Escherichia coli K-12 Transcriptomics for Assessing the Mechanism of Action of High-Power Ultrasound
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Preparation of Inoculum
2.2. Preparation of the Working Culture
2.3. Ultrasound Treatments
2.4. RNA Extraction
2.5. RNA-Seq
2.6. Bioinformatics
3. Results
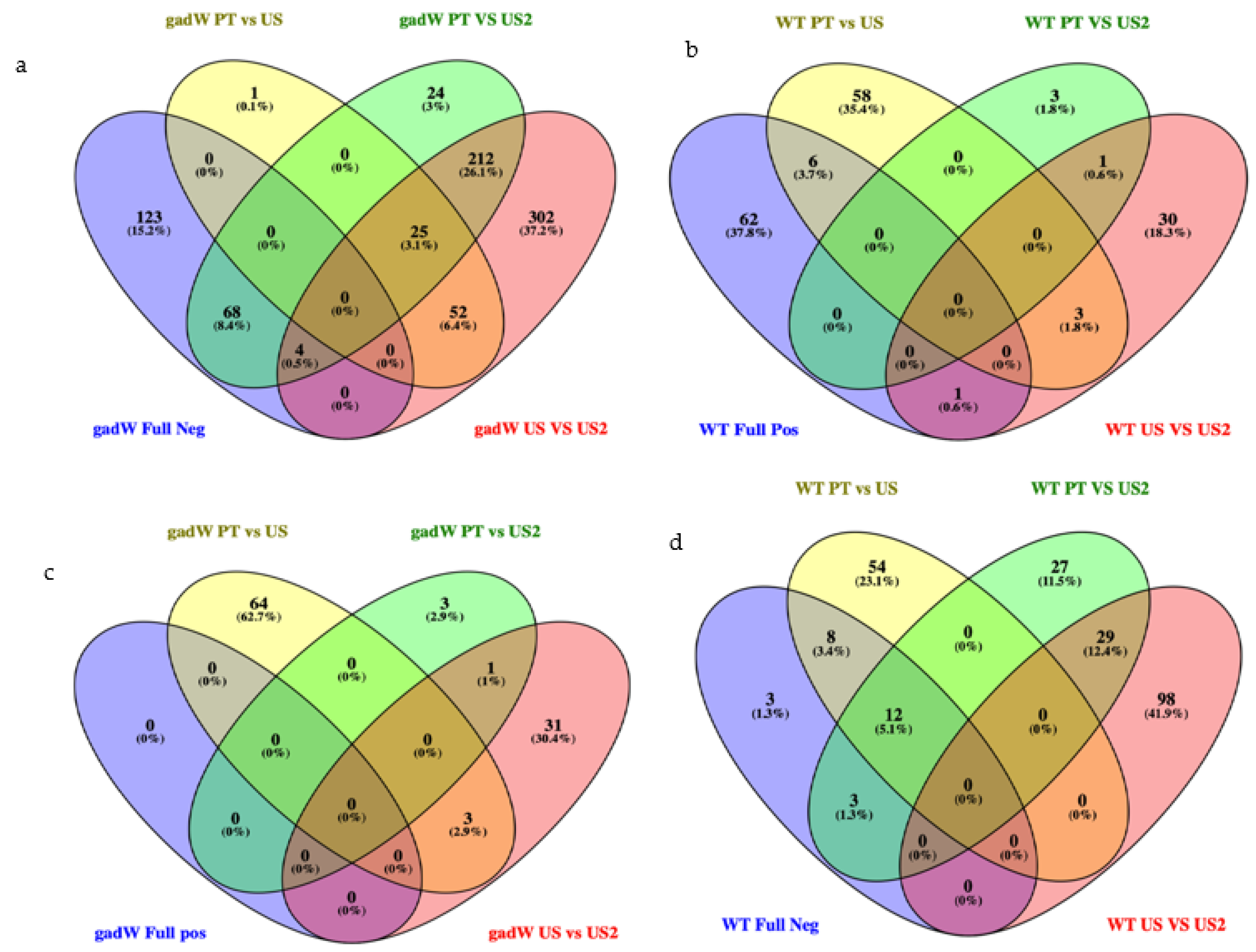
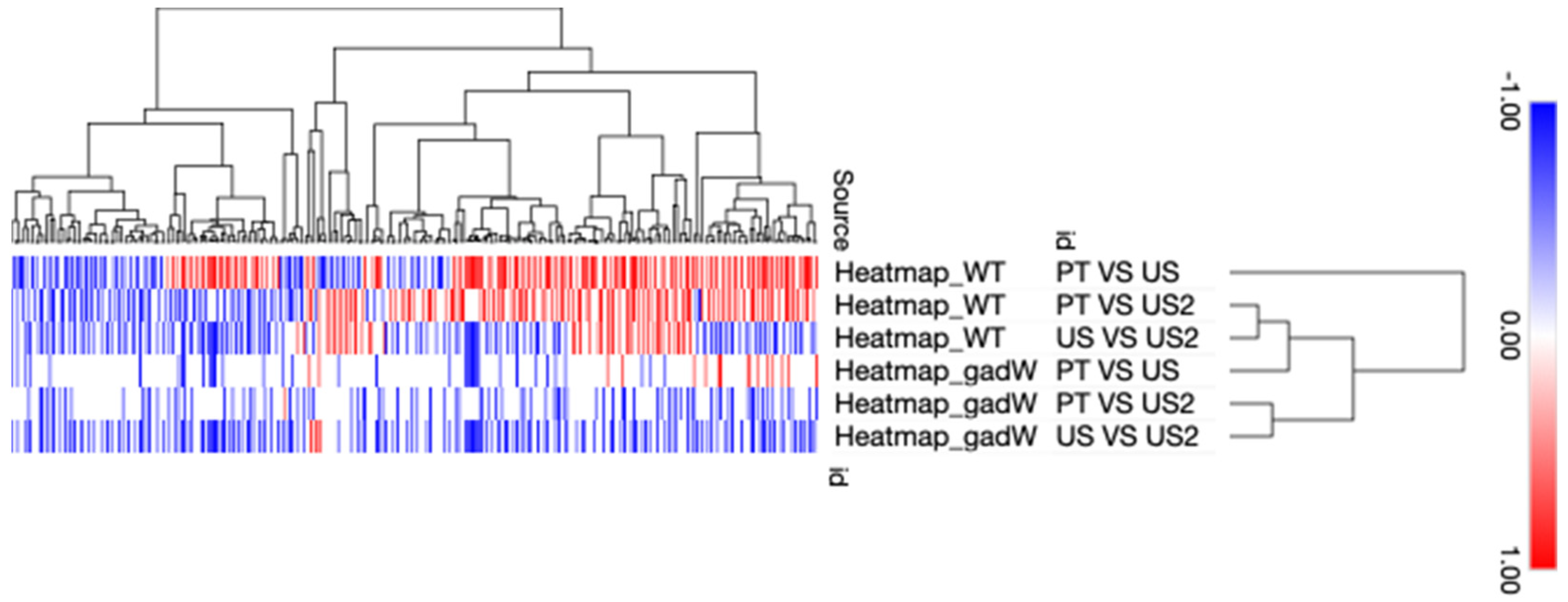
3.1. Differentially Expressed Genes (DEGs) and Function Enrichment Analysis
3.2. Gene Ontology Analysis of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cesaro, A.; Belgiorno, V. Removal of Endocrine Disruptors from Urban Wastewater by Advanced Oxidation Processes (AOPs): A Review. Open Biotechnol. J. 2016, 10, 151–172. [Google Scholar] [CrossRef]
- Han, C.; Andersen, J.; Pillai, S.; Fagan, R.; Falaras, P.; Byrne, J.; Dunlop, P.; Choi, H.; Jiang, W.; O’Shea, K.; et al. Chapter green nanotechnology: Development of nanomaterials for environmental and energy applications. ACS Symp. Ser. 2013, 1124, 201–229. [Google Scholar] [CrossRef]
- Gibson, J.H.; Yong, D.H.N.; Farnood, R.R.; Seto, P. A Literature Review of Ultrasound Technology and Its Application in Wastewater Disinfection. Water Qual. Res. J. 2008, 43, 23–35. [Google Scholar] [CrossRef]
- Sango, D.M.; Abela, D.; McElhatton, A.; Valdramidis, V. Assisted ultrasound applications for the production of safe foods. J. Appl. Microbiol. 2014, 116, 1067–1083. [Google Scholar] [CrossRef]
- Madge, B.; Jensen, J. Disinfection of wastewater using a 20-kHz ultrasound unit. Water Env. Res. 2002, 74, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Oyib, D.H. Ultrasound in Water Treatment: Suppressing Algal Growth and Biofilm Formation. 2010. Available online: http://www.iwaponline.com/w21/01103/w21011030052.htm (accessed on 9 June 2013).
