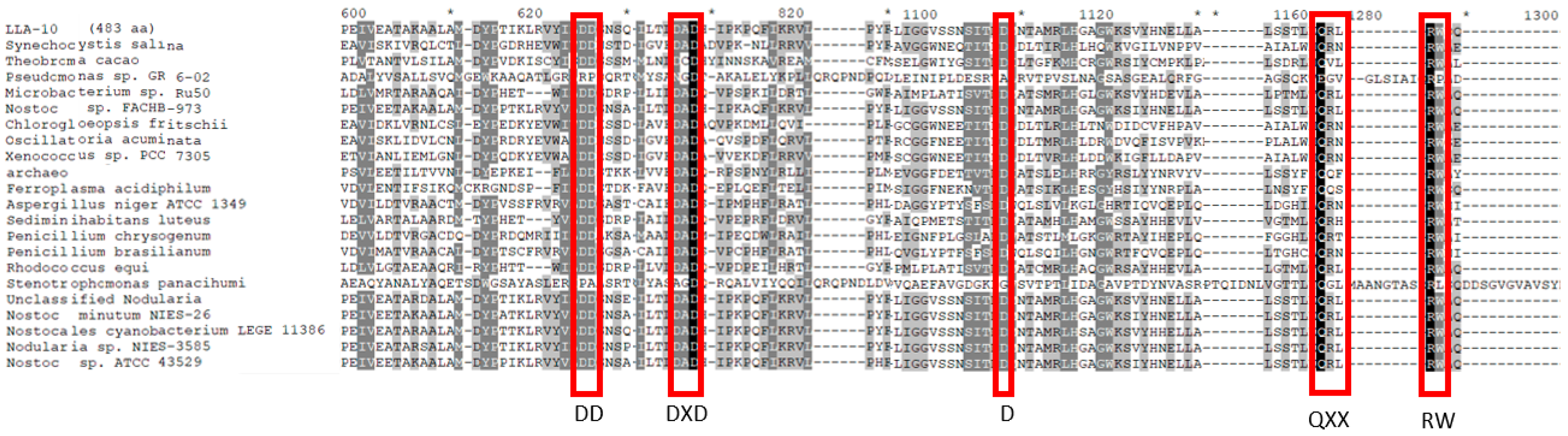
3.1.2. Cellulose Synthase Catalytic Domains Alignment
3.1.3. Alignment of the Catalytic Region of Cellulose Synthase from Strain LLA-10
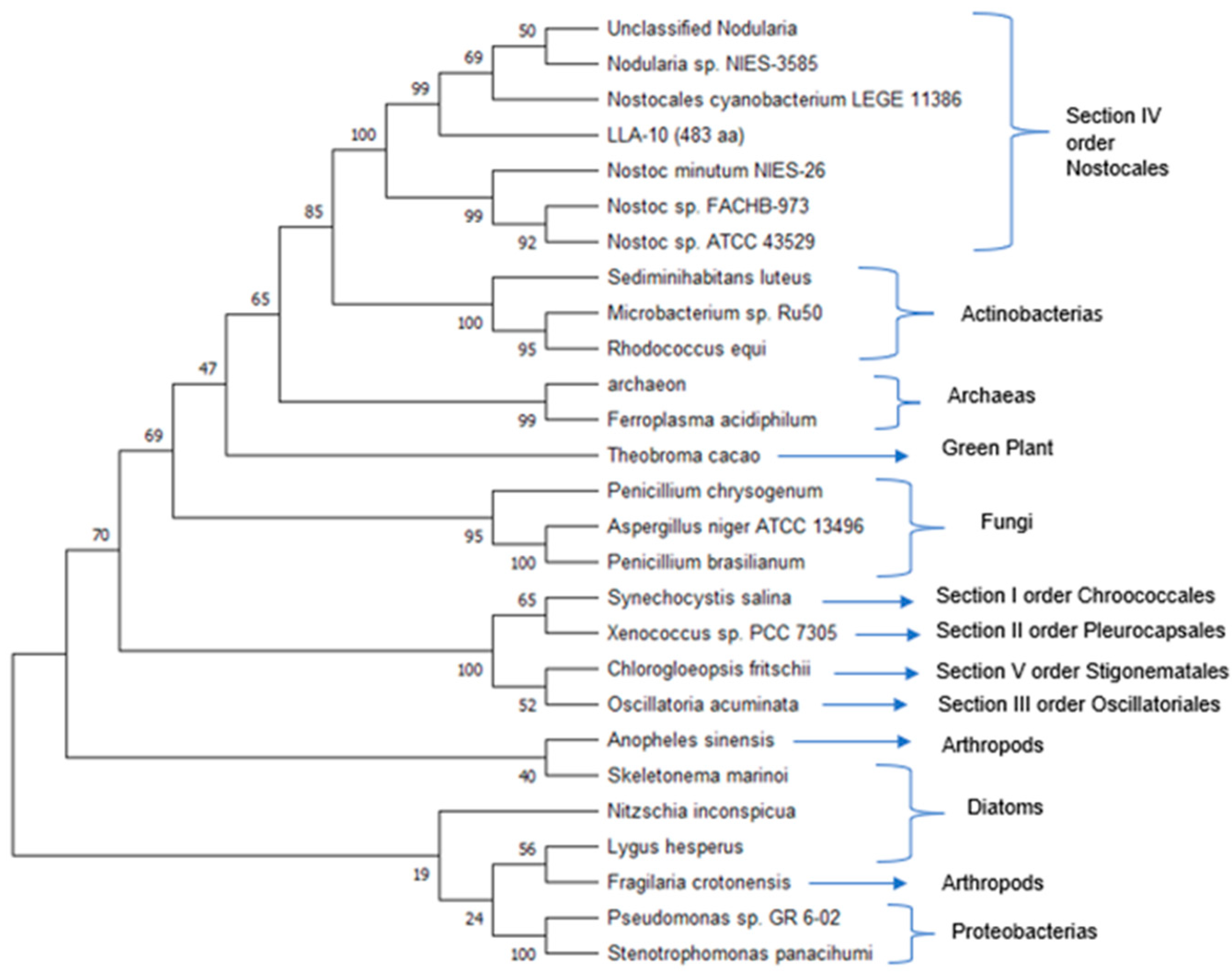
3.1.4. Phylogenetic Analysis with the Catalytic Region of Cellulose Synthase of Cyanobacterium Strain LLA-10
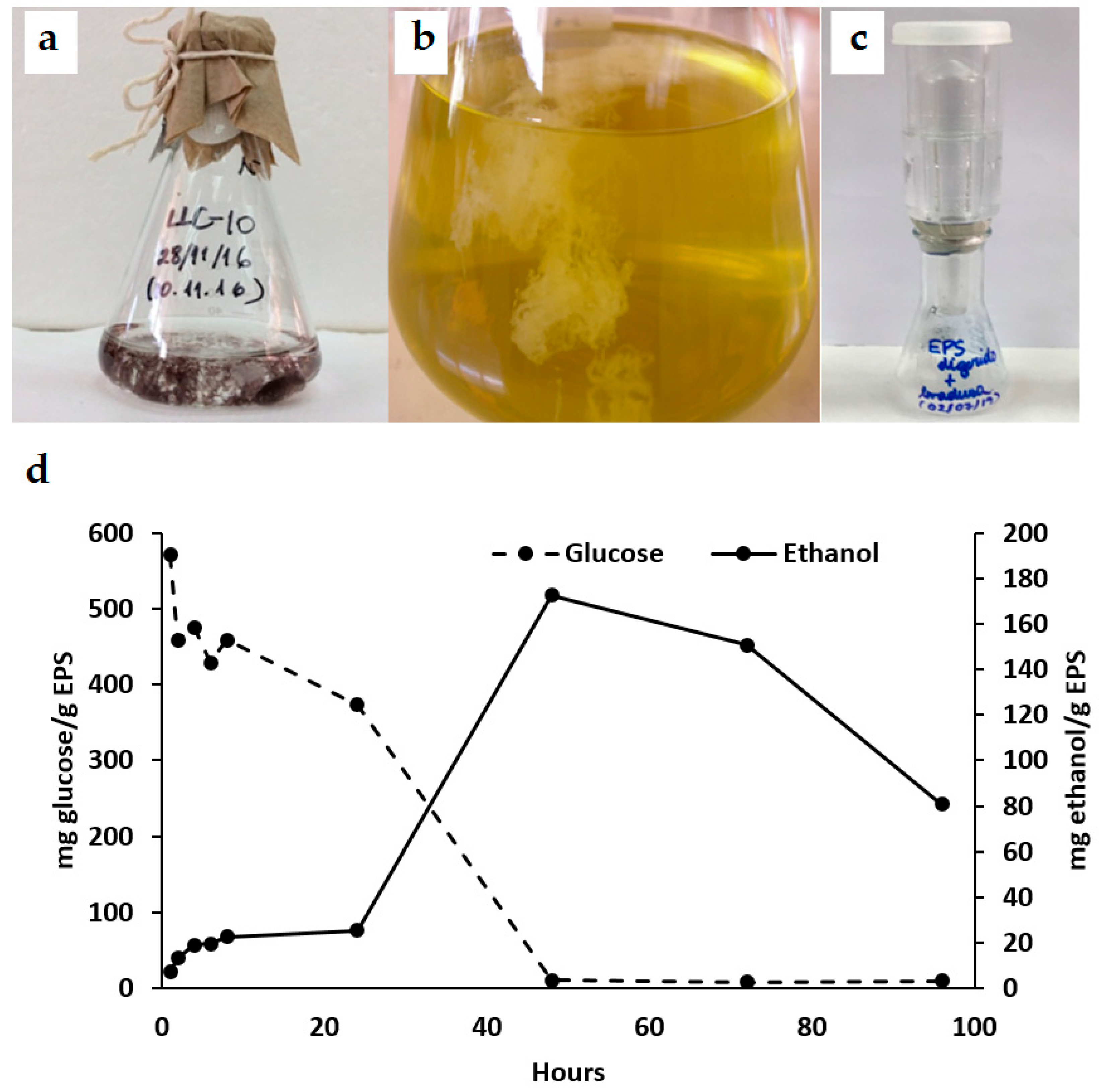
3.2. Enzymatic EPS Degradation and Fermentation
3.2.1. EPS Content among Cyanobacteria Strains
3.2.2. Fermentation and Ethanol Production from Cyanobacteria EPS
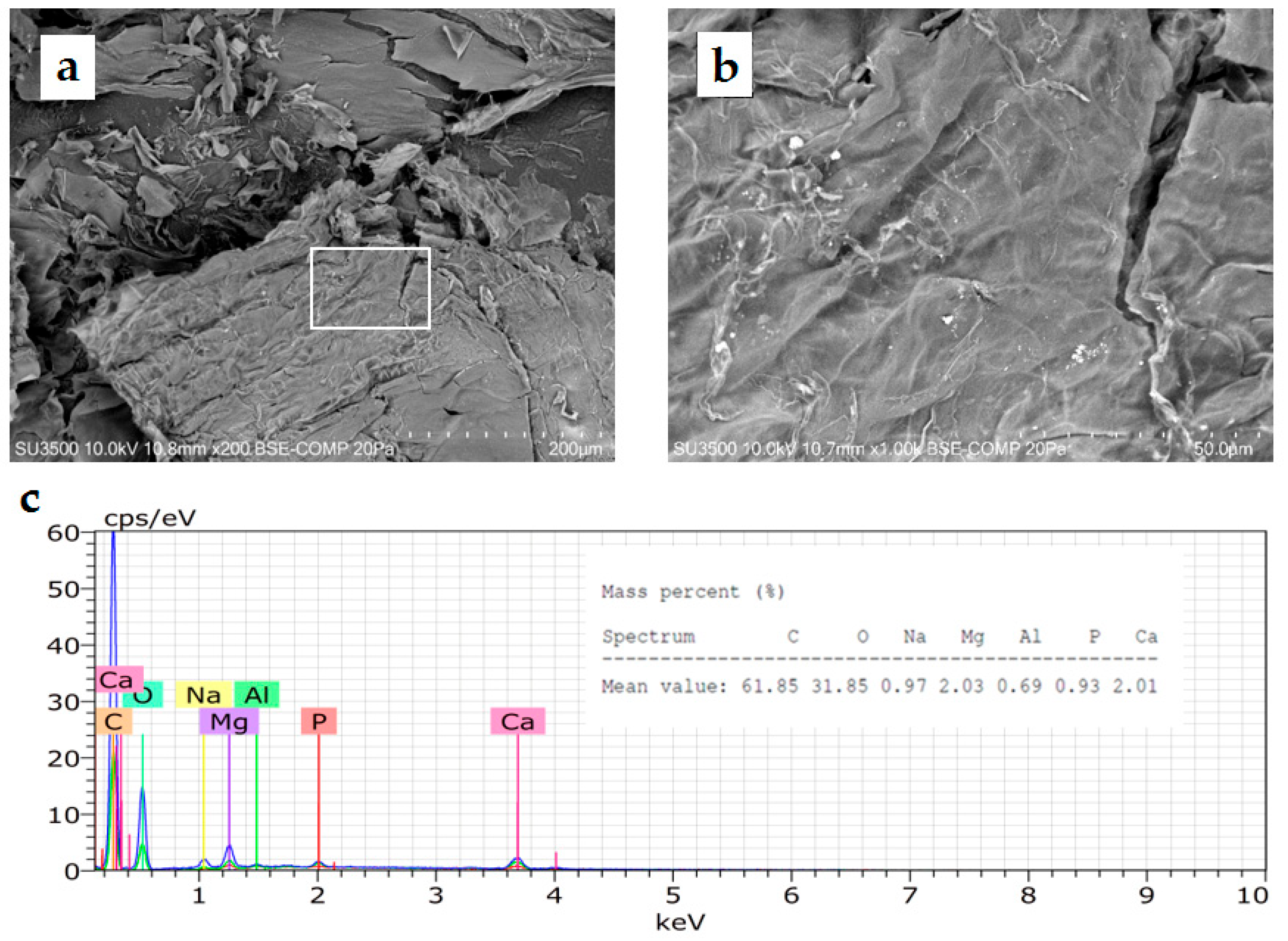
3.2.3. Microscopy Analysis of Cyanobacteria EPS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Philippis, R.; Vincenzini, M. Exocellular polysaccharides from cyanobacteria and their possible applications. FEMS Microbiol. Rev. 1998, 22, 151–175. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, Y.; Paulsen, B.; Klaveness, D. Studies on Polysaccharides from Three Edible Species of Nostoc (Cyanobacteria) with Different Colony Morphologies: Comparison of Monosaccharide Compositions and Viscosities of Polysaccharides from Field Colonies and Suspension Cultures. J. Phycol. 