Abstract
Escherichia coli is a key indicator of food hygiene, and its monitoring in meat samples points to the potential presence of antimicrobial-resistant strains capable of causing infections in humans, encompassing resistance profiles categorized as serious threats by the Centers for Disease Control and Prevention (CDC), such as Extended-Spectrum Beta-Lactamase (ESBL)—a problem with consequences for animal, human, and environmental health. The objective of the present work was to isolate and characterize ESBL-producing E. coli strains from poultry, pork, and beef meat samples, with a characterization of their virulence and antimicrobial resistance profiles. A total of 450 meat samples (150 chicken, 150 beef, and 150 pork) were obtained from supermarkets and subsequently cultured in medium supplemented with cefotaxime. The isolated colonies were characterized biochemically, followed by antibiogram testing using the disk diffusion technique. Further classification involved biofilm formation and the presence of antimicrobial resistance genes (blaCTX-M, AmpC-type, mcr-1, and fosA3), and virulence genes (eaeA, st, bfpA, lt, stx1, stx2, aggR, iss, ompT, hlyF, iutA, iroN, fyuA, cvaC, and hylA). Statistical analysis was performed via the likelihood-ratio test. In total, 168 strains were obtained, with 73% originating from chicken, 22% from pork, and 17% from beef samples. Notably, strains exhibited greater resistance to tetracycline (51%), ciprofloxacin (46%), and fosfomycin (38%), apart from β-lactams. The detection of antimicrobial resistance in food-isolated strains is noteworthy, underscoring the significance of antimicrobial resistance as a global concern. More than 90% of the strains were biofilm producers, and strains carrying many ExPEC genes were more likely to be biofilm formers (OR 2.42), which increases the problem since the microorganisms have a greater chance of environment persistence and genetic exchange. Regarding molecular characterization, bovine samples showed a higher prevalence of blaCTX-M-1 (OR 6.52), while chicken strains were more likely to carry the fosA3 gene (OR 2.43, CI 1.17–5.05) and presented between 6 to 8 ExPEC genes (OR 2.5, CI 1.33–5.01) compared to other meat samples. Concerning diarrheagenic E. coli genes, two strains harbored eae. It is important to highlight these strains, as they exhibited both biofilm-forming capacities and multidrug resistance (MDR), potentially enabling colonization in diverse environments and causing infections. In conclusion, this study underscores the presence of β-lactamase-producing E. coli strains, mainly in poultry samples, compared to beef and pork samples. Furthermore, all meat sample strains exhibited many virulence-associated extraintestinal genes, with some strains harboring diarrheagenic E. coli (DEC) genes.
1. Introduction
Detection of resistance [1] and virulence [2] genes is pivotal in the characterization of microorganisms in food [3,4,5,6,7,8]. In addition, the ability of these microorganisms to survive in different environments, exhibiting heightened resistance to antimicrobial action [9,10], and their facile exchange of genetic material [11,12] contributes to effective monitoring and assists in the development of strategies to decrease the potential transmission of pathogens from food to humans.
Beef, pork, and poultry are among the main sources of protein in the human diet [13,14] and are correlated with most foodborne illnesses [15], thus serving as essential indicators of food safety [16,17]. Bacteria are responsible for 35.8% of foodborne illness cases in Brazil, according to the Brazilian Ministry of Health [18], and in the United States, they cause average annual costs of US $17.5 billion [19,20].
Food contamination may occur at any stage of the production chain, such as processing, washing, distribution, marketing, and even during home preparation [21,22,23]. Evisceration and skinning/plucking are critical points in the slaughter process [24,25,26]. Utensils and the handlers’ hands also contribute to spreading bacteria on the surface of carcasses and cross-contamination between them [23].
A method employed for assessing food contamination is through the analysis of hygiene indicator microorganisms [2,27], for example, Escherichia coli. This bacterium, a beneficial microorganism present in the gastrointestinal tracts of humans and animals, may harbor genes that confer pathogenic characteristics [28]. This microorganism is the major responsible for water and foodborne disease outbreaks in Brazil [29]. It can be classified into different pathotypes, depending on its isolation source, virulence factors, and associated clinical symptoms [28].
E. coli presents various diarrheagenic pathotypes (DEC), characterized by the presence of specific virulence genes. These include enteroinvasive E. coli (EIEC) a non-lactose fermenting pathotype, harboring the ipaH gene located on invasion plasmid H enteropathogenic; E. coli (EPEC) with the eae gene encoding intimin and maybe the bfp gene; enterotoxigenic E. coli (ETEC) identified by the st gene, which codes for a thermostable toxin, and the lt gene, responsible for a thermolabile toxin; Shiga toxin-producing E. coli (STEC) carrying the stx gene, which encodes the Shiga toxin; and enteroaggregative E. coli (EAEC) that presents aggregative adhesion in lineage cells, and often aggR gene, encoding aggregate adhesion fimbriae. These pathotypes can lead to a spectrum of clinical outcomes, ranging from mild diarrhea to conditions such as hemolytic uremic syndrome (HUS) and, in severe cases, death [28,30].
Extraintestinal pathogenic E. coli (ExPEC) has the potential to cause diverse infections, including meningitis, pneumonia, and urinary tract infections. Several genes facilitate the characterization of the virulence profiles in these strains, such as iroN (salmochelin siderophore receptor), fyuA (yersiniabactin siderophore receptor), iutA (aerobactin siderophore receptor), ompT (episomal outer membrane protein), hlyA (hemolysin A), hlyF (hemolysin F), iss (serum resistance-associated protein), and cvaC (colicin V gene) [28,31]. Identifying the pathotypes of ExPEC present in meat samples is crucial [2], since these strains may be clonally related to those isolated from human infections [23,32].
Antimicrobial resistance in microorganisms isolated from meat samples poses a high risk to consumers [2,33]. Strains producing Extended-spectrum β-lactamase (ESBL), classified as serious threats by the Centers for Disease Control and Prevention (CDC), are responsible for over 35,000 annual deaths and two million infections [34]. Notably, during the COVID-19 pandemic, these numbers have surged by 32% [35]. ESBL-producing E. coli strains have been previously isolated from pork, poultry, beef, and other sources of animal protein [4,36,37,38,39,40,41]. Therefore, surveillance is pivotal in the context of the One Health framework.
Although the relationship between animal and human health dates back to the 19th century [42], the complexity of this connection led to the concept of ‘One World, One Health’ in 2003. This concept highlights the interconnection of humans, animals, plants, and their shared environment [43,44]. The existence of environments hosting diverse species raises pertinent health concerns [45], including issues related to food safety and antimicrobial resistance [46,47].
This study aimed to isolate and characterize ESBL-producing Escherichia coli strains obtained from chicken, pork, and beef meat samples, encompassing both phenotypic and genotypic evaluation of their virulence and antimicrobial resistance profiles.
2. Materials and Methods
2.1. Meat Samples
Over a consecutive five-month period (from August to December 2019), a total of 450 meat samples, comprising 150 each of chicken, beef, and pork, were acquired from supermarkets within the region of Londrina, Paraná State, Brazil. These samples, both commercial and refrigerated, were stored under refrigeration until processing.
