The Regulatory Functions of the Multiple Alternative Sigma Factors RpoE, RpoHI, and RpoHII Depend on the Growth Phase in Rhodobacter sphaeroides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids, Strains, Media, and Culture Conditions
2.2. Expression and Purification of His6-Tagged RpoHI or RpoHII Protein
2.3. Electrophoresis Mobility Shift Assay
2.4. Construction of Reporter Plasmid
2.5. Construction of pos19 Deletion Strain
2.6. Overexpression Plasmid Construction
2.7. Diparental Conjugation
2.8. β-Galactosidase Assay
2.9. Quantitative Real-Time RT PCR
2.10. Data Analysis
3. Results
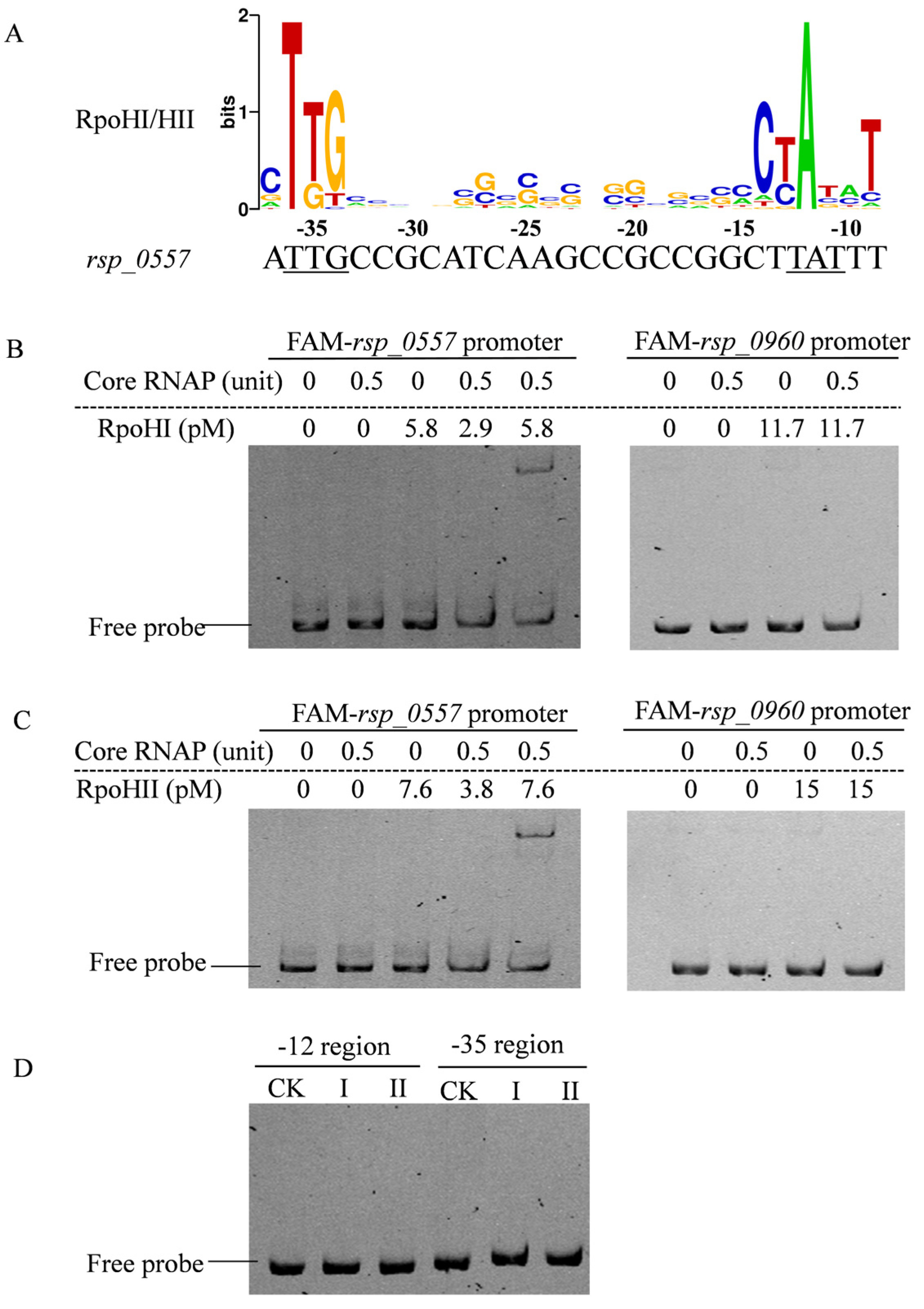
3.1. The rsp_0557 Is Regulated by RpoHI/RpoHII in R. sphaeroides
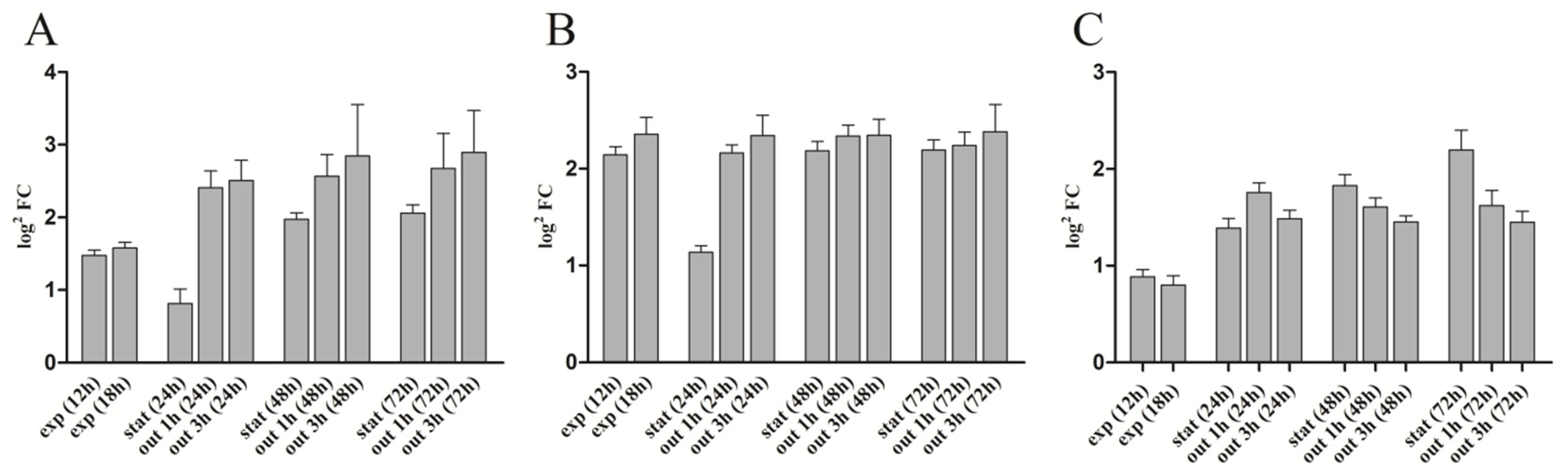
3.2. Growth Phase-Dependent Expression of rsp_0557
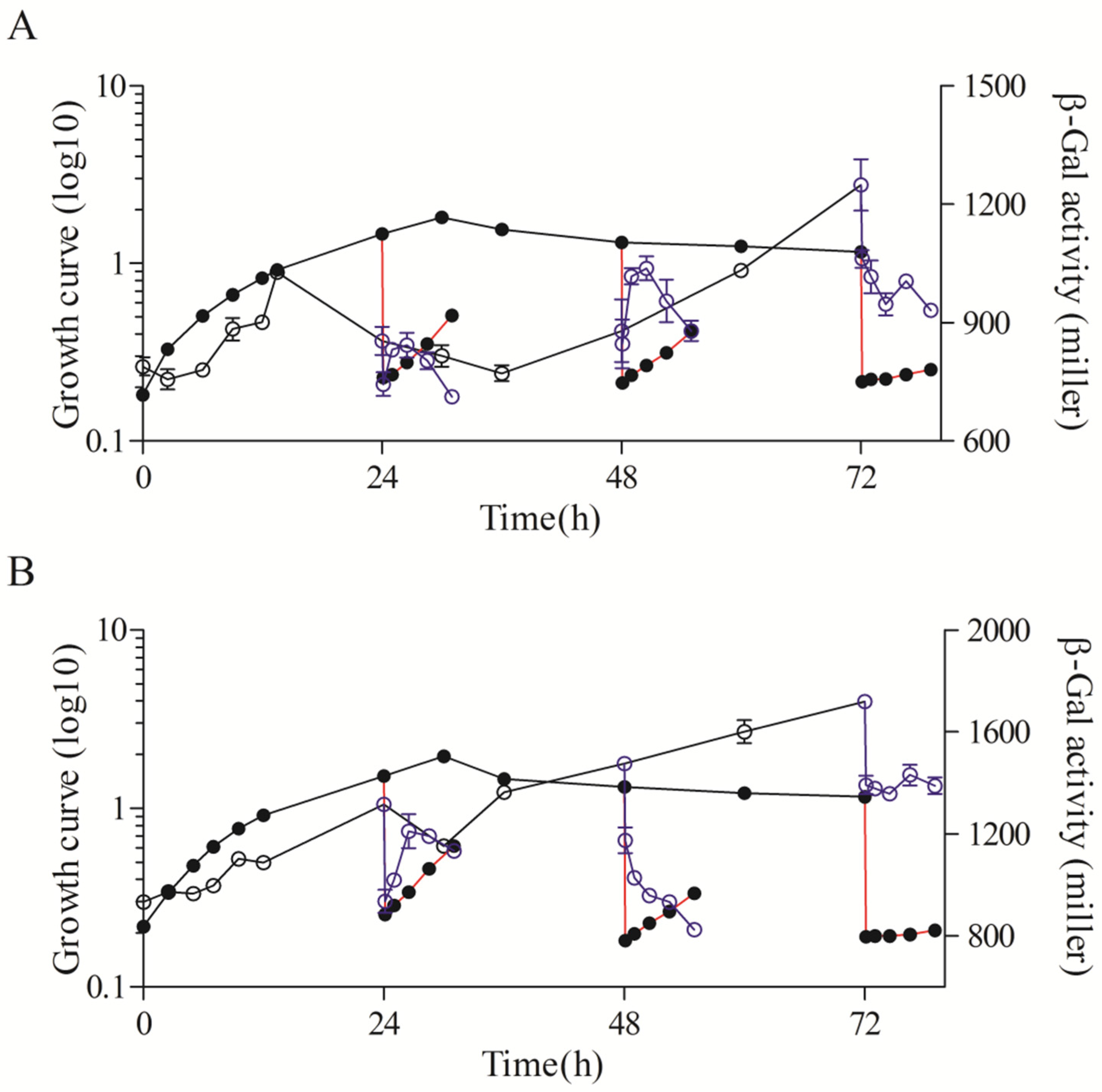
3.3. Major Roles of RpoHI/RpoHII in the Transcription of rsp_0557 in Different Growth Phases
3.4. Major Role of RpoE in the Transcription of rsp_0557 in Different Growth Phases
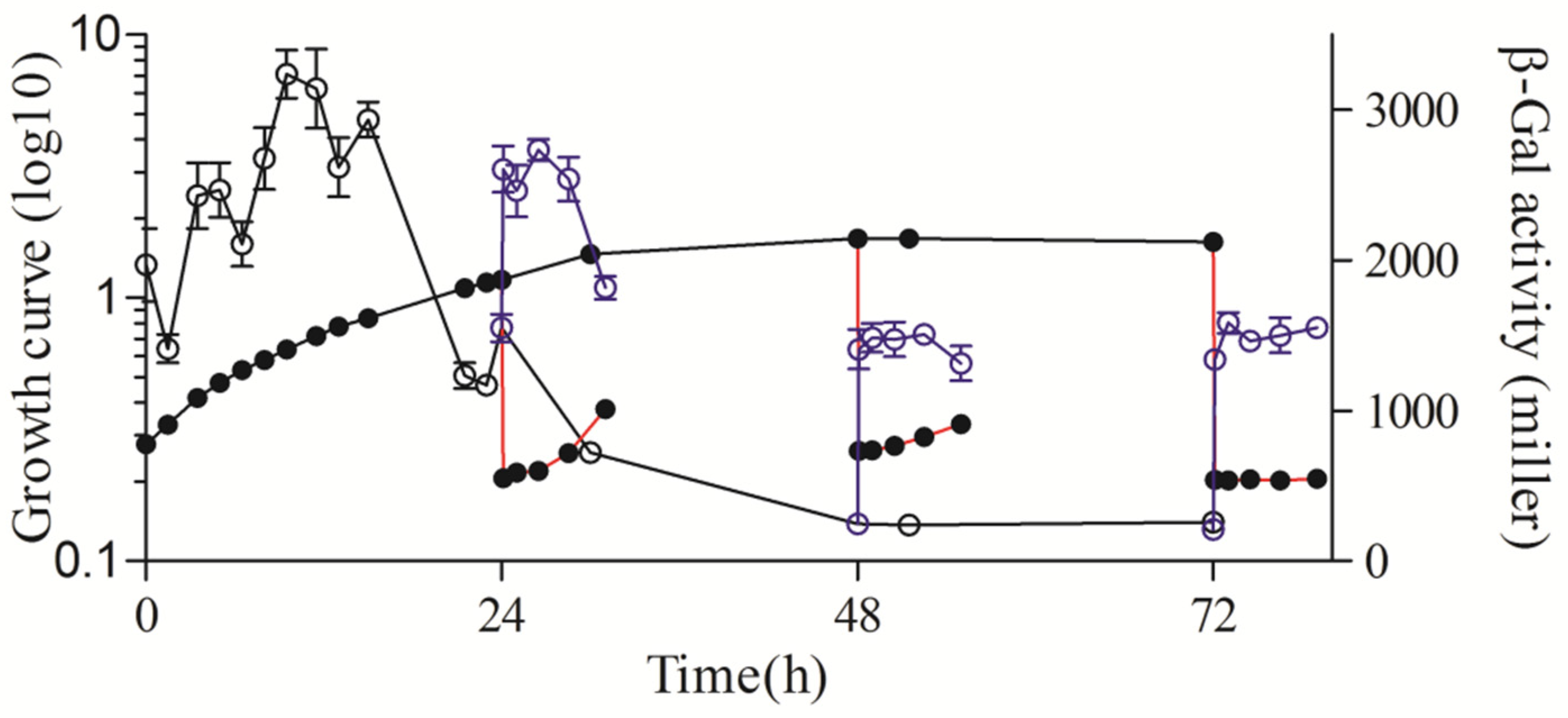
3.5. sRNA Pos19 Is Involved in Regulating the Expression of rsp_0557
3.6. ChrR Participates in Maintaining the Stability of rsp_0557 Expression Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llorens, J.M.N.; Tormo, A.; Martinez-Garcia, E. Stationary phase in Gram-negative bacteria. FEMS Microbiol. Rev. 2010, 34, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Hengge-Aronis, R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002, 66, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Schellhorn, H.E.; Stones, V.L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J. Bacteriol. 1992, 174, 4769–4776. [Google Scholar] [CrossRef] [PubMed]
- Mika, F.; Hengge, R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Gene Dev. 2005, 19, 2770–2781. [Google Scholar] [CrossRef] [PubMed]