1998, 34, 962–968. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Decho, A. Biochemical characterization of cyanobacterial extracellular polymers (EPS) from modern marine stromatolites (Bahamas). Prep. Biochem. Biotechnol. 2000, 30, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Zille, A.; Micheletti, E.; Moradas-Ferreira, P.; De Philippis, R.; Tamagnini, P. Complexity of cyanobacterial exopolysaccharides: Composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 2009, 33, 917–994. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Bilger, W.; Scherer, S. UV-B-Induced synthesis of photoprotective pigments and extracellular polysaccharides in the terrestrial cyanobacterium Nostoc commune. J. Bacteriol. 1997, 179, 1940–1945. [Google Scholar] [CrossRef] [PubMed]
- Helm, R.; Huang, Z.; Edwards, D.; Leeson, H.; Peery, W.; Potts, M. Structural Characterization of the Released Polysaccharide of Desiccation-Tolerant Nostoc commune DRH-1. J. Bacteriol. 2000, 182, 974–982. [Google Scholar] [CrossRef]
- Rossi, F.; De Philippis, R. Review Role of Cyanobacterial Exopolysaccharides in Phototrophic Biofilms and in Complex Microbial Mats. Life 2015, 5, 1218–1238. [Google Scholar] [CrossRef]
- Nobles, D.; Romanovicz, D.; Brown, M. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001, 127, 529–542. [Google Scholar] [CrossRef]
- Nobles, D.; Brown, M. The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose 2004, 11, 437–448. [Google Scholar] [CrossRef]
- Pereira, S.; Mota, R.; Vieira, C.P.; Vieira, J.; Tamagnini, P. Phylum-wide analysis of genes/proteins related to the last steps of assembly and export of extracellular polymeric substances (EPS) in cyanobacteria. Sci. Rep. 2015, 5, 14835. [Google Scholar] [CrossRef]
- Abidi, W.; Torres-Sánchez, L.; Siroy, A.; Krasteva, P.V. Weaving of bacterial cellulose by the Bcs secretion systems. FEMS Microbiol. Rev. 2022, 46, fuab051. [Google Scholar] [CrossRef]
- Delmer, D. Cellulose Biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 245–276. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Ortiz, M.; Membrillo, J. Mecanismos moleculares de la síntesis de celulosa en bacterias. TIP Rev. Espec. Cienc. Químico-Biológicas 2004, 7, 26–34. [Google Scholar]