2.2. Isolation and Biochemical Identification of E. coli
Meat samples were macerated and incubated in buffered peptone water (1:10—Difco, Detroit, MI, USA) at 37 °C for 18–24 h. After incubation, they were seeded onto MacConkey agar (Difco, Detroit, MI, USA) supplemented with cefotaxime (CTX) at a final concentration of 8 μg/mL. One lactose-fermenting colony per sample was subjected to biochemical identification using triple-sugar iron agar, indole production, Simmons citrate, urease production, lysine decarboxylation, and sorbitol and cellobiose fermentation tests [48].
These colonies were stored at −20 °C in Brain Heart Infusion (BHI) broth (Himedia Laboratories Pvt. Ltd., Mumbai, India), also supplemented with cefotaxime (8 μg/mL) and 20% glycerol until further processing.
2.3. Antibiotic Susceptibility Testing
Antimicrobial sensitivity was determined using the disk diffusion method [49], with seven distinct classes of antimicrobials: (1) β-lactams: ampicillin (AMP, 10 μg), amoxicillin-clavulanic acid (AMC, 10/20 μg), cefazolin (CFZ, 30 μg), cefoxitin, (CFO, 30 μg), ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), aztreonam (ATM, 30 μg), and imipenem (IPM, 30 μg); (2) quinolones: ciprofloxacin (CIP, 5 μg); (3) sulfonamides: sulfamethoxazole + trimethoprim (SXT, 1.25/23.75 μg); (4) tetracyclines: tetracycline (TET, 30 μg); (5) aminoglycosides: gentamicin (CN, 10 μg); (6) amphenicols: chloramphenicol (C, 30 μg); and (7) fosfomycin: fosfomycin/trometamol (FOT, 200 μg; Oxoid Ltd., Basingstoke, Hants, United Kingdom). The reference strain E. coli ATCC 25,922 was used as a control. Results were interpreted according to CLSI guidelines [50].
Multidrug resistance was characterized by resistance to three or more distinct classes of antimicrobial agents, excluding β-lactams [51].
2.4. Evaluation of Biofilm Formation
All isolates were submitted to quantitative biofilm formation assessment according to Stepanović et al. [52], using 96-well polystyrene plates and crystal violet staining. Absorbance readings (A) were measured at a wavelength of 540 nm using a spectrophotometer (LMR-961 plate reader—Loccus, ABIMAQ, São Paulo, Brazil) [53].
E. coli 042 (O44:H18) ATCC strain served as a positive biofilm formation control, while E. coli 25922 was the negative control. TSB broth (BD Difco, Franklin, NJ, USA) was used as a blank. Tests were performed in triplicate, and biofilms were categorized as absent, weak, moderate, strong, or very strong.
2.5. Molecular Identification of Resistance Genes
The following genes associated with β-lactam resistance were investigated: blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, and blaCTX-M-25 [54], as well as AmpC-type production (MOX, FOX, EBC, ACC, DHA, and CIT) [55]. Moreover, the colistin resistance-coding gene (mcr-1) [56] and the fosfomycin resistance gene (fosA3) [57] were investigated.
2.6. Molecular Identification of Virulence Genes
The presence of genes related to DEC (eaeA, st, bfpA, lt, stx1, stx2 [58], and aggR [59]), and genes associated with ExPEC (iss, ompT, hlyF, iutA, iroN [60], hylA, fyuA, and cvaC [31]) was evaluated (Figure S4).
All polymerase chain reaction (PCR) amplicons were visualized using 1.5% agarose gels stained with GelRed (Biotium, Hayward, CA, USA).
After amplification of eae gene, by PCR, the products were purified with PureLink™ PCR Purification Kit (Invitrogen® Life Technologies, Eugene, OR, USA) and sequenced using ABI3500 Genetic Analyzer sequencer, using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied BioSystems®, Foster City, CA, USA).
The sequences were analyzed by Chromas (2.6.6) software and ClustalX (2.1), and using BLAST, the sequences were compared with the NCBI database.
2.7. Statistical Analysis
The odds ratio (OR) calculations were performed using the epiDisplay package (version 3.5.0.2). In this analysis, categorical data following a binomial distribution were modeled by logistic regression using the stats package (version 4.3.0). The variables were analyzed for their predictive values in OR calculation, and the statistical significance was determined via the likelihood-ratio test (LR-test) with a significance level of 5%.
For the analysis of ExPEC genes, isolates were grouped according to the number of genes they harbored: ExPEC 1 (0 to 2 genes), ExPEC 2 (3 to 5 genes), and ExPEC 3 (6 to 8 genes).
3. Results
3.1. Isolation and Identification of Strains Using Biochemical Methods
Out of the initial 450 samples (150 samples from each type of meat), subsequent to processing and identification, a total of 168 (37%) E. coli strains were isolated. Among these, 109/150 (73%—Figure S3) strains originated from chicken samples, 33/150 (22%—Figure S2) from pork samples, and 26/150 (17%—Figure S1) strains from beef samples.
Notably, chicken-derived strains were more likely to be ESBL producers, displaying the capacity of growth in culture media with third-generation cephalosporins, when compared to pork strains (OR 9.99, CI 5.74–17.80), and beef strains (OR 13.42, CI 7.50–24.73).
3.2. Antibiotic Susceptibility Testing
Besides -lactams, other noteworthy antimicrobial resistance patterns can be highlighted, with 51% (85/168) of strains exhibiting resistance to tetracycline, 46% (78/168) to ciprofloxacin, and 38% (64/168) to fosfomycin, while 100% of strains were susceptible to imipenem (Figure 1).
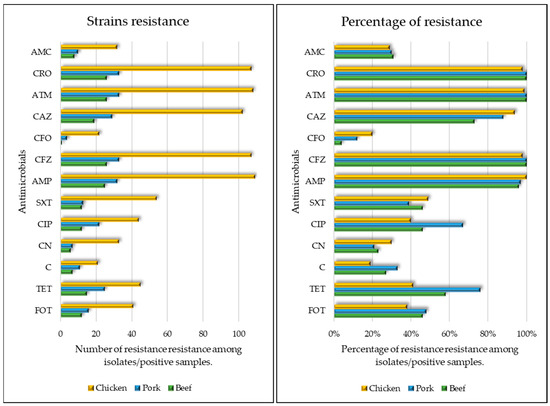
Figure 1.
Comparative antimicrobial susceptibility profile across beef, chicken, and pork samples. On the left a figure relating to the number of isolates per meat sample and on the right a figure relating to the percentage of resistance per meat sample. FOT—fosfomycin-trometamol; TET—tetracycline; SXT—trimethoprim-sulfamethoxazole; C—chloramphenicol; CN—gentamicin; CIP—ciprofloxacin; AMC—amoxicillin-clavulanic acid; AMP—ampicillin; CFZ—cefazolin; CFO—cefoxitin; CRO—ceftriaxone; CAZ—ceftazidime; ATM—aztreonam.