- Naddeo, V.; Landi, M.; Belgiorno, V.; Napoli, R. Wastewater disinfection by combination of ultrasound and ultraviolet irradiation. J. Hazard. Mater. 2009, 168, 925–929. [Google Scholar] [CrossRef]
- Vasilyak, L.M. Ultrasound application in systems for the disinfection of water. Surf. Eng. Appl. Electrochem. 2010, 46, 489–493. [Google Scholar] [CrossRef]
- Drakopoulou, S.; Terzakis, S.; Fountoulakis, M.; Mantzavinos, D.; Manios, T. Ultrasound-induced inactivation of gram-negative and gram-positive bacteria in secondary treated municipal wastewater. Ultrason. Sonochem. 2009, 16, 629–634. [Google Scholar] [CrossRef]
- Antoniadis, A.; Poulios, I.; Nikolakaki, E.; Mantzavinos, D. Sonochemical disinfection of municipal wastewater. J. Hazard. Mater. 2007, 146, 492–495. [Google Scholar] [CrossRef]
- Broekman, S.; Pohlmann, O.; Beardwood, E.; de Meulenaer, E.C. Ultrasonic treatment for microbiological control of water systems. Ultrason. Sonochem. 2010, 17, 1041–1048. [Google Scholar] [CrossRef]
- Spiteri, D.; Chot-Plassot, C.; Sclear, J.; Karatzas, K.; Scerri, C.; Valdramidis, V. Ultrasound processing of liquid system(s) and its antimicrobial mechanism of action. Lett. Appl. Microbiol. 2017, 65, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M. Understanding the Regulation of Acid Resistance in E. coli Using Whole Genome Techniques; University of Birmingham: Birmingham, UK, 2011. [Google Scholar]
- Nath, A.; Mukhim, K.; Swer, T.; Dutta, D.; Verma, N.; Deka, B.; Gangwar, B. A Review on the Application of Nanotechnology in Food Processing and Packaging. J. Food Prod. Dev. Packag. 2014, 1, 7–21. [Google Scholar]
- Mahapatra, A.K.; Muthukumarappan, K.; Julson, J.L. Applications of Ozone, Bacteriocins and Irradiation in Food Processing: A Review. Crit. Rev. Food Sci. Nutr. 2005, 45, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Perni, S.; Shama, G.; Hobman, J.L.; Lund, P.A.; Kershaw, C.J.; Hidalgo-Arroyo, G.A.; Penn, C.W.; Deng, X.T.; Walsh, J.L.; Kong, M.G. Probing bactericidal mechanisms induced by cold atmospheric plasmas with Escherichia coli mutants. Appl. Phys. Lett. 2007, 90, 073902. [Google Scholar] [CrossRef]
- Laroussi, M. Sterilization of contaminated matter with an atmospheric pressure plasma. IEEE Trans. Plasma Sci. 1996, 24, 1188–1191. [Google Scholar] [CrossRef]
- Patil, S.; Bourke, P.; Kelly, B.; Frías, J.M.; Cullen, P. The effects of acid adaptation on Escherichia coli inactivation using power ultrasound. Innov. Food Sci. Emerg. Technol. 2009, 10, 486–490. [Google Scholar] [CrossRef]
- Leyer, G.J.; Wang, L.L.; A Johnson, E. Acid adaptation of Escherichia coli O157:H7 increases survival in acidic foods. Appl. Environ. Microbiol. 1995, 61, 3752–3755. [Google Scholar] [CrossRef]
- Berry, E.D.; Cutter, C.N. Effects of Acid Adaptation of Escherichia coli O157:H7 on Efficacy of Acetic Acid Spray Washes to Decontaminate Beef Carcass Tissue. Appl. Environ. Microbiol. 2000, 66, 1493–1498. [Google Scholar] [CrossRef]
- Šeputienė, V.; Daugelavičius, A.; Sužiedėlis, K.; Sužiedėlienė, E. Acid response of exponentially growing Escherichia coli K-12. Microbiol. Res. 2006, 161, 65–74. [Google Scholar] [CrossRef]