- Matthysse, A.G.; Deschet, K.; Williams, M.; Marry, M.; White, A.R.; Smith, W.C. A functional cellulose synthase from ascidian epidermis. Proc. Natl. Acad. Sci. USA 2018, 101, 986–991. [Google Scholar] [CrossRef]
- Grenville-Briggs, L.J.; Anderson, V.L.; Fugelstad, J.; Avrova, A.O.; Bouzenzana, J.; Williams, A.; Wawra, S.; Whisson, S.C.; Birch, P.R.; Bulone, V.; et al. Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell 2008, 20, 720–738. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Kao, M.R.; Saldivar, R.K.; Díaz-Moreno, S.M.; Xing, X.; Furlanetto, V.; Yayo, J.; Divne, C.; Vilaplana, F.; Abbott, D.W.; et al. The Gram-positive bacterium Romboutsia ilealis harbors a polysaccharide synthase that can produce (1,3;1,4)-β-D-glucans. Nat. Commun. 2023, 14, 4526. [Google Scholar] [CrossRef]
- Doblin, M.; Kurek, I.; Jacob-Wilk, D.; Delmer, D. Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiol. 2002, 43, 1407–1420. [Google Scholar] [CrossRef]
- Saxena, I.; Brown, M.J. Cellulose Biosynthesis: Current views and evolving concepts. Ann. Bot. 2005, 96, 9–21. [Google Scholar] [CrossRef]