Furthermore, strains isolated from chicken samples presented a higher resistance rate to ceftazidime (OR 1.52, CI 1.21–1.91), while beef-derived strains displayed lower resistance to ceftazidime (OR 0.72, IC 0.60–0.87). Additionally, strains from pork were more resistant to tetracycline (OR 1.22, IC 1.07–1.40) and ciprofloxacin (OR 1.13, IC 1.01–1.28), when compared to the other antimicrobials.
Moreover, among all the strains, 45% (75/168) displayed resistance to three or more classes of antimicrobials, excluding β-lactams.
3.3. Evaluation of Biofilm Formation
It can be observed that only 6% (11/168) of all strains did not produce biofilms, while 46% (78/169) showed moderate biofilm production. Regarding different meat samples, 100% of the strains isolated from beef were biofilm producers (Table 1).

Table 1.
Comparison of biofilm production profiles among isolates from beef, chicken, and pork samples.
Additionally, strains harboring six to eight ExPEC genes (OR 2.42, CI 1.5–87) had a higher proportion of biofilm-producing strains compared to non-biofilm-producing strains.
3.4. Molecular Identification of Resistance Genes
Regarding β-lactam resistance genes, they presented a lower prevalence of blaCTX-M-9 (OR 0.02, CI 0–0.37) than other genes within the blaCTX-M group (Figure 2). No isolates harboring the blaCTX-M-25 and mcr-1 genes were observed.
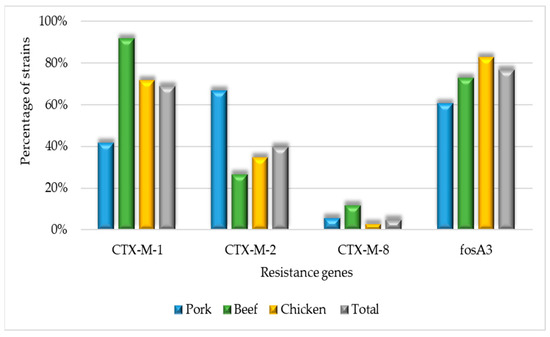
Figure 2.
Molecular identification of resistance gene percentage across beef, chicken, and pork strains.
Strains from chicken were more likely to harbor the fosA3 gene (OR 2.43, CI 1.17–5.05), while those from pork were less frequently associated with this gene (OR 0.37, CI 0.16–0.83) compared to other meat source. Beef-derived strains were more likely to carry the blaCTX-M-1 gene (OR 6.52, CI 1.48–28.72).
Among the Amp-C genes, it was observed that only the CIT gene was present in three strains, and no strains carried the MOX, FOX, EBC, ACC, or DHA genes. All the strains with the CIT gene were from chicken sources and displayed characteristics such as biofilm production, MDR, and the presence of fosA3, iss, ompT, hlyF, iroN, and iutA genes.
3.5. Molecular Identification of ExPEC Virulence Genes
The hlyA gene was not detected in any strain. The most prevalent gene was ompT, detected in 76% (127/168) of the strains, followed by hlyF, iutA, iroN, iss, cvaC, and fyuA (Figure 3).
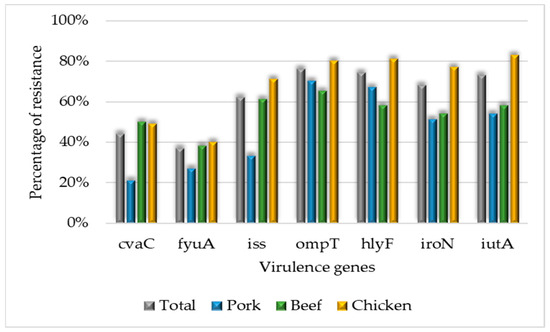
Figure 3.
Percentage of virulence genes detected in beef, chicken, and pork strains.
Pork strains presented more ExPEC 1 (42%) and ExPEC 2 (33%). Chicken strains presented more ExPEC genes (six to eight ExPEC genes, ExPEC 3) (OR 2.5, CI 1.33–5.01) than other meat samples. Remarkably, strains carrying three to five ExPEC genes exhibited more resistance to CIP (OR 1.15, CI 1.01–1.32) than other antimicrobials (Table 2).

Table 2.
Comparison of number of ExPEC genes profiles among isolates from beef, chicken, and pork samples.
3.6. Features of Diarrheogenic E. coli (DEC) Strains
The results obtained for DEC genes demonstrated that all strains were negative for stx, aggr, lt, and st.
Additionally, two strains harbored the eae gene (confirmed by genetic sequencing) but lacked the bfp gene. One of these strains originated from pork, while the other was isolated from chicken. Both strains were MDR, harbored the blaCTX-M-2 and fosA3 genes, and were classified as biofilm producers.
4. Discussion
In this study, we analyzed 450 samples, comprising 150 each from beef, pork, and chicken origins. The culture medium used in the initial isolation was supplemented with a third-generation cephalosporin (cefotaxime). Notably, poultry meat samples presented the highest number of E. coli strains, with more than 70% of samples yielding positive isolates, when compared to pork strains (OR 9.99, IC 5.74–17.80) and beef strains (OR 13.42, IC 7.50–24.73). Previous research conducted in the same region of the present study identified a high prevalence of MDR E. coli strains, ESBL producers, and carrying significant antimicrobial resistance genes in poultry production, as observed by Gazal et al. [61] and Menck–Costa et al. [48].
The detection of β-lactamase-producing strains is a major concern within the One Health approach [34,62,63]. According to the bulletin from the online notification system for hospital infections in Paraná [64], Escherichia coli was the fifth most frequently detected microorganism in diseases, with 31% of cases involving ESBL-producing strains. In our study, upon analysis of meat samples, we detected that 37% presented third-generation cephalosporins-resistant strains. But we alert that for chicken meat, 73% of carcass samples presented strains capable of growing in the presence of cefotaxime. It is worth highlighting that we are facing a pandemic of antimicrobial resistance [65].
Among the strains in this study, aside from being ESBL producers (exhibiting resistance to third-generation cephalosporins), over 45% were resistant to three or more classes of antimicrobials other than β-lactams. We detected greater resistance to tetracycline and ciprofloxacin, consistent with findings reported by Koga et al. [36] in strains isolated from chicken carcasses in Paraná in 2013, which showed high resistance rates to tetracycline and nalidixic acid. This suggests the persistence of a similar resistance profile over time.
The resistance profile to other antimicrobials among ESBL-producing strains was expected since the plasmids carrying CTX-M group genes often carry resistance determinants to other antimicrobials, as observed by Azargun et al. [66]. These researchers detected that approximately two-thirds of their ESBL-producing Enterobacteriaceae strains were resistant to quinolones. Similarly, Menck–Costa et al. [48] pointed out the potential of these strains to carry resistance to other antimicrobials, including quinolones, and correlated ESBL strains with the presence of the fosA3 gene. These findings illustrate the importance of detecting and monitoring ESBL strains, especially in meat, as their presence can lead to transmission to humans through contaminated food, resulting in infections that may be difficult to treat [1,67].