- Majdalani, N.; Chen, S.; Murrow, J.; John, K.; Gottesman, S. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol. Microbiol. 2001, 39, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Majdalani, N.; Cunning, C.; Sledjeski, D.; Elliott, T.; Gottesman, S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 12462–12467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Altuvia, S.; Tiwari, A.; Argaman, L.; Hengge-Aronis, R.; Storz, G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998, 17, 6061–6068. [Google Scholar] [CrossRef]
- Azam, T.A.; Iwata, A.; Nishimura, A.; Ueda, S.; Ishihama, A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999, 181, 6361–6370. [Google Scholar] [CrossRef]
- Jin, D.J.; Cagliero, C.; Zhou, Y.N. Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 2012, 36, 269–287. [Google Scholar] [CrossRef]
- Landgraf, J.R.; Wu, J.; Calvo, J.M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 1996, 178, 6930–6936. [Google Scholar] [CrossRef]
- Costanzo, A.; Ades, S.E. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 2006, 188, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Blom, E.J.; Ridder, A.N.; Lulko, A.T.; Roerdink, J.B.; Kuipers, O.P. Time-resolved transcriptomics and bioinformatic analyses reveal intrinsic stress responses during batch culture of Bacillus subtilis. PLoS ONE 2011, 6, e27160. [Google Scholar] [CrossRef] [PubMed]
- Sonenshein, A.L. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 2005, 8, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Wolz, C. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol. 2014, 304, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Aretha, F.; Herrou, J.; Willett, J.; Crosson, S. General Stress Signaling in the Alphaproteobacteria. Annu. Rev. Genet. 2015, 49, 603. [Google Scholar]
