Comprehensive Genomic Characterization of Cronobacter sakazakii Isolates from Infant Formula Processing Facilities Using Whole-Genome Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates and Sample Preparation
2.2. DNA Extraction
2.3. WGS and Bioinformatic Analysis
2.4. Detection of Virulence and Antimicrobial Resistance Genes
2.5. Antimicrobial Susceptibility Test
2.6. Detection of Plasmids and Prophages
3. Results and Discussion
3.1. General Features of C. Sakazakii Strains
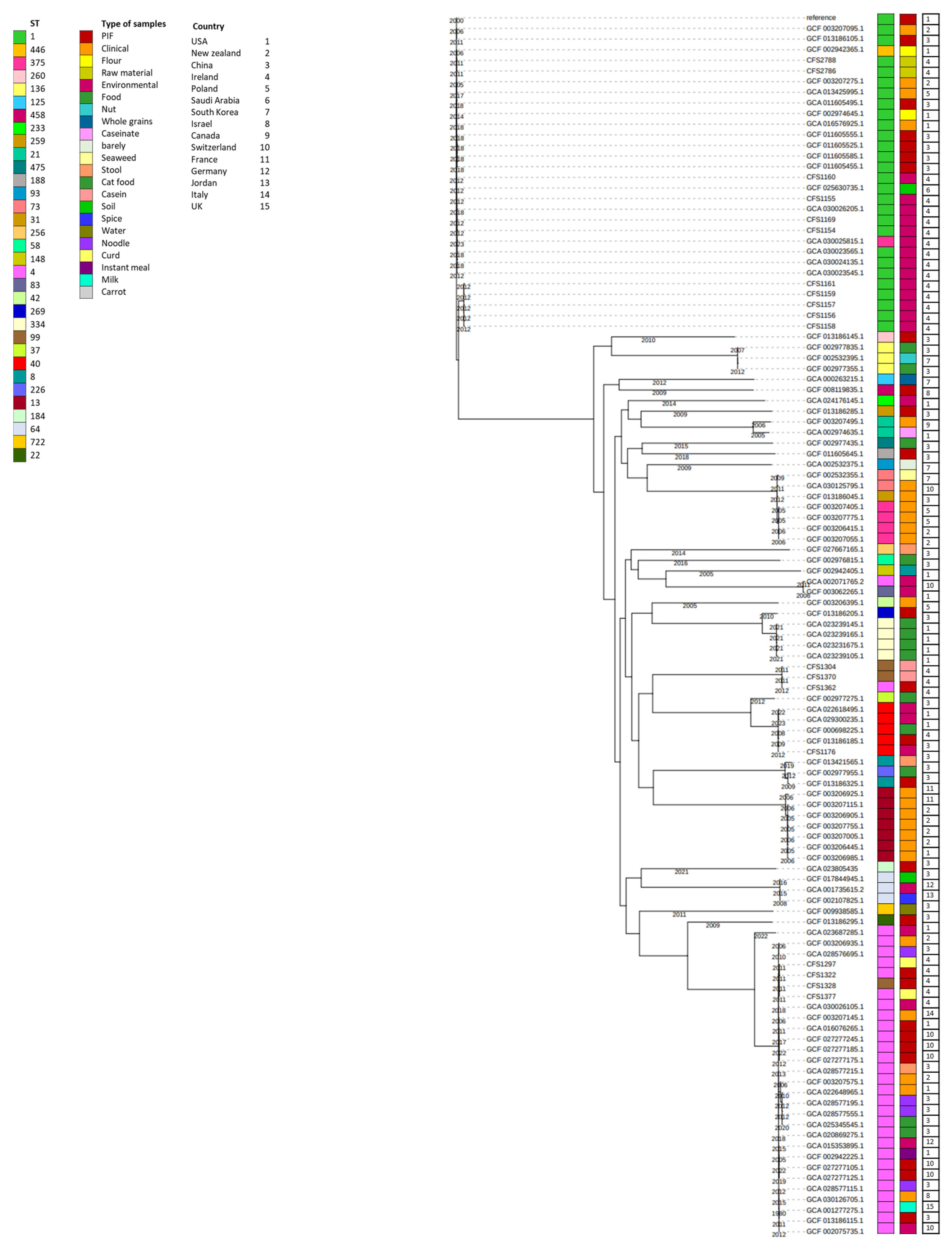
3.2. Phylogenetic Analysis
3.3. Virulence Factors Identified in the Selected Isolates
3.3.1. Virulence Factors Associated with Environmental Persistence
Adaptation to High Temperature
Adaptation to Acidic Environments, Osmotic Pressure, and Desiccation
3.3.2. Virulence Factors Associated with Invasion and Persistence in the Host Cell
Cell Attachment and Invasion
- 1-Flagellar and chemotaxis proteins
- 2-Outer membrane proteins (OMPs)
- 3-Lipopolysaccharides (LPSs)
- 4-Macrophage survival
- 5-Anti-Toxins
- 6-Utilization of Sialic Acid
- 7-Iron Acquisition System
- 8-Zinc Acquisition System
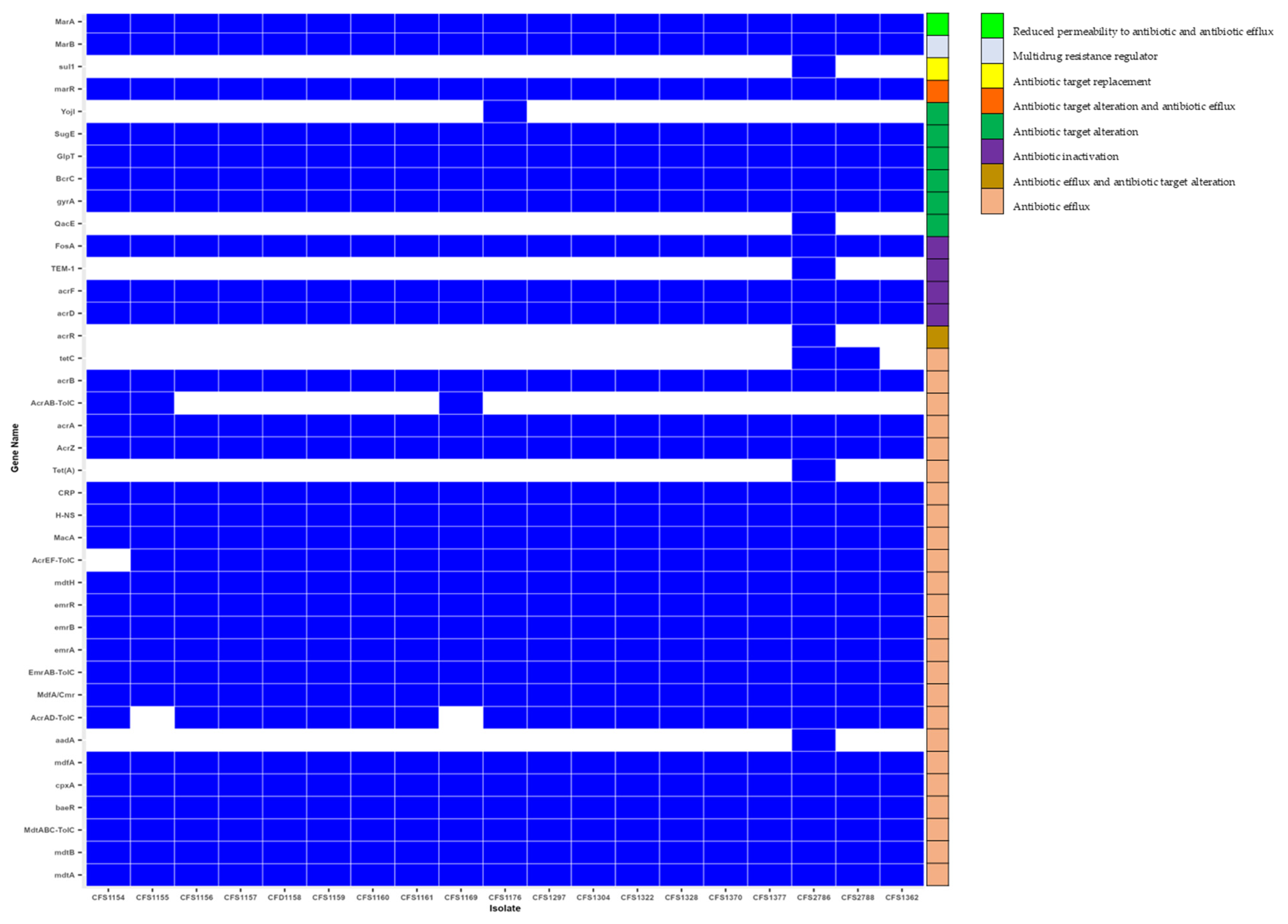
3.4. Antimicrobial Resistance Genes
3.5. Antibiotic Resistance Profile
3.6. Detection of Plasmids and Mobile Genetic Elements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Joseph, S.; Forsythe, S.J. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front. Microbiol. 2012, 3, 397. [Google Scholar] [CrossRef] [PubMed]
- Fei, P.; Man, C.; Lou, B.; Forsythe, S.J.; Chai, Y.; Li, R.; Niu, J.; Jiang, Y. Genotyping and source tracking of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and an infant formula production factory in China. Appl. Environ. Microbiol. 2015, 81, 5430–5439. [Google Scholar] [CrossRef]
- Holý, O.; Parra-Flores, J.; Lepuschitz, S.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Mancilla-Rojano, J.; Ruppitsch, W.; Forsythe, S. Molecular characterization of Cronobacter sakazakii strains isolated from powdered milk. Foods 2020, 10, 20. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Riffo, F.; Lepuschitz, S.; Maury-Sintjago, E.; Rodríguez-Fernández, A.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Mancilla-Rojano, J.; Troncoso, M. Profiling the virulence and antibiotic resistance genes of Cronobacter sakazakii strains isolated from powdered and dairy formulas by whole-genome sequencing. Front. Microbiol. 