In our study, the fosA3 gene was most frequently detected in chicken meat samples (OR 2.43, CI 1.17–5.05) and least commonly in pork meat samples (OR 0.37, CI 0.16–0.83). Moreover, the high detection of resistance to fosfomycin in strains of animal origin is concerning, as this antimicrobial is considered the last therapeutic option against untreated urinary tract infections in humans [68,69]. Thus, selective antimicrobial use restricted to veterinary medicine or human medicine could minimize the problem of resistance in the animal-human axis [1].
It is essential to highlight that although this study identified a limited number of strains carrying DEC genes (specifically genes of atypical EPEC), these strains were classified as biofilm producers and displayed an MDR profile. Enteropathogenic E. coli (EPEC) can be characterized as typical or atypical. Typical EPEC strains possess the EAF plasmid, which includes the eae gene and is associated with the bundle-forming pilus (bfp) gene. This pathotype is known to cause diarrhea, particularly in children, while the atypical pathotype is frequently related to endemic outbreaks in underdeveloped countries [70]. Nevertheless, it is worth noting that the atypical EPEC, which lacks the bfp gene, is more commonly implicated in outbreaks in Brazil [29] and is rare in contaminated food sources [70,71].
The detection and prevention of DEC genes in such samples can help monitor the prevalence of DEC strains in the food supply. Furthermore, it is pivotal for the development of effective strategies to reduce their transmission to humans. Parussolo et al. [72] isolated strains of EPEC from raw milk in Brazil. These strains exhibited resistance to third-generation cephalosporins and carried the blaTEM gene. The detection of DEC strains with ESBL genes in food samples increases the importance of surveillance efforts. DEC, which mainly affects developing countries [73], poses challenges for treatment owing to its antimicrobial resistance profile. It is interesting to point out that both strains harboring the eae gene did not present any ExPEC gene.
In this study, ExPEC 3—strains harboring six to eight genes of extraintestinal pathogenic E. coli were most detected in chicken meat (OR 2.5, CI 1.33–5.01). These strains are known to carry genes associated with Avian Pathogenic Escherichia coli [74,75]. The episomal outer membrane protease gene, ompT, which cleaves colicins, was the most prevalent among these strains. This gene has previously been detected in strains responsible for urinary tract infections in humans [76,77]. Soncini et al. [32] pointed out the existence of a clonal relationship between strains isolated from humans and those isolated from animal protein, highlighting the importance of monitoring strains originating from meat sources.
Furthermore, strains classified as ExPEC 3, possessing a high number of virulence genes, displayed greater biofilm-forming capacity (OR 2.42, CI 1–5.87), which worsened the problem since these microorganisms have an increased likelihood of persisting in various environments, facilitating genetic exchange [1]. Thus, they represent a substantial risk for causing extraintestinal infections in humans, as demonstrated by Cyoia et al. [4]. The possible exchange of antimicrobial resistance genes from environmental bacteria to bacteria associated with human infections remains a significant concern [1,23].
In the geographic region where this study was carried out raw consumption of beef, pork, and chicken is uncommon, but it is important to emphasize that cooking these food items effectively eliminates the microorganisms but does not eliminate their genetic material, with the potential capacity for incorporation by other viable microorganisms [78,79]. Additionally, these microorganisms can be spread horizontally within multiple environments, such as kitchens, butcher shops, and refrigerators, potentially contaminating other food. Importantly, some of these contaminated foods may not be cooked before consumption [80,81,82]. Consequently, the detection and characterization of E. coli strains isolated from meat samples are important for monitoring and crucial to maintaining public health and safety, reducing economic losses in the food industry, and ultimately enhancing food safety protocols.
Among ESBL strains, the blaCTX-M-1 gene was the most prevalent, consistent with findings from prior studies [48,83] and corroborated by a European study [84]. Remarkably, the predominance of CTX-M-1 genes was observed in beef samples, although beef is not typically regarded as a primary indicator of antimicrobial resistance [40,85,86,87]. Conversely, pork strains presented more blaCTX-M-2 genes, highlighting the critical importance of detecting and characterizing strains that produce β-lactamase enzymes, as they provide insights into the epidemiological distribution of these genes. Such insights hold significant relevance in the context of antimicrobial resistance, an issue that has consequences for animal, human, and environmental health.
The detection of AmpC-producing strains is pivotal as it evaluates resistance to β-lactams. The first AmpC, the CMY-1 gene, was reported by Bauernfeind in 1989 [88]. Over time, with increasing selection pressure, many AmpC β-lactamase genes, surpassing 220 types, have been reported. AmpC genes have been correlated with food-producing animals [89]. However, in our study, we detected only a limited number of AmpC-producing strains, all isolated from chicken. These strains exhibited multidrug resistance (MDR), biofilm-producing capacity, and harbored various resistance-associated genes, including fosA3, iss, ompT, hlyF, iroN, and iutA. Therefore, strains with a relevant and worrying profile of antimicrobial resistance, as well as a high percentage of ExPEC genes, pose a risk for the occurrence of infections in humans, as discussed previously in this study.
All strains isolated in this study tested negative for carbapenem resistance, corroborating that carbapenem-resistant Enterobacteriaceae are not commonly found in non-human bacterial sources [38,83,90]. Moreover, the strains were also negative for the blaCTX-M-25 gene, which is commonly not detected in animal samples in Brazil [36,48,61]. Similarly, the mcr-1 gene, responsible for colistin resistance, an important antimicrobial in human clinical use [91], was not detected in any strain.
Notably, the hlyA gene, associated with alpha-hemolysin production and induction of kidney damage, was not detected in the strains. This gene is frequently isolated from patients with pyelonephritis [92] but has not been previously detected in meat strains during the same period [93].
According to Aslam et al. [94], antimicrobial resistance in samples of animal origin magnifies the emergence of superbugs. Several measures must be implemented to minimize this public health risk. These measures involve reducing the use of antimicrobials in animal production, ensuring meticulous care during animal slaughtering [95], imposing restrictions on the use of specific antimicrobials in animal production, highlighting the importance of these actions within the framework of the One Health approach, and ensuring the use of antimicrobials only under the guidance of professionals trained in the One Health approach.
The present study comprehensively characterizes E. coli strains resistant to third-generation cephalosporins (ESBL) in relation to relevant virulence and antimicrobial resistance genes. It underscores the public health risk these microorganisms pose in beef, chicken, and pork meat and highlights the importance of ongoing monitoring efforts.
5. Conclusions
Our study demonstrates the presence of β-lactamase-producing E. coli strains, mainly in poultry samples, when comparing different meat sources. Moreover, among all meat samples, ExPEC strains harboring many virulence genes, and some strains carrying DEC genes were identified. Notably, the samples exhibited a significant resistance profile to several classes of antimicrobials. These data corroborate existing literature and underscore the need for and importance of ongoing monitoring efforts for strains within meat samples.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11112712/s1, Figure S1: Profile of E. coli strains isolated from beef meat samples; Figure S2: Profile of E. coli strains isolated from pork meat samples; Figure S3: Profile of E. coli strains isolated from chicken meat samples. Table S1: Primers used for the detection of diarrheagenic E. coli (DEC) and extraintestinal pathogenic E. coli (ExPEC) genes, as well as antimicrobial resistance-related genes.
