SARS-CoV-2 Papain-like Protease Negatively Regulates the NLRP3 Inflammasome Pathway and Pyroptosis by Reducing the Oligomerization and Ubiquitination of ASC
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Reanalysis of Single-Cell RNA Sequencing Data
2.3. Cell Culture and Stimulation
2.4. Plasmid Construction and Transfection
2.5. Lentivirus Production and Infection
2.6. Co-IP Assay and Co-IP Ubiquitination Assay
2.7. Lactate Dehydrogenase (LDH) Release Assay
2.8. ASC Oligomerization
2.9. IL-1β Estimation
2.10. Dual-Luciferase Reporter Assay
2.11. Immunofluorescence
2.12. Statistical Analysis
3. Results
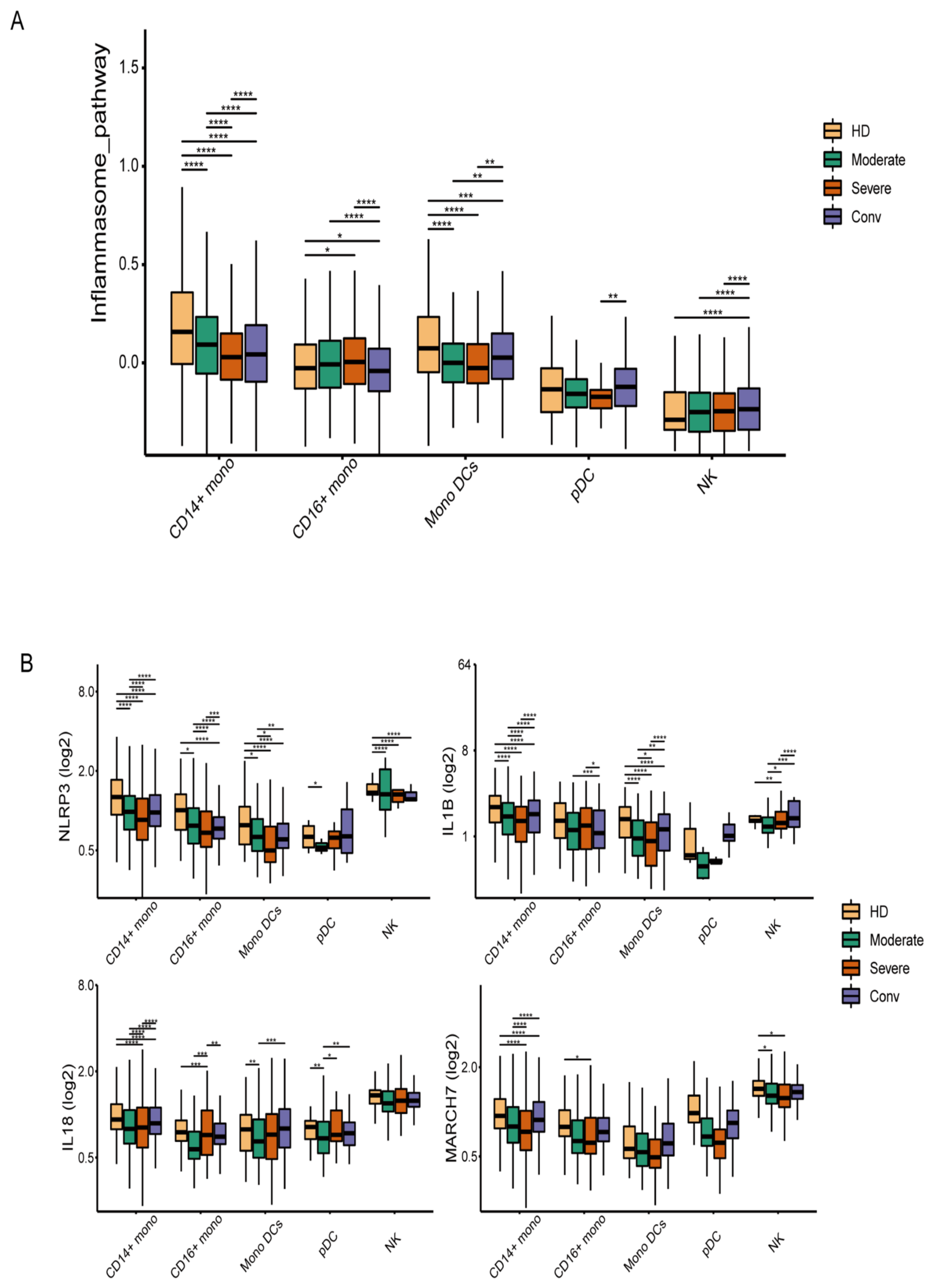
3.1. The Inflammasome Pathway Is Downregulated in the Innate Immune Cells of COVID-19 Patients