2021, 12, 694922. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Acuña, S.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Chavarría-Sepulveda, P.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Mancilla-Rojano, J. Genomic Characterization of Cronobacter spp. and Salmonella spp. Strains Isolated From Powdered Infant Formula in Chile. Front. Microbiol. 2022, 13, 884721. [Google Scholar] [CrossRef]
- Phair, K.; Pereira, S.G.; Kealey, C.; Fanning, S.; Brady, D.B. Insights into the mechanisms of Cronobacter sakazakii virulence. Microb. Pathog. 2022, 169, 105643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.J.; Bin, W.; Jing, Y.; Liang, H.Y.; Dong, S.G.; Ming, Z. Examine the Correlation between Heat Shock Protein IbpA and Heat Tolerance in Cronobacter sakazakii. Biomed. Environ. Sci. 2017, 30, 606–610. [Google Scholar] [PubMed]
- Niu, H.; Yang, M.; Qi, Y.; Liu, Y.; Wang, X.; Dong, Q. Heat shock in Cronobacter sakazakii induces direct protection and cross-protection against simulated gastric fluid stress. Food Microbiol. 2022, 103, 103948. [Google Scholar] [CrossRef]
- Heijden, J.v.d.; Reynolds, L.A.; Deng, W.; Mills, A.; Scholz, R.; Imami, K.; Foster, L.J.; Duong, F.; Finlay, B.B. Salmonella Rapidly Regulates Membrane Permeability To Survive Oxidative Stress. mBio 2016, 7, e01238-16. [Google Scholar] [CrossRef]
- Šulskis, D. The Structural Modularity and Inherent Dynamics of the DegP Protease together with its Intertwined Role in the Bacterial Periplasm. Ph.D. Thesis, University of Gothenburg, Gothenburg, Sweden, 2021. [Google Scholar]
- Zheng, J.; Wu, Y.; Lin, Z.; Wang, G.; Jiang, S.; Sun, X.; Tu, H.; Yu, Z.; Qu, D. ClpP participates in stress tolerance, biofilm formation, antimicrobial tolerance, and virulence of Enterococcus faecalis. BMC Microbiol. 2020, 20, 30. [Google Scholar] [CrossRef]
- Osaili, T.; Forsythe, S. Desiccation resistance and persistence of Cronobacter species in infant formula. Int. J. Food Microbiol. 2009, 136, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.; Cummins, C.; Hunt, M.; Wong, V.; Reuter, S.; Holden, M.; Fookes, M.; Keane, J.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Gan, X.; Li, M.; Yan, S.; Wang, X.; Wang, W.; Li, F. Genomic Landscape and Phenotypic Assessment of Cronobacter sakazakii Isolated From Raw Material, Environment, and Production Facilities in Powdered Infant Formula Factories in China. Front. Microbiol. 2021, 12, 686189. [Google Scholar] [CrossRef]