- Saxena, I.; Brown, M.J. Identification of cellulose synthase(s) in higher plants: Sequence analysis of processive β-glycosyltransferases with the common motif D,D,D,35Q(R,Q)XRW. Cellulose 1997, 4, 33–49. [Google Scholar] [CrossRef]
- Özçimen, D.; İnan, B. An Overview of Bioethanol Production from Algae. Available online: https://www.intechopen.com/books/biofuels-status-and-perspective/an-overview-of-bioethanol-production-from-algae (accessed on 28 July 2023). [CrossRef]
- Sharma, N.; Rai, A.; Stal, L. Cyanobacteria and Economic Perspective, 1st ed.; John Wiley & Sons: Chichester, UK, 2014; pp. 181–193. [Google Scholar]
- Shiru, J.; Hainfeng, Y.; Yongxian, L.; Yujie, D. Characterization of Extracellular Polysaccharides from Nostoc flagelliforme Cells in Liquid Suspension Culture. Biotechnol. Bioprocess. Eng. 2007, 12, 271–275. [Google Scholar] [CrossRef]
- De Farias Silva, C.; Bertucco, A. Bioethanol from microalgae and cyanobacteria: A review and technological outlook. Process Biochem. 2016, 51, 1833–1842. [Google Scholar] [CrossRef]
- Algenol. Available online: https://www.algenol.com/ (accessed on 3 October 2023).