Author Contributions
Conceptualization: M.F.M.-C., A.A.S.B., M.S.S., L.P.M., S.P.D.R. and G.N.; Methodology: M.F.M.-C., A.A.S.B., M.S.S., B.Q.d.S., C.E.C., H.Y.K., L.P.M., L.J. and M.d.S.; Validation: R.K.T.K.; Formal analysis: R.K.T.K. and L.P.M.; Investigation: M.F.M.-C., M.S.S., B.Q.d.S. and C.E.C.; Resources: R.K.T.K., A.A.S.B., S.P.D.R. and G.N.; Data curation: M.F.M.-C.; Writing—original draft preparation: M.F.M.-C. and R.K.T.K. Writing—review and editing: M.F.M.-C. and R.K.T.K.; Supervision: R.K.T.K., S.P.D.R. and G.N.; Project administration: R.K.T.K.; Funding acquisition: R.K.T.K. and S.P.D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Microbiology Postgraduate Program of Universidade Estadual de Londrina and in part by the National Council for Scientific and Technological Development CNPq (409335/2021-5 Chamada CNPq/MCTI/FNDCT Nº 18/2021—Faixa B—Grupos Consolidados and 305972/2022-7 to RK). MFMC was funded by a Ph.D. scholarship from “CP 13/2020—PROGRAMA DE PESQUISA E INOVAÇÃO FUNDAÇÃO ARAUCÁRIA.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Acknowledgments
We would like to thank all the laboratory groups that indirectly contributed to the development of this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Larsson, D.G.J.; Flach, C.-F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020, 2020, e4674235. [Google Scholar] [CrossRef] [PubMed]
- Beshiru, A.; Igbinosa, I.H.; Igbinosa, E.O. Prevalence of Antimicrobial Resistance and Virulence Gene Elements of Salmonella Serovars rom Ready-to-Eat (RTE) Shrimps. Front. Microbiol. 2019, 10, 1613. [Google Scholar] [CrossRef] [PubMed]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; de Brito, K.C.T.; de Brito, B.G.; Nakazato, G.; Kobayashi, R.K.T. Distribution of ExPEC Virulence Factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli Isolated from Commercialized Chicken Carcasses. Front. Microbiol. 2019, 9, 3254. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Kumar, D.; Hussain, S.; Pathak, A.; Shukla, M.; Prasanna Kumar, V.; Anisha, P.N.; Rautela, R.; Upadhyay, A.K.; Singh, S.P. Prevalence, Antimicrobial Resistance and Virulence Genes Characterization of Nontyphoidal Salmonella Isolated from Retail Chicken Meat Shops in Northern India. Food Control 2019, 102, 104–111. [Google Scholar] [CrossRef]
- Schirone, M.; Visciano, P.; Tofalo, R.; Suzzi, G. Editorial: Foodborne Pathogens: Hyg\iene and Safety. Front. Microbiol. 2019, 10, 1974. [Google Scholar] [CrossRef]
- Maguire, M.; Kase, J.A.; Roberson, D.; Muruvanda, T.; Brown, E.W.; Allard, M.; Musser, S.M.; González-Escalona, N. Precision long-read metagenomics sequencing for food safety by detection and assembly of Shiga toxin-producing Escherichia coli in irrigation water. PLoS ONE 2021, 16, e0245172. [Google Scholar] [CrossRef]
- Hull, D.M.; Harrell, E.; Harden, L.; Thakur, S. Multidrug Resistance and Virulence Genes Carried by Mobile Genomic Elements in Salmonella Enterica Isolated from Live Food Animals, Processed, and Retail Meat in North Carolina, 2018–2019. Int. J. Food Microbiol. 2022, 378, 109821. [Google Scholar] [CrossRef]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The Interconnection between Biofilm Formation and Horizontal Gene Transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bhattacharjee, S.; Paul, S.; Nath, S.; Paul, S. Biofilm—A Syntrophic Consortia of Microbial Cells: Boon or Bane? Appl. Biochem. Biotechnol. 2022, 195, 5583–5604. [Google Scholar] [CrossRef]
- Jacques, M.; Malouin, F. One Health—One Biofilm. Vet. Res. 2022, 53, 51. [Google Scholar] [CrossRef] [PubMed]
- Chitlapilly Dass, S.; Wang, R. Biofilm through the Looking Glass: A Microbial Food Safety Perspective. Pathogens 2022, 11, 346. [Google Scholar] [CrossRef]
- Mazhangara, I.R.; Chivandi, E.; Mupangwa, J.F.; Muchenje, V. The Potential of Goat Meat in the Red Meat Industry. Sustainability 2019, 11, 3671. [Google Scholar] [CrossRef]
- Macho-González, A.; Garcimartín, A.; López-Oliva, M.E.; Bastida, S.; Benedí, J.; Ros, G.; Nieto, G.; Sánchez-Muniz, F.J. Can Meat and Meat-Products Induce Oxidative Stress? Antioxidants 2020, 9, 638. [Google Scholar] [CrossRef] [PubMed]
- Warmate, D.; Onarinde, B.A. Food Safety Incidents in the Red Meat Industry: A Review of Foodborne Disease Outbreaks Linked to the Consumption of Red Meat and Its Products, 1991 to 2021. Int. J. Food Microbiol. 2023, 398, 110240. [Google Scholar] [CrossRef] [PubMed]
- Soice, E.; Johnston, J. How Cellular Agriculture Systems Can Promote Food Security. Front. Sustain. Food Syst. 2021, 5, 753996. [Google Scholar] [CrossRef]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of Foodborne Illnesses, Hospitalizations, and Deaths to Food Commodities by Using Outbreak Data, United States, 1998–2008. Emerg. Infect. Dis. J. 2013, 19, 407–415. [Google Scholar] [CrossRef]
- Finger, J.A.F.F.; Baroni, W.S.G.V.; Maffei, D.F.; Bastos, D.H.M.; Pinto, U.M. Overview of Foodborne Disease Outbreaks in Brazil from 2000 to 2018. Foods 2019, 8, 434. [Google Scholar] [CrossRef]
- Scharff, R.L. Food Attribution and Economic Cost Estimates for Meat- and Poultry-Related Illnesses. J. Food Prot. 2020, 83, 959–967. [Google Scholar] [CrossRef]
- Hoffmann, S.; Ahn, J.-W. Updating Economic Burden of Foodborne Diseases Estimates for Inflation and Income Growth. Available online: http://www.ers.usda.gov/publications/pub-details/?pubid=102639 (accessed on 7 July 2023).