- Tan, Y.S.; Zhang, R.K.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Microbial Adaptation to Enhance Stress Tolerance. Front. Microbiol. 2022, 13, 888746. [Google Scholar] [CrossRef]
- Wu, R.A.; Yuk, H.G.; Liu, D.; Ding, T. Recent advances in understanding the effect of acid-adaptation on the cross-protection to food-related stress of common foodborne pathogens. Crit. Rev. Food Sci. Nutr. 2022, 62, 7336–7353. [Google Scholar] [CrossRef]
- Fei, Y.-Y.; Gai, J.-Y.; Zhao, T.-J. Identification of regulated genes conferring resistance to high concentrations of glyphosate in a new strain of Enterobacter. FEMS Microbiol. Lett. 2013, 349, 135–143. [Google Scholar] [CrossRef]
- Singh, N.; Goel, G.; Raghav, M. Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 2015, 6, 433–440. [Google Scholar] [CrossRef]
- Jang, H.; Gopinath, G.R.; Eshwar, A.; Srikumar, S.; Nguyen, S.; Gangiredla, J.; Patel, I.R.; Finkelstein, S.B.; Negrete, F.; Woo, J.; et al. The Secretion of Toxins and Other Exoproteins of Cronobacter: Role in Virulence, Adaption, and Persistence. Microorganisms 2020, 8, 229. [Google Scholar] [CrossRef]
- Maerani, M.; Dewanti-Hariyadi, R.; Nurjanah, S. Expression of stress regulator and virulence genes of Cronobacter sakazakii strain Yrt2a as a response to acid stress. Food Sci. Biotechnol. 2020, 29, 1273–1279. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, X.; Zhang, M.; Ling, N.; Zeng, H.; Gao, J.; Jiao, R.; Wu, Q.; Zhang, J. Potential factors involved in virulence of Cronobacter sakazakii isolates by comparative transcriptome analysis. J. Dairy Sci. 2017, 100, 8826–8837. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, H.; Kim, K.P.; Yoon, H.; Kang, D.H.; Ryu, S. Hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 2015, 83, 2089–2098. [Google Scholar] [CrossRef]
- Aly, M.A.; Domig, K.J.; Kneifel, W.; Reimhult, E. Whole genome sequencing-based comparison of food isolates of Cronobacter sakazakii. Front. Microbiol. 2019, 10, 1464. [Google Scholar] [CrossRef] [PubMed]