- Tosun, H. The Effect of Acid Adaptation Conditions on Acid Tolerance Response of Escherichia coli O157: H7. Turkish J. Biol. 2014, 29, 197–202. [Google Scholar]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Transcriptomic analysis of Escherichia coli MG1655 cells exposed to pulsed electric fields. Innov. Food Sci. Emerg. Technol. 2015, 29, 78–86. [Google Scholar] [CrossRef]
- Wecke, T.; Mascher, T. Antibiotic research in the age of omics: From expression profiles to interspecies communication. J. Antimicrob. Chemother. 2011, 66, 2689–2704. [Google Scholar] [CrossRef]
- Carruthers, M.D.; Minion, C. Transcriptome analysis of Escherichia coli O157:H7 EDL933 during heat shock. FEMS Microbiol. Lett. 2009, 295, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-H.; Kim, J.; Kim, S.-M.; Kim, S.; Lee, J.-C.; Ahn, J.-M.; Cho, J.-Y. σB-dependent protein induction in Listeria monocytogenes during vancomycin stress. FEMS Microbiol. Lett. 2010, 308, 94–100. [Google Scholar] [CrossRef]
- Seo, S.W.; Kim, D.; O’brien, E.J.; Szubin, R.; Palsson, B.O. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 2015, 6, 7970. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; A Kohanski, M.; Collins, J.J. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 2009, 12, 482–489. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G., III; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The Complete Genome Sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nakahigashi, K.; Nakamichi, T.; Yoshino, M.; Takai, Y.; Touda, Y.; Furubayashi, A.; Kinjyo, S.; Dose, H.; Hasegawa, M.; et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009, 5, 335. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Smith, D.K.; Kassam, T.; Singh, B.; Elliott, J.F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 1992, 174, 5820–5826. [Google Scholar] [CrossRef]
- Feehily, C.; Karatzas, K. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2012, 114, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, R.; Karatzas, K. Stress adaptation of Listeria monocytogenes in acidic ready-to-eat products A2—Kotzekidou, Parthena. In Food Hygiene and Toxicology in Ready to Eat Foods; Academic Press: San Diego, FL, USA, 2016; Chapter 10; pp. 167–182. [Google Scholar]
- Klepsch, M.; Kovermann, M.; Löw, C.; Balbach, J.; Permentier, H.; Fusetti, F.; de Gier, J.; Slotboom, D.; Berntsson, R.P.-A. Escherichia coli Peptide Binding Protein OppA Has a Preference for Positively Charged Peptides. J. Mol. Biol. 2011, 414, 75–85. [Google Scholar] [CrossRef]
- Coleman, S.T.; Fang, T.K.; Rovinsky, S.A.; Turano, F.J.; Moye-Rowley, W.S. Expression of a Glutamate Decarboxylase Homologue Is Required for Normal Oxidative Stress Tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 244–250. [Google Scholar] [CrossRef]
- Boura, M.; Brensone, D.; Karatzas, K.A. A novel role for the glutamate decarboxylase system in Listeria monocytogenes; protection against oxidative stress. Food Microbiol. 2019, 85, 103284. [Google Scholar] [CrossRef] [PubMed]
- Janausch, I.; Zientz, E.; Tran, Q.; Kröger, A.; Unden, G. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta (BBA) Bioenerg. 2001, 1553, 39–56. [Google Scholar] [CrossRef]