- Remes, B.; Rische-Grahl, T.; Müller, K.M.H.; Klug, G. An RpoHI-dependent response promotes outgrowth after extended stationary phase in the Alphaproteobacterium Rhodobacter sphaeroides. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Nuss, A.M.; Glaeser, J.; Berghoff, B.A.; Klug, G. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J. Bacteriol. 2010, 192, 2613–2623. [Google Scholar] [CrossRef]
- Dufour, Y.S.; Imam, S.; Koo, B.M.; Green, H.A.; Donohue, T.J. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet. 2012, 8, e1002929. [Google Scholar] [CrossRef]
- Nuss, A.M.; Glaeser, J.; Klug, G. RpoHII activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J. Bacteriol. 2009, 191, 220–230. [Google Scholar] [CrossRef]
- Anthony, J.R.; Newman, J.D.; Donohue, T.J. Interactions between the Rhodobacter sphaeroides ECF sigma factor, sigma(E), and its anti-sigma factor, ChrR. J. Mol. Biol. 2004, 341, 345–360. [Google Scholar] [CrossRef]
- Nuss, A.M.; Adnan, F.; Weber, L.; Berghoff, B.A.; Glaeser, J.; Klug, G. DegS and RseP homologous proteases are involved in singlet oxygen dependent activation of RpoE in Rhodobacter sphaeroides. PLoS ONE 2013, 8, e79520. [Google Scholar] [CrossRef] [PubMed]
- Braatsch, S.; Gomelsky, M.; Kuphal, S.; Klug, G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 2002, 45, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Billenkamp, F.; Peng, T.; Berghoff, B.A.; Klug, G. A cluster of four homologous small RNAs modulates C1 metabolism and the pyruvate dehydrogenase complex in Rhodobacter sphaeroides under various stress conditions. J. Bacteriol. 2015, 197, 1839–1852. [Google Scholar] [CrossRef] [PubMed]
- Peuser, V.; Remes, B.; Klug, G. Role of the Irr protein in the regulation of iron metabolism in Rhodobacter sphaeroides. PLoS ONE 2012, 7, e42231. [Google Scholar] [CrossRef] [PubMed]