- Srikumar, S.; Cao, Y.; Yan, Q.; Van Hoorde, K.; Nguyen, S.; Cooney, S.; Gopinath, G.R.; Tall, B.D.; Sivasankaran, S.K.; Lehner, A. RNA sequencing-based transcriptional overview of xerotolerance in Cronobacter sakazakii SP291. Appl. Environ. Microbiol. 2019, 85, e01993-18. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Dever, K.; Sivasankaran, S.K.; Nguyen, S.V.; Macori, G.; Naithani, A.; Gopinath, G.R.; Tall, B.; Lehner, A.; Stephan, R.; et al. Alterations in the Transcriptional Landscape Allow Differential Desiccation Tolerance in Clinical Cronobacter sakazakii. Appl. Environ. Microbiol. 2021, 87, e0083021. [Google Scholar] [CrossRef]
- Alsonosi, A.M. Identification of Physiological and Virulence Traits of Clinical Strains of Cronobacter Malonaticus. Ph.D. Thesis, Nottingham Trent University, Nottingham, UK, 2017. [Google Scholar]
- Ye, Y.; Gao, J.; Jiao, R.; Li, H.; Wu, Q.; Zhang, J.; Zhong, X. The membrane proteins involved in virulence of Cronobacter sakazakii virulent G362 and attenuated L3101 isolates. Front. Microbiol. 2015, 6, 1238. [Google Scholar] [CrossRef]
- Yan, Q.; Power, K.A.; Cooney, S.; Fox, E.; Gopinath, G.R.; Grim, C.J.; Tall, B.D.; McCusker, M.P.; Fanning, S. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: A persistent isolate cultured from a powdered infant formula production facility. Front. Microbiol. 2013, 4, 256. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Jiang, X.; Forsythe, S.; Zhang, D.; Shen, Y.; Ding, Y.; Wang, J.; Zhang, J.; Wu, Q.; Ye, Y. Food Safety Risks and Contributing Factors of Cronobacter spp. Engineering 2022, 12, 128–138. [Google Scholar] [CrossRef]
- Yan, X.; Gurtler, J.; Fratamico, P.; Hu, J.; Gunther IV, N.W.; Juneja, V.; Huang, L. Comprehensive approaches to molecular biomarker discovery for detection and identification of Cronobacter spp.(Enterobacter sakazakii) and Salmonella spp. Appl. Environ. Microbiol. 2011, 77, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Eshwar, A.K.; Tasara, T.; Stephan, R.; Lehner, A. Influence of FkpA variants on survival and replication of Cronobacter spp. in human macrophages. Res. Microbiol. 2015, 166, 186–195. [Google Scholar] [CrossRef]
- Bartoli, J.; Viala, J.P.; Bouveret, E. SlyA transcriptional regulator is not directly affected by ppGpp levels. Front. Microbiol. 2020, 11, 1856. [Google Scholar] [CrossRef]
- Grim, C.; Kothary, M.; Gopinath, G.; Jarvis, K.; Beaubrun, J.J.-G.; McClelland, M.; Tall, B.; Franco, A. Identification and characterization of Cronobacter iron acquisition systems. Appl. Environ. Microbiol. 2012, 78, 6035–6050. [Google Scholar] [CrossRef]