- Ramond, E.; Gesbert, G.; Rigard, M.; Dairou, J.; Dupuis, M.; Dubail, I.; Meibom, K.; Henry, T.; Barel, M.; Charbit, A. Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Francisella Phagosomal Escape. PLoS Pathog. 2014, 10, e1003893. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, M.; Weme, R.G.J.D.O.; Duurkens, R.H.; Slotboom, D.J. Biochemical Characterization of the C 4 -Dicarboxylate Transporter DctA from Bacillus subtilis. J. Bacteriol. 2010, 192, 2900–2907. [Google Scholar] [CrossRef]
- Sá-Pessoa, J.; Paiva, S.; Ribas, D.; Silva, I.J.; Viegas, S.C.; Arraiano, C.M.; Casal, M. SATP (YaaH), a succinate–acetate transporter protein in Escherichia coli. Biochem. J. 2013, 454, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Karinou, E.; Hoskisson, P.A.; Strecker, A.; Unden, G.; Javelle, A. The E. coli dicarboxylic acid transporters DauA act as a signal transducer by interacting with the DctA uptake system. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Lo, B.; Khalil, T.; Sanwal, M. Transport of Succinate in Escherichia. Biol. Chem. 1972, 247, 6323–6332. [Google Scholar] [CrossRef]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, D.; Hu, S.; Xiao, X.; Yu, Y.; Li, X. Transcriptomic analysis by RNA-seq of Escherichia coli O157:H7 response to prolonged cold stress. LWT 2018, 97, 17–24. [Google Scholar] [CrossRef]
- Al Mamun, A.A.M.; Lombardo, M.-J.; Shee, C.; Lisewski, A.M.; Gonzalez, C.; Lin, D.; Nehring, R.B.; Saint-Ruf, C.; Gibson, J.L.; Frisch, R.L.; et al. Identity and Function of a Large Gene Network Underlying Mutagenic Repair of DNA Breaks. Science 2012, 338, 1344–1348. [Google Scholar] [CrossRef]
- Brown, L.; Elliott, T. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 1996, 178, 3763–3770. [Google Scholar] [CrossRef] [PubMed]
- Muffler, A.; Fischer, D.; Hengge-Aronis, R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. J. Bone Jt. Surg. 1996, 10, 1143–1151. [Google Scholar] [CrossRef]
- Guisbert, E.; Rhodius, V.A.; Ahuja, N.; Witkin, E.; Gross, C.A. Hfq Modulates the σ E -Mediated Envelope Stress Response and the σ 32 -Mediated Cytoplasmic Stress Response in Escherichia coli. J. Bacteriol. 2007, 189, 1963–1973. [Google Scholar] [CrossRef]
- King, T.; Lucchini, S.; Hinton, J.C.D.; Gobius, K. Transcriptomic Analysis of Escherichia coli O157:H7 and K-12 Cultures Exposed to Inorganic and Organic Acids in Stationary Phase Reveals Acidulant- and Strain-Specific Acid Tolerance Responses. Appl. Environ. Microbiol. 2010, 76, 6514–6528. [Google Scholar] [CrossRef]
- Karp, P.D.; Weaver, D.; Paley, S.; Fulcher, C.; Kubo, A.; Kothari, A.; Krummenacker, M.; Subhraveti, P.; Weerasinghe, D.; Gama-Castro, S.; et al. The EcoCyc Database. EcoSal Plus 2014, 6. [Google Scholar] [CrossRef]
- Allen, M.J.; White, G.F.; Morby, A.P. The response of Escherichia coli to exposure to the biocide polyhexamethylene biguanide. Microbiology 2006, 152, 989–1000. [Google Scholar] [CrossRef]
- Kelly, T.; Stachula, S.; Raetz, C.; Anderson, M. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)- glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J. Biol. Chem. 1993, 268, 19866–19874. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn Diagrams. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 30 November 2018).