- Alga Energy. Available online: https://www.algaenergy.es/ (accessed on 3 October 2023).
- Biofuel Systems. Available online: https://www.biofuelsystems.com/ (accessed on 3 October 2023).
- Allen, M.; Arnon, D. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 1955, 30, 366–372. [Google Scholar] [CrossRef]
- Atschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic Local Alignment Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Tamura, J.; Masatoshi, D.; Masatoshi, N.; Kumar, N. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Jarman, T.; Deavin, L.; Slocombe, S.; Righelato, R. Investigation of the effect of environmental conditions on the rate of exopolysaccharide synthesis in Azotobacter vinelandii. J. Gen. Microbiol. 1978, 107, 59–64. [Google Scholar] [CrossRef]
- Trinder, P. Determination of glucose in blood using glucose oxidase with on alternative oxygen receptor. Ann. Clin. Biochem. 1969, 6, 24–27. [Google Scholar] [CrossRef]
- Kaplan, N.; Ciotti, M. Enzymatic determination of ethanol. Methods Enzymol. 1957, 3, 253–255. [Google Scholar] [CrossRef]
- Galetović, A.; Araya, J.; Gómez-Silva, B. Biochemical composition and toxicity of edible colonies of the cyanobacterium Nostoc sp. Llayta. Rev. Chil Nutr 2017, 44, 360–370. [Google Scholar] [CrossRef]
- Rivera, M.; Galetović, A.; Licuime, R.; Gómez-Silva, B. A Microethnographic and Ethnobotanical Approach to Llayta Consumption among Andes Feeding Practices. Foods 2018, 7, 202. [Google Scholar] [CrossRef]
- Galetović, A.; Azevedo, J.; Castelo-Branco, R.; Oliveira, F.; Gómez-Silva, B.; Vasconcelos, V. Absence of Cyanotoxins in Llayta, Edible Nostocaceae Colonies from the Andes Highlands. Toxins 2020, 12, 382. [Google Scholar] [CrossRef] [PubMed]
- Saxena, I.M.; Brown, M.J.; Dandekar, T. Structure--function characterization of cellulose synthase: Relationship to other glycosyltransferases. Phytochemistry 2001, 57, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Saxena, I.M.; Brown, M.J. Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J. Bacteriol. 1995, 177, 5276–5283. [Google Scholar] [CrossRef][Green Version]
- Busch, C.; Hofmann, F.; Selzer, J.; Munro, S.; Jeckel, D.; Aktories, K. A Common Motif of Eukaryotic Glycosyltransferases Is Essential for the Enzyme Activity of Large Clostridial Cytotoxins. J. Biol. Chem. 1998, 273, 19566–19572. [Google Scholar] [CrossRef] [PubMed]
- Souza, D. Produção de Celulose Bacteriana: Identificação do Operon bcs e Produção de Biofilme Celulósico por Chromobacterium Violaceum, Dissertação Mestrado, Universidade Federal de Santa Catarina. Available online: http://repositorio.ufsc.br/xmlui/handle/123456789/87780 (accessed on 4 July 2023).