- Neupane, D.P.; Kumar, S.; Yukl, E.T. Two ABC transporters and a periplasmic metallochaperone participate in zinc acquisition in Paracoccus denitrificans. Biochemistry 2018, 58, 126–136. [Google Scholar] [CrossRef]
- Carvalho, G.G.; Calarga, A.P.; Teodoro, J.R.; Queiroz, M.M.; Astudillo-Trujillo, C.A.; Levy, C.E.; Brocchi, M.; Kabuki, D.Y. Isolation, comparison of identification methods and antibiotic resistance of Cronobacter spp. in infant foods. Food Res. Int. 2020, 137, 109643. [Google Scholar] [CrossRef] [PubMed]
- Holý, O.; Alsonosi, A.; Hochel, I.; Röderová, M.; Zatloukalova, S.; Mlynárčik, P.; Kolář, M.; Petrželová, J.; Alazraq, A.; Chmelař, D. Antibiotic Susceptibility of spp. Isolated from Clinical Samples. Pol. J. Microbiol. 2019, 68, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Li, M.; Xu, J.; Yan, S.; Wang, W.; Li, F. Emerging of Multidrug-Resistant Cronobacter sakazakii Isolated from Infant Supplementary Food in China. Microbiol. Spectr. 2022, 10, e0119722. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.H.; Bo, Y.; Xiang, Y.; Zhang, Z.; Zhang, T.; Zeng, Y.C.; Cui, Z.G.; Huo, X.X. Two cases of multi-antibiotic resistant Cronobacter spp. infections of infants in China. Biomed. Environ. Sci. 2017, 30, 601–605. [Google Scholar]
- Pakbin, B.; Brück, W.M.; Allahyari, S.; Rossen, J.W.A.; Mahmoudi, R. Antibiotic Resistance and Molecular Characterization of Cronobacter sakazakii Strains Isolated from Powdered Infant Formula Milk. Foods 2022, 11, 1093. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.V.; Vasconcellos, L.; da Silva, I.C.; de Mello Medeiros, V.; Forsythe, S.J.; Brandão, M.L.L. Multi-locus sequence typing and antimicrobial susceptibility profile of Cronobacter sakazakii and Cronobacter malonaticus isolated from corn-based farinaceous foods commercialized in Brazil. Food Res. Int. 2020, 129, 108805. [Google Scholar] [CrossRef]
- Fei, P.; Jiang, Y.; Feng, J.; Forsythe, S.J.; Li, R.; Zhou, Y.; Man, C. Antibiotic and Desiccation Resistance of Cronobacter sakazakii and C. malonaticus Isolates from Powdered Infant Formula and Processing Environments. Front. Microbiol. 2017, 8, 316. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Aguirre, J.; Juneja, V.; Jackson, E.E.; Cruz-Córdova, A.; Silva-Sanchez, J.; Forsythe, S. Virulence and antibiotic resistance profiles of Cronobacter sakazakii and Enterobacter spp. involved in the diarrheic hemorrhagic outbreak in Mexico. Front. Microbiol. 2018, 9, 2206. [Google Scholar] [CrossRef]