- Grutzner, J.; Remes, B.; Eisenhardt, K.; Scheller, D.; Kretz, J.; Madhugiri, R.; McIntosh, M.; Klug, G. sRNA-mediated RNA processing regulates bacterial cell division. Nucleic Acids Res. 2021, 49, 7035–7052. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Berghoff, B.A.; Eisenhardt, B.D.; Remes, B.; Klug, G. Characteristics of Pos19-A small coding RNA in the oxidative stress response of Rhodobacter sphaeroides. PLoS ONE 2016, 11, e0163425. [Google Scholar] [CrossRef] [PubMed]
- Francez-Charlot, A.; Kaczmarczyk, A.; Fischer, H.M.; Vorholt, J.A. The general stress response in Alphaproteobacteria. Trends Microbiol. 2015, 23, 164–171. [Google Scholar] [CrossRef]
- Li, Q.; Peng, T.; Klug, G. The PhyR homolog RSP_1274 of Rhodobacter sphaeroides is involved in defense of membrane stress and has a moderate effect on RpoE (RSP_1092) activity. BMC Microbiol. 2018, 18, 18. [Google Scholar] [CrossRef]
- Börner, J.; Friedrich, T.; Bartkuhn, M.; Klug, G. Ribonuclease E strongly impacts bacterial adaptation to different growth conditions. RNA Biol. 2023, 20, 120–135. [Google Scholar] [CrossRef]
- Remes, B.; Berghoff, B.A.; Förstner, K.U.; Klug, G. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genom. 2014, 15, 794. [Google Scholar] [CrossRef]
- Su, A.; Chi, S.; Li, Y.; Tan, S.; Qiang, S.; Chen, Z.; Meng, Y. Metabolic redesign of Rhodobacter sphaeroides for lycopene production. J. Agric. Food Chem. 2018, 66, 5879–5885. [Google Scholar] [CrossRef]
- Qu, Y.; Su, A.; Li, Y.; Meng, Y.; Chen, Z. Manipulation of the regulatory genes ppsR and prrA in Rhodobacter sphaeroides enhances lycopene production. J. Agric. Food Chem. 2021, 69, 4134–4143. [Google Scholar] [CrossRef] [PubMed]