- Brown, M.J.; Saxena, I.; Kudlicka, K. Cellulose biosynthesis in higher plants. Trends Plant Sci. 1996, 1, 149–156. [Google Scholar] [CrossRef]
- Kawano, Y.; Saotome, T.; Ochiai, Y.; Katayama, M.; Narikawa, R.; Ikeuchi, M. Cellulose Accumulation and a Cellulose Synthase Gene are Responsible for Cell Aggregation in the Cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol. 2011, 52, 957–958. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008, 178, 240–242. [Google Scholar] [CrossRef]
- Yin, Y.; Huang, J.; Xu, Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009, 9, 99. [Google Scholar] [CrossRef]
- Chase, W.R.; Zhaxybayeva, O.; Rocha, J.; Cosgrove, D.J.; Shapiro, L.R. Global cellulose biomass, horizontal gene transfers and domain fusions drive microbial expansin evolution. New Phytol. 2020, 226, 921–938. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, P.K.; Pattnaik, R.; Kumar, S.; Ojha, S.K.; Srichandan, H.; Parhi, P.K.; Jyothi, R.K.; Sarangi, P.K. Biochemistry, Synthesis, and Applications of Bacterial Cellulose: A Review. Front. Bioeng. Biotechnol. 2022, 8, 780409. [Google Scholar] [CrossRef] [PubMed]
- Möllers, K.; Cannella, D.; Jørgensen, H.; Frigaard, N.U. Cyanobacterial biomass as carbohydrate and nutrient feedstock for bioethanol production by yeast fermentation. Biotechnol. Biofuels 2014, 7, 64. [Google Scholar] [CrossRef]
- Markou, G.; Angelidaki, I.; Nerantzis, E.; Georgakakis, D. Bioethanol production by carbohydrate-enriched biomass of Arthrospira (Spirulina) platensis. Energies 2013, 6, 3937–3950. [Google Scholar] [CrossRef]
- Pyo, D.; Kim, T.; Yoo, J. Efficient extraction of bioethanol from freshwater cyanobacteria using supercritical fluid pretreatment. Bull. Korean Chem. Soc. 2013, 34, 379–383. [Google Scholar] [CrossRef][Green Version]
- Aikawa, S.; Inokuma, K.; Wakai, S.; Sasaki, K.; Ogino, C.; Chang, J.; Hasunuma, T.; Kondo, A. Direct and highly productive conversion of cyanobacteria Arthrospira platensis to ethanol with CaCl2 addition. Biotechnol. Biofuels 2018, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, S.; Joseph, A.; Yamada, R.; Izumi, Y.; Yamagishi, T.; Matsuda, F.; Kawai, H.; Chang, J.; Hasunumabch, T.; Kondo, A. Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis processes. Energy Environ. Sci. 2013, 6, 1844–1849. [Google Scholar] [CrossRef]
- Kehr, J.-C.; Dittmann, E. Review Biosynthesis and Function of Extracellular Glycans in Cyanobacteria. Life 2015, 5, 164–180. [Google Scholar] [CrossRef]

| Strain | EPS (g L−1) |
|---|---|
| LLA-10 | 2.65 ± 0.13 |
| LLC-10 | 7.11 ± 0.02 |
| RPU-10 | 1.84 ± 0.05 |
| LCHI-10 | 1.73 ± 0.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galetović, A.; Peña, G.; Fernández, N.; Urrutia, M.; Flores, N.; Gómez-Silva, B.; Di Ruggiero, J.; Shene, C.; Bustamante, M. Cellulose Synthase in Atacama Cyanobacteria and Bioethanol Production from Their Exopolysaccharides. Microorganisms 2023, 11, 2668. https://doi.org/10.3390/microorganisms11112668
Galetović A, Peña G, Fernández N, Urrutia M, Flores N, Gómez-Silva B, Di Ruggiero J, Shene C, Bustamante M. Cellulose Synthase in Atacama Cyanobacteria and Bioethanol Production from Their Exopolysaccharides. Microorganisms. 2023; 11(11):2668. https://doi.org/10.3390/microorganisms11112668
Chicago/Turabian StyleGaletović, Alexandra, Gabriel Peña, Nicole Fernández, Milton Urrutia, Nataly Flores, Benito Gómez-Silva, Jocelyne Di Ruggiero, Carolina Shene, and Mariela Bustamante. 2023. "Cellulose Synthase in Atacama Cyanobacteria and Bioethanol Production from Their Exopolysaccharides" Microorganisms 11, no. 11: 2668. https://doi.org/10.3390/microorganisms11112668
APA StyleGaletović, A., Peña, G., Fernández, N., Urrutia, M., Flores, N., Gómez-Silva, B., Di Ruggiero, J., Shene, C., & Bustamante, M. (2023). Cellulose Synthase in Atacama Cyanobacteria and Bioethanol Production from Their Exopolysaccharides. Microorganisms, 11(11), 2668. https://doi.org/10.3390/microorganisms11112668