- Eshetea, B.B. Characterization of the Resistome and Bacteriophages in Kitfo, an Ethiopian Beef Steak Tartar. Ph.D. Thesis, Howard University, Washington, DC, USA, 2021. [Google Scholar]
- Li, L.; Olsen, R.H.; Wang, C.; Song, A.; Xiao, J.; Meng, H.; Ronco, T.; Shi, L. First report of two foodborne Salmonella enterica subsp. enterica serovar Bovismorbificans isolates carrying a novel mega-plasmid harboring blaDHA-1 and qnrB4 genes. Int. J. Food Microbiol. 2021, 360, 109439. [Google Scholar] [CrossRef]
- Torrez Lamberti, M.F.; Terán, L.C.; Lopez, F.E.; de las Mercedes Pescaretti, M.; Delgado, M.A. Genomic and proteomic characterization of two strains of Shigella flexneri 2 isolated from infants’ stool samples in Argentina. BMC Genom. 2022, 23, 495. [Google Scholar] [CrossRef]
- Jang, H.; Chase, H.R.; Gangiredla, J.; Grim, C.J.; Patel, I.R.; Kothary, M.H.; Jackson, S.A.; Mammel, M.K.; Carter, L.; Negrete, F. Analysis of the molecular diversity among Cronobacter species isolated from filth flies using targeted PCR, pan genomic DNA microarray, and whole genome sequencing analyses. Front. Microbiol. 2020, 11, 561204. [Google Scholar] [CrossRef]
- Batrich, M.; Maskeri, L.; Schubert, R.; Ho, B.; Kohout, M.; Abdeljaber, M.; Abuhasna, A.; Kholoki, M.; Psihogios, P.; Razzaq, T. Pseudomonas diversity within urban freshwaters. Front. Microbiol. 2019, 10, 195. [Google Scholar] [CrossRef]
- van Zyl, L.J.; Abrahams, Y.; Stander, E.A.; Kirby-McCollough, B.; Jourdain, R.; Clavaud, C.; Breton, L.; Trindade, M. Novel phages of healthy skin metaviromes from South Africa. Sci. Rep. 2018, 8, 12265. [Google Scholar] [CrossRef]
- Yu, Y.; Xie, Z.; Yang, J.; Yang, R.; Li, Y.; Zhu, Y.; Zhao, Y.; Yang, Q.; Chen, J.; Alwathnani, H.A.; et al. Citrobacter portucalensis Sb-2 contains a metalloid resistance determinant transmitted by Citrobacter phage Chris1. J. Hazard. Mater. 2023, 443 Pt. A, 130184. [Google Scholar] [CrossRef]

| Isolates | Antibiotics | MIC * (µg) | Antibiotic Resistance Profile |
|---|---|---|---|
| CFS1154 | FAZ ** | 8 | R *** |
| MIN | 4 | R | |
| CFS1155 | MIN | 4 | R |
| CFS1156 | FAZ | 8 | R |
| MIN | 2 | R | |
| CFS1157 | FAZ | 8 | R |
| MIN | 4 | R | |
| CFS1158 | FAZ | 16 | R |
| MIN | 4 | R | |
| CFS1159 | FAZ | 8 | R |
| MIN | 2 | R | |
| CFS1160 | FAZ | 8 | R |
| MIN | 2 | R | |
| CFS1161 | FAZ | 8 | R |
| MIN | 2 | R | |
| CFS1169 | FAZ | 8 | R |
| MIN | 2 | R | |