- Datsenko, K.; Wanner, B. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
| Strain with Treatment | Replicate 1 | Replicate 2 | ||
|---|---|---|---|---|
| Unique Reads | Duplicate Reads | Unique Reads | Duplicate Reads | |
| WT PT | 1,907,618 | 9,782,951 | 1,313,674 | 10,376,895 |
| WT US | 1,638,411 | 8,729,411 | 2,259,703 | 8,107,832 |
| WT US2 | 2,510,105 | 7,998,899 | 2,896,851 | 7,612,563 |
| ΔgadW PT | 1,903,140 | 8,570,279 | 2,348,814 | 8,124,605 |
| ΔgadW US | 2,247,793 | 8,586,961 | 2,727,471 | 8,107,283 |
| ΔgadW US2 | 2,466,568 | 7,854,287 | 2,819,178 | 7,501,677 |
| Fold Change log2FC | p-Value | ||||||
|---|---|---|---|---|---|---|---|
| DEGs | PT vs. US | PT vs. US2 | US vs. US2 | PT vs. US | PT vs. US2 | US vs. US2 | Annotation |
| rpoS | 0.510 | 0.466 | −0.076 | <0.0001 | <0.0001 | 0.19477937 | General stress |
| sodC | 1.182 | 1.013 | −0.165 | <0.0001 | <0.0001 | 0.3052228 | Oxidase gene |
| sodA | 1.164 | −0.170 | −1.365 | <0.0001 | <0.0001 | <0.0001 | Oxidase gene |
| SodB | 0.438 | 0.270 | −0.195 | <0.0001 | 0.0297 | 0.0469 | Oxidase gene |
| yifE | 0.381 | −0.959 | −1.367 | 0.00155 | <0.0001 | <0.0001 | Stress-induced mutagenesis |
| nuoG | 0.521 | 0.449 | −0.099 | <0.0001 | <0.0001 | 0.326 | Stress-induced mutagenesis |
| hemL | 0.654 | −0.202 | −0.883 | <0.0001 | 0.0941 | <0.0001 | Stress-induced mutagenesis |
| hdfR | 0.713 | 0.496 | −0.250 | <0.0001 | <0.0001 | 0.0479 | negatively expresses flagellar master |
| cspC | 0.811 | −0.246 | −1.101 | <0.0001 | 0.0495 | <0.0001 | Stress-induced mutagenesis |
| hfq | 0.855 | −0.151 | −1.039 | <0.0001 | 0.00530 | <0.0001 | Stress-induced mutagenesis |
| sdhB | 1.552 | 0.334 | −1.214 | <0.0001 | 0.0503 | <0.0001 | Stress-induced mutagenesis |
| lpxD | 0.265 | 0.247 | −0.060 | 0.0468 | 0.0795 | 0.601 | lipid biosynthesis |
| dctA | 1.254 | 0.522 | −0.761 | <0.0001 | <0.0001 | <0.0001 | required for dicarboxylate transport |
| speG | −1.003 | −1.061 | <0.0001 | <0.0001 | Protection against polyamine toxicity | ||
| cpxP | 0.397 | −0.164 | −0.595 | <0.0001 | <0.0001 | <0.0001 | Auxiliary protein in the CpX two-component envelope stress response system |
| lpxC | 0.399 | −0.436 | <0.0001 | <0.0001 | Develops new antibacterial agents | ||
| Biological Process | Molecular Function | Cellular Component | |||||
|---|---|---|---|---|---|---|---|
| Strain | Treatment | Positive Expression | Negative Expression | Positive Expression | Negative Expression | Positive Expression | Negative Expression |
| WT | PT vs. US | 396 | N/A | N/A | N/A | N/A | N/A |
| PT vs. US2 | 301 | N/A | 11 | N/A | 52 | 85 | |
| US vs. US2 | 41 | 220 | 14 | N/A | 15 | 90 | |
| gadW | PT vs. US | 104 | 281 | N/A | 75 | N/A | 481 |
| PT vs. US2 | N/A | 2585 | N/A | 1186 | N/A | 2582 | |
| US vs. US2 | N/A | 4714 | N/A | 1634 | N/A | 4122 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spiteri, D.; Griffin, S.; Karatzas, K.A.; Scerri, C.; Valdramidis, V.P. Escherichia coli K-12 Transcriptomics for Assessing the Mechanism of Action of High-Power Ultrasound. Microorganisms 2023, 11, 2768. https://doi.org/10.3390/microorganisms11112768
Spiteri D, Griffin S, Karatzas KA, Scerri C, Valdramidis VP. Escherichia coli K-12 Transcriptomics for Assessing the Mechanism of Action of High-Power Ultrasound. Microorganisms. 2023; 11(11):2768. https://doi.org/10.3390/microorganisms11112768
Chicago/Turabian StyleSpiteri, David, Sholeem Griffin, Kimon Andreas Karatzas, Christian Scerri, and Vasilis P. Valdramidis. 2023. "Escherichia coli K-12 Transcriptomics for Assessing the Mechanism of Action of High-Power Ultrasound" Microorganisms 11, no. 11: 2768. https://doi.org/10.3390/microorganisms11112768
APA StyleSpiteri, D., Griffin, S., Karatzas, K. A., Scerri, C., & Valdramidis, V. P. (2023). Escherichia coli K-12 Transcriptomics for Assessing the Mechanism of Action of High-Power Ultrasound. Microorganisms, 11(11), 2768. https://doi.org/10.3390/microorganisms11112768