- Abdissa, R.; Haile, W.; Fite, A.T.; Beyi, A.F.; Agga, G.E.; Edao, B.M.; Tadesse, F.; Korsa, M.G.; Beyene, T.; Beyene, T.J.; et al. Prevalence of Escherichia coli O157:H7 in Beef Cattle at Slaughter and Beef Carcasses at Retail Shops in Ethiopia. BMC Infect. Dis. 2017, 17, 277. [Google Scholar] [CrossRef]
- Machado-Moreira, B.; Richards, K.; Brennan, F.; Abram, F.; Burgess, C.M. Microbial Contamination of Fresh Produce: What, Where, and How? Compr. Rev. Food Sci. Food Saf. 2019, 18, 1727–1750. [Google Scholar] [CrossRef]
- Enciso-Martínez, Y.; González-Aguilar, G.A.; Martínez-Téllez, M.A.; González-Pérez, C.J.; Valencia-Rivera, D.E.; Barrios-Villa, E.; Ayala-Zavala, J.F. Relevance of Tracking the Diversity of Escherichia coli Pathotypes to Reinforce Food Safety. Int. J. Food Microbiol. 2022, 374, 109736. [Google Scholar] [CrossRef] [PubMed]
- Arguello, H.; Carvajal, A.; Collazos, J.A.; García-Feliz, C.; Rubio, P. Prevalence and Serovars of Salmonella Enterica on Pig Carcasses, Slaughtered Pigs and the Environment of Four Spanish Slaughterhouses. Food Res. Int. 2012, 45, 905–912. [Google Scholar] [CrossRef]
- Abraham, S.; O’Dea, M.; Sahibzada, S.; Hewson, K.; Pavic, A.; Veltman, T.; Abraham, R.; Harris, T.; Trott, D.J.; Jordan, D. Escherichia coli and Salmonella spp. Isolated from Australian Meat Chickens Remain Susceptible to Critically Important Antimicrobial Agents. PLoS ONE 2019, 14, e0224281. [Google Scholar] [CrossRef]
- Santos, R.A.; Garcia, R.G.; Gandra, E.R.S.; Burbarelli, M.F.C.; Muchon, J.L.; Caldara, F.R. Carcass Washing as an Alternative to Trimming-Is It Possible to Use Carcass Washing as an Alternative to Trimming in Commercial Broiler Slaughterhouses in Brazil? Braz. J. Poult. Sci. 2020, 22, eRBCA. [Google Scholar] [CrossRef]
- Santeramo, F.G.; Lamonaca, E. Objective Risk and Subjective Risk: The Role of Information in Food Supply Chains. Food Res. Int. 2021, 139, 109962. [Google Scholar] [CrossRef] [PubMed]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The Population Genetics of Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef] [PubMed]
- MS, 2020. Boletim Epidemiológico da Secretaria de Vigilância em Saúde. Ministério da Saúde. v. 51. n.32. 2020. Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/edicoes/2020/boletim-epidemiologico-svs-32.pdf/view (accessed on 1 September 2023).
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L. Extended Virulence Genotypes of Escherichia coli Strains from Patients with Urosepsis in Relation to Phylogeny and Host Compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef]
- Soncini, J.G.M.; Cerdeira, L.; Sano, E.; Koga, V.L.; Tizura, A.T.; Tano, Z.N.; Nakazato, G.; Kobayashi, R.K.T.; Aires, C.A.M.; Lincopan, N.; et al. Genomic Insights of High-Risk Clones of ESBL-Producing Escherichia coli Isolated from Community Infections and Commercial Meat in Southern Brazil. Sci. Rep. 2022, 12, 9354. [Google Scholar] [CrossRef]
- Raphael, E.; Glymour, M.M.; Chambers, H.F. Trends in Prevalence of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Isolated from Patients with Community- and Healthcare-Associated Bacteriuria: Results from 2014 to 2020 in an Urban Safety-Net Healthcare System. Antimicrob. Resist. Infect. Control 2021, 10, 118. [Google Scholar] [CrossRef]
- CDC. Centers for Disease Control and Prevention (U.S.) Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention (U.S.): Atlanta, GA, USA, 2019.
- CDC COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; National Center for Emerging and Zoonotic Infectious Diseases: Atlanta, GA, USA, 2022.
- Koga, V.L.; Rodrigues, G.R.; Scandorieiro, S.; Vespero, E.C.; Oba, A.; de Brito, B.G.; de Brito, K.C.T.; Nakazato, G.; Kobayashi, R.K.T. Evaluation of the Antibiotic Resistance and Virulence of Escherichia coli Strains Isolated from Chicken Carcasses in 2007 and 2013 from Paraná, Brazil. Foodborne Pathog. Dis. 2015, 12, 479–485. [Google Scholar] [CrossRef]
- Kürekci, C.; Osek, J.; Aydın, M.; Tekeli, İ.O.; Kurpas, M.; Wieczorek, K.; Sakin, F. Evaluation of Bulk Tank Raw Milk and Raw Chicken Meat Samples as Source of ESBL Producing Escherichia coli in Turkey: Recent Insights. J. Food Saf. 2019, 39, e12605. [Google Scholar] [CrossRef]
- Sahin, S.; Mogulkoc, M.N.; Kürekci, C. Disinfectant and Heavy Metal Resistance Profiles in Extended Spectrum β-Lactamase (ESBL) Producing Escherichia coli Isolates from Chicken Meat Samples. Int. J. Food Microbiol. 2022, 377, 109831. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Oh, T.; Baek, S.-Y. Multidrug Resistance, Biofilm Formation, and Virulence of Escherichia coli Isolates from Commercial Meat and Vegetable Products. Foodborne Pathog. Dis 2018, 15, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in Prevalence and Characteristics of ESBL/pAmpC Producing E. coli in Food in Germany. Vet. Microbiol 2019, 233, 52–60. [Google Scholar] [CrossRef]
- Uhland, F.C.; Li, X.-Z.; Mulvey, M.R.; Reid-Smith, R.; Sherk, L.M.; Ziraldo, H.; Jin, G.; Young, K.M.; Reist, M.; Carson, C.A. Extended Spectrum β-Lactamase-Producing Enterobacterales of Shrimp and Salmon Available for Purchase by Consumers in Canada—A Risk Profile Using the Codex Framework. Antibiotics 2023, 12, 1412. [Google Scholar] [CrossRef]
- Kahn, L.H.; Kaplan, B.; Steele, J.H. Confronting Zoonoses through Closer Collaboration between Medicine and Veterinary Medicine (as ‘One Medicine’). Vet. Ital. 2007, 43, 5–19. [Google Scholar]
- Mackenzie, J.S.; Jeggo, M. The One Health Approach—Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef] [PubMed]
- Wildlife Conservation Society One World-One Health: Construindo Pontes Interdisciplinares. 2004. Available online: http://www.oneworldonehealth.org/sept2004/owoh_sept04.html (accessed on 1 September 2023).
- CDC. The Biggest Antibiotic-Resistant Threats in the U.S. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 9 July 2023).