- Hübner, P.; Willison, J.C.; Vignais, P.M.; Bickle, T.A. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 1991, 173, 2993–2999. [Google Scholar] [CrossRef] [PubMed]
- Grutzner, J.; Billenkamp, F.; Spanka, D.T.; Rick, T.; Monzon, V.; Forstner, K.U.; Klug, G. The small DUF1127 protein CcaF1 from Rhodobacter sphaeroides is an RNA-binding protein involved in sRNA maturation and RNA turnover. Nucleic Acids Res. 2021, 49, 3003–3019. [Google Scholar] [CrossRef] [PubMed]
- Müller, K. Small Regulatory RNAs Promoting the Oxidative Stress Response and Adaptive Metabolic Changes in Rhodobacter sphaeroides. Ph.D. Thesis, Universitaet Giessen, Giessen, Germany, 2016. [Google Scholar]
- Eisenhardt, K.M.H.; Remes, B.; Grutzner, J.; Spanka, D.T.; Jager, A.; Klug, G. A complex network of sigma factors and sRNA StsR regulates stress responses in R. sphaeroides. Int. J. Mol. Sci. 2021, 22, 7557. [Google Scholar] [CrossRef] [PubMed]
- Lange, R.; Hengge-Aronis, R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Gene Dev. 1994, 8, 1600–1612. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Polen, T.; Heuveling, J.; Wendisch, V.F.; Hengge, R. Genome-wide analysis of the general stress response network in Escherichia coli: SigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005, 187, 1591–1603. [Google Scholar] [CrossRef] [PubMed]




| Strain or Plasmid | Description or Relevant Features | Source/Reference |
|---|---|---|
| E. coli strains | ||
| DH5α | For Sub-Cloning | Weidi |
| BL21 (DE3) | For protein expression | TIANGEN |
| S17-1 | For diparental conjugation | Lab preservation |
| R. sphaeroide strains | ||
| 2.4.1 | Wild type | Lab preservation |
| TF18 | rpoE chrR mutation inWT, Tpr | Lab preservation |
| ΔrpoHI | WT rpoHI::Kmr cassette | Lab preservation |
| ΔrpoHII | WT rpoHII::Spr cassette | Lab preservation |