| CFS1176 | FAZ | 16 | R |
| MIN | 2 | R | |
| A/S 2 | 8/4 | R | |
| CFS1297 | FAZ | 4 | R |
| MIN | 4 | R | |
| CFS1304 | FAZ | 16 | R |
| MIN | 4 | R | |
| CIP | 2 | R | |
| CFS1322 | FAZ | 8 | R |
| MIN | 4 | R | |
| TAZ | 2 | I | |
| AXO | 1 | S | |
| CFS1362 | FAZ | 8 | R |
| MIN | 4 | R | |
| CFS1328 | FAZ | 8 | R |
| MIN | 4 | R | |
| CFS1370 | FAZ | 8 | R |
| MIN | 4 | R | |
| CFS1377 | FAZ | 8 | R |
| MIN | 2 | R | |
| CFS2786 | AMP | 16 | R |
| FAZ | 16 | R | |
| C/T | 4 | R | |
| CIP | 2 | R | |
| MIN | 4 | R | |
| CZA | 16/4 | R | |
| NIT | 64 | R | |
| A/S 2 | 16/8 | R | |
| CFS2788 | FAZ | 8 | R |
| MIN | 4 | R | |
| CIP | 2 | R |
| Isolate | Plasmid | %Identity | Query/Template Length | Contig | Position in Contigs | Accession No. |
|---|---|---|---|---|---|---|
| CFS1156 | Col(pHAD28) | 94.96 | 119/131 | NODE_38_length_3676_cov_857.909693 | 1716...1834 | KU674895 |
| CFS1157 | Col(pHAD28) | 94.96 | 119/131 | NODE_41_length_3676_cov_857.909693 | 1716...1834 | KU674895 |
| CFS1158 | Col(pHAD28) | 94.96 | 119/131 | NODE_41_length_3676_cov_857.909693 | 1716...1834 | KU674895 |
| CFS1160 | Col(pHAD28) | 94.96 | 119/131 | NODE_37_length_3672_cov_866.208737 | 1610...1728 | KU674895 |
| CFS1176 | Col(pHAD28) | 94.96 | 119/131 | NODE_106_length_2518_cov_463.535120 | 1949...2067 | KU674895 |
| CFS1362 | Col(pHAD28) | 94.96 | 119/131 | NODE_30_length_3676_cov_5.826291 | 1705...1823 | KU674895 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mousavi, Z.E.; Koolman, L.; Macori, G.; Fanning, S.; Butler, F. Comprehensive Genomic Characterization of Cronobacter sakazakii Isolates from Infant Formula Processing Facilities Using Whole-Genome Sequencing. Microorganisms 2023, 11, 2749. https://doi.org/10.3390/microorganisms11112749
Mousavi ZE, Koolman L, Macori G, Fanning S, Butler F. Comprehensive Genomic Characterization of Cronobacter sakazakii Isolates from Infant Formula Processing Facilities Using Whole-Genome Sequencing. Microorganisms. 2023; 11(11):2749. https://doi.org/10.3390/microorganisms11112749
Chicago/Turabian StyleMousavi, Zeinab Ebrahimzadeh, Leonard Koolman, Guerrino Macori, Séamus Fanning, and Francis Butler. 2023. "Comprehensive Genomic Characterization of Cronobacter sakazakii Isolates from Infant Formula Processing Facilities Using Whole-Genome Sequencing" Microorganisms 11, no. 11: 2749. https://doi.org/10.3390/microorganisms11112749