- Silva, A.; Silva, V.; Pereira, J.E.; Maltez, L.; Igrejas, G.; Valentão, P.; Falco, V.; Poeta, P. Antimicrobial Resistance and Clonal Lineages of Escherichia coli from Food-Producing Animals. Antibiotics 2023, 12, 1061. [Google Scholar] [CrossRef]
- Grudlewska-Buda, K.; Bauza-Kaszewska, J.; Wiktorczyk-Kapischke, N.; Budzyńska, A.; Gospodarek-Komkowska, E.; Skowron, K. Antibiotic Resistance in Selected Emerging Bacterial Foodborne Pathogens—An Issue of Concern? Antibiotics 2023, 12, 880. [Google Scholar] [CrossRef]
- Menck-Costa, M.F.; Baptista, A.A.S.; de Souza Gazal, L.E.; Justino, L.; Sanches, M.S.; de Souza, M.; Nishio, E.K.; Queiroz dos Santos, B.; Cruz, V.D.; Berbert, J.V.M.; et al. High-Frequency Detection of fosA3 and blaCTX-M-55 Genes in Escherichia coli from Longitudinal Monitoring in Broiler Chicken Farms. Front. Microbiol. 2022, 13, 846116. [Google Scholar] [CrossRef] [PubMed]
- Twenty-Fifth Informational Supplement CLSI Document M100; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- CLSI Guideline M100-S29; Suggested Grouping of US-FDA Approved Antimicrobial Agents That Should Be Considered for Routine Testing and Reporting on Nonfastidious Organisms by Clinical Laboratories. 29th ed. Clinical and Laboratory Institute: Wayne, PA, USA, 2019.
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A Modified Microtiter-Plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Yang, X.; Guo, W.; Wang, J.; Chen, G. Inclusion Complex of Docetaxel with Sulfobutyl Ether β-Cyclodextrin: Preparation, In Vitro Cytotoxicity and In Vivo Safety. Polymers 2020, 12, 2336. [Google Scholar] [CrossRef]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for Rapid Detection of Genes Encoding CTX-M Extended-Spectrum (Beta)-Lactamases. J. Antimicrob. Chemother. 2006, 57, 154–155. [Google Scholar] [CrossRef]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC Beta-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Sato, N.; Kawamura, K.; Nakane, K.; Wachino, J.-I.; Arakawa, Y. First Detection of Fosfomycin Resistance Gene fosA3 in CTX-M-Producing Escherichia coli Isolates from Healthy Individuals in Japan. Microb. Drug Resist. Larchmt. 2013, 19, 477–482. [Google Scholar] [CrossRef]
- Aranda, K.R.S.; Fagundes-Neto, U.; Scaletsky, I.C.A. Evaluation of Multiplex PCRs for Diagnosis of Infection with Diarrheagenic Escherichia coli and Shigella spp. J. Clin. Microbiol. 2004, 42, 5849–5853. [Google Scholar] [CrossRef]
- Toma, C.; Lu, Y.; Higa, N.; Nakasone, N.; Chinen, I.; Baschkier, A.; Rivas, M.; Iwanaga, M. Multiplex PCR Assay for Identification of Human Diarrheagenic Escherichia coli. J. Clin. Microbiol. 2003, 41, 2669–2671. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of Minimal Predictors of Avian Pathogenic Escherichia coli Virulence for Use as a Rapid Diagnostic Tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef] [PubMed]
- de Souza Gazal, L.E.; Medeiros, L.P.; Dibo, M.; Nishio, E.K.; Koga, V.L.; Gonçalves, B.C.; Grassotti, T.T.; de Camargo, T.C.L.; Pinheiro, J.J.; Vespero, E.C.; et al. Detection of ESBL/AmpC-Producing and Fosfomycin-Resistant Escherichia coli From Different Sources in Poultry Production in Southern Brazil. Front. Microbiol. 2021, 11, 604544. [Google Scholar] [CrossRef]
- Yair, Y.; Gophna, U. Pandemic Bacteremic Escherichia coli Strains: Evolution and Emergence of Drug-Resistant Pathogens. In Escherichia coli, a Versatile Pathogen; Current Topics in Microbiology and Immunology; Frankel, G., Ron, E.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 416, pp. 163–180. ISBN 978-3-319-99663-9. [Google Scholar]
- Daga, A.P.; Koga, V.L.; Soncini, J.G.M.; de Matos, C.M.; Perugini, M.R.E.; Pelisson, M.; Kobayashi, R.K.T.; Vespero, E.C. Escherichia coli Bloodstream Infections in Patients at a University Hospital: Virulence Factors and Clinical Characteristics. Front. Cell. Infect. Microbiol. 2019, 9, 191. [Google Scholar] [CrossRef]
- SONIH. Boletim Epidemiológico das IRAS. Secretaria do Estado da saúde do Paraná. 2019. Available online: https://www.saude.pr.gov.br/sites/default/arquivos_restritos/files/documento/2021-02/boletim_sonih_resumido_15_05_2020.pdf (accessed on 1 September 2023).
- Laxminarayan, R. The Overlooked Pandemic of Antimicrobial Resistance. Lancet Lond. Engl. 2022, 399, 606–607. [Google Scholar] [CrossRef]
- Azargun, R.; Gholizadeh, P.; Sadeghi, V.; Hosainzadegan, H.; Tarhriz, V.; Memar, M.Y.; Pormohammad, A.; Eyvazi, S. Molecular Mechanisms Associated with Quinolone Resistance in Enterobacteriaceae: Review and Update. Trans. R. Soc. Trop. Med. Hyg. 2020, 114, 770–781. [Google Scholar] [CrossRef]
- Dsani, E.; Afari, E.A.; Danso-Appiah, A.; Kenu, E.; Kaburi, B.B.; Egyir, B. Antimicrobial Resistance and Molecular Detection of Extended Spectrum β-Lactamase Producing Escherichia coli Isolates from Raw Meat in Greater Accra Region, Ghana. BMC Microbiol. 2020, 20, 253. [Google Scholar] [CrossRef]
- WHO. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- WHO. The 2019 WHO AWaRe Classification of Antibiotics for Evaluation and Monitoring of Use. 2019. Available online: https://apps.who.int/iris/handle/10665/327957 (accessed on 11 August 2023).