| ΔrpoHIΔrpoHII | WT rpoHIIrpoHI::Kmr.Spr cassette | Lab preservation |
| ΔChrR | WT chrR::Tpr cassette | Lab preservation |
| ΔPos19 | Pos19 markerless deletion mutant | This study |
| Plasmids | ||
| pJET1.2/blunt cloning vector | Apr, 2.97 kb | Thermo |
| pBBRIMCS5-lacZ | Gmr | Lab preservation |
| pBBRI-0557-lacZ | Expression of rsp_0557; Gmr | This study |
| pK18mobsacB | suicide vector, sacB (sucrose sensitivity), Kmr | Lab preservation |
| pK18mobsacB::pos19 | For pos19 deletion in 2.4.1 | This study |
| pET30b | For protein expression | Lab preservation |
| pET30-RpoHI | RpoHI protein expression vector | This study |
| pET30-RpoHII | RpoHII protein expression vector | This study |
| pRK16S | Overexpression vector controlled by 16S promoter | Lab preservation |
| pRKPos19 | pos19 overexpression vector | This study |
| Name | Sequence 5′-3′ | Purpose |
|---|---|---|
| RpoHI-E-f | CATATGAGCACTTACACCAGCCTTCCC | EMSA |
| RpoHI-E-r | GCGGCCGCGGCGGGGATCGTCATG | EMSA |
| RpoHII-E-f | CATATGGCACTGGACGGATATACCGATC | EMSA |
| RpoHII-E-r | GCGGCCGCTAGGAGGAAGTGATGCACCTCC | EMSA |
| 0557-EMSA-F | TGCCTGCAGGTCGACGATCCGCACGGCGCCAC | EMSA |
| 0557-EMSA-R | CTCGAAGGCGGCCATGG | EMSA |
| 0557-35M-F | GAACCAAGTTCCGACATCAAGC | EMSA |
| 0557-35M-R | GTCGGAACTTGGTTCCGGA | EMSA |
| 0557-12M-F | GCCGGCTAAATTGCCTGTTC | EMSA |
| 0557-12M-R | CGTCTGAACAGGCAATTTAGCC | EMSA |
| 0960-EMSA-F | TGCCTGCAGGTCGACGATTCCGACGGAAAACAGGATCC | EMSA |
| 0960-EMSA-R | GCTTGCCTCCTGTAGGGCC | EMSA |
| FAM Fluorescent primer | TGCCTGCAGGTCGACGAT | EMSA |
| RSP_0557-f | GAATTCCCGCACGGCG | Cloning |
| RSP_0557-r | AAGCTTCTCGAAGGCGGC | Cloning |
| 0019-XbaI-up | TCTAGACGATCAGGCGACGG | Cloning-knockout |
| 0019-up | GGCCTTCTCCGGGGCACAGCTTACGCAGGGTCG | Cloning-knockout |