- Govindarajan, D.K.; Viswalingam, N.; Meganathan, Y.; Kandaswamy, K. Adherence Patterns of Escherichia coli in the Intestine and Its Role in Pathogenesis. Med. Microecol. 2020, 5, 100025. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef]
- Parussolo, L.; Sfaciotte, R.A.P.; Dalmina, K.A.; Melo, F.D.; Costa, U.M.; Ferraz, S.M. Detection of Virulence Genes and Antimicrobial Resistance Profiles of Escherichia coli Isolates from Raw Milk and Artisanal Cheese in Southern Brazil. Semina Ciênc. Agrár. 2019, 40, 163. [Google Scholar] [CrossRef]
- Mare, A.D.; Ciurea, C.N.; Man, A.; Tudor, B.; Moldovan, V.; Decean, L.; Toma, F. Enteropathogenic Escherichia coli—A Summary of the Literature. Gastroenterol. Insights 2021, 12, 28–40. [Google Scholar] [CrossRef]
- Johar, A.; Al-Thani, N.; Al-Hadidi, S.H.; Dlissi, E.; Mahmoud, M.H.; Eltai, N.O. Antibiotic Resistance and Virulence Gene Patterns Associated with Avian Pathogenic Escherichia coli (APEC) from Broiler Chickens in Qatar. Antibiotics 2021, 10, 564. [Google Scholar] [CrossRef]
- Johnson, T.J.; Miller, E.A.; Flores-Figueroa, C.; Munoz-Aguayo, J.; Cardona, C.; Fransen, K.; Lighty, M.; Gonder, E.; Nezworski, J.; Haag, A.; et al. Refining the Definition of the Avian Pathogenic Escherichia coli (APEC) Pathotype through Inclusion of High-Risk Clonal Groups. Poult. Sci. 2022, 101, 102009. [Google Scholar] [CrossRef] [PubMed]
- Desloges, I.; Taylor, J.A.; Leclerc, J.-M.; Brannon, J.R.; Portt, A.; Spencer, J.D.; Dewar, K.; Marczynski, G.T.; Manges, A.; Gruenheid, S.; et al. Identification and Characterization of OmpT-like Proteases in Uropathogenic Escherichia coli Clinical Isolates. MicrobiologyOpen 2019, 8, e915. [Google Scholar] [CrossRef] [PubMed]
- Hasanli, L.; Dagi, H.T.; Arslan, U. Investigation of Antibiotic Susceptibility and Virulence Genes in Escherichia coli Strains Isolated from Blood and Urine Samples. J. Pediatr. Infect. Dis. 2022, 17, 98–105. [Google Scholar] [CrossRef]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in Natural Environments: Features, Relevance and Applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef]
- Calderón-Franco, D.; van Loosdrecht, M.C.M.; Abeel, T.; Weissbrodt, D.G. Free-Floating Extracellular DNA: Systematic Profiling of Mobile Genetic Elements and Antibiotic Resistance from Wastewater. Water Res. 2021, 189, 116592. [Google Scholar] [CrossRef]
- Braz, V.S.; Melchior, K.; Moreira, C.G. Escherichia coli as a Multifaceted Pathogenic and Versatile Bacterium. Front. Cell. Infect. Microbiol. 2020, 10, 548492. [Google Scholar] [CrossRef]
- USDA. Food Safety Education Month: Preventing Cross-Contamination | Food Safety and Inspection Service 2022. Available online: http://www.fsis.usda.gov/news-events/events-meetings/food-safety-education-month-preventing-cross-contamination (accessed on 7 October 2023).
- Tropea, A. Microbial Contamination and Public Health: An Overview. Int. J. Environ. Res. Public Health 2022, 19, 7441. [Google Scholar] [CrossRef]
- Clemente, L.; Manageiro, V.; Correia, I.; Amaro, A.; Albuquerque, T.; Themudo, P.; Ferreira, E.; Caniça, M. Revealing mcr-1-Positive ESBL-Producing Escherichia coli Strains among Enterobacteriaceae from Food-Producing Animals (Bovine, Swine and Poultry) and Meat (Bovine and Swine), Portugal, 2010–2015. Int. J. Food Microbiol. 2019, 296, 37–42. [Google Scholar] [CrossRef]
- Schill, F.; Abdulmawjood, A.; Klein, G.; Reich, F. Prevalence and Characterization of Extended-Spectrum β-Lactamase (ESBL) and AmpC β-Lactamase Producing Enterobacteriaceae in Fresh Pork Meat at Processing Level in Germany. Int. J. Food Microbiol. 2017, 257, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Giammanco, A.; Cardamone, C.; Oliveri, G.; Mascarella, C.; Capra, G.; Fasciana, T. Extra-Intestinal Fluoroquinolone-Resistant Escherichia coli Strains Isolated from Meat. BioMed Res. Int. 2018, 2018, 8714975. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Randall, L.P.; Lodge, M.P.; Elviss, N.C.; Lemma, F.L.; Hopkins, K.L.; Teale, C.J.; Woodford, N. Evaluation of Meat, Fruit and Vegetables from Retail Stores in Five United Kingdom Regions as Sources of Extended-Spectrum Beta-Lactamase (ESBL)-Producing and Carbapenem-Resistant Escherichia coli. Int. J. Food Microbiol. 2017, 241, 283–290. [Google Scholar] [CrossRef]
- Bauernfeind, A.; Chong, Y.; Lee, K. Plasmid-Encoded AmpC Beta-Lactamases: How Far Have We Gone 10 Years after the Discovery? Yonsei Med. J. 1998, 39, 520–525. [Google Scholar] [CrossRef]
- Koga, V.L.; Maluta, R.P.; da Silveira, W.D.; Ribeiro, R.A.; Hungria, M.; Vespero, E.C.; Nakazato, G.; Kobayashi, R.K.T. Characterization of CMY-2-Type Beta-Lactamase-Producing Escherichia coli Isolated from Chicken Carcasses and Human Infection in a City of South Brazil. BMC Microbiol. 2019, 19, 174. [Google Scholar] [CrossRef]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-Resistant Enterobacteriaceae in Wildlife, Food-Producing, and Companion Animals: A Systematic Review. Clin. Microbiol. Infect. 2018, 24, 1241–1250. [Google Scholar] [CrossRef]
- Li, M.; Yang, F.; Lu, Y.; Huang, W. Identification of Enterococcus faecalis in a Patient with Urinary-Tract Infection Based on Metagenomic next-Generation Sequencing: A Case Report. BMC Infect. Dis. 2020, 20, 467. [Google Scholar] [CrossRef]
- Wang, C.; Li, Q.; Lv, J.; Sun, X.; Cao, Y.; Yu, K.; Miao, C.; Zhang, Z.-S.; Yao, Z.; Wang, Q. Alpha-Hemolysin of Uropathogenic Escherichia coli Induces GM-CSF-Mediated Acute Kidney Injury. Mucosal Immunol. 2020, 13, 22–33. [Google Scholar] [CrossRef]
- Sanches, M.S.; Rodrigues da Silva, C.; Silva, L.C.; Montini, V.H.; Lopes Barboza, M.G.; Migliorini Guidone, G.H.; Dias de Oliva, B.H.; Nishio, E.K.; Faccin Galhardi, L.C.; Vespero, E.C.; et al. Proteus mirabilis from Community-Acquired Urinary Tract Infections (UTI-CA) Shares Genetic Similarity and Virulence Factors with Isolates from Chicken, Beef and Pork Meat. Microb. Pathog. 2021, 158, 105098. [Google Scholar] [CrossRef]
- Aslam, M.; Diarra, M.S.; Checkley, S.; Bohaychuk, V.; Masson, L. Characterization of Antimicrobial Resistance and Virulence Genes in Enterococcus spp. Isolated from Retail Meats in Alberta, Canada. Int. J. Food Microbiol. 2012, 156, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Dhaka, P.; Chantziaras, I.; Vijay, D.; Bedi, J.S.; Makovska, I.; Biebaut, E.; Dewulf, J. Can Improved Farm Biosecurity Reduce the Need for Antimicrobials in Food Animals? A Scoping Review. Antibiotics 2023, 12, 893. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).