| 0019-down | GACCCTGCGTAAGCTGTGCCCCGGAGAAGGCCC | Cloning-knockout |
| 0019-BamHI-down | GGATCCTGCGCCCTCAG | Cloning-knockout |
| pPos19-F | GGATCCCGATCAACCCAAGCAGAA | Overexpression |
| pPos19-R | GAATTCGGATGTCCCGCTCAGG | Overexpression |
| rpoZ_RT-f | ATCGCGGAAGAGACCCAGAG | RT-PCR |
| rpoZ_RT-r | GAGCAGCGCCATCTGATCCT | RT-PCR |
| rpoHI_RT-f | GATCGCCAAGGATCT | RT-PCR |
| rpoHI_RT-r | CTGGTCGCTGTCTTCA | RT-PCR |
| rpoHII_RT-f | GCCGATGAACGACCTGAT | RT-PCR |
| rpoHII_RT-r | AAGAACAGCGCCTTCTGG | RT-PCR |
| rpoE_RT-f | GTCTGGCAGAAGGCTCAT | RT-PCR |
| rpoE_RT-r | GTTCTCCTGCTGCATCTC | RT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zheng, M.; Tang, Z.; Zhong, S.; Bu, T.; Li, Q. The Regulatory Functions of the Multiple Alternative Sigma Factors RpoE, RpoHI, and RpoHII Depend on the Growth Phase in Rhodobacter sphaeroides. Microorganisms 2023, 11, 2678. https://doi.org/10.3390/microorganisms11112678
Zhang J, Zheng M, Tang Z, Zhong S, Bu T, Li Q. The Regulatory Functions of the Multiple Alternative Sigma Factors RpoE, RpoHI, and RpoHII Depend on the Growth Phase in Rhodobacter sphaeroides. Microorganisms. 2023; 11(11):2678. https://doi.org/10.3390/microorganisms11112678
Chicago/Turabian StyleZhang, Jing, Meijia Zheng, Zizhong Tang, Shanpu Zhong, Tongliang Bu, and Qingfeng Li. 2023. "The Regulatory Functions of the Multiple Alternative Sigma Factors RpoE, RpoHI, and RpoHII Depend on the Growth Phase in Rhodobacter sphaeroides" Microorganisms 11, no. 11: 2678. https://doi.org/10.3390/microorganisms11112678
APA StyleZhang, J., Zheng, M., Tang, Z., Zhong, S., Bu, T., & Li, Q. (2023). The Regulatory Functions of the Multiple Alternative Sigma Factors RpoE, RpoHI, and RpoHII Depend on the Growth Phase in Rhodobacter sphaeroides. Microorganisms, 11(11), 2678. https://doi.org/10.3390/microorganisms11112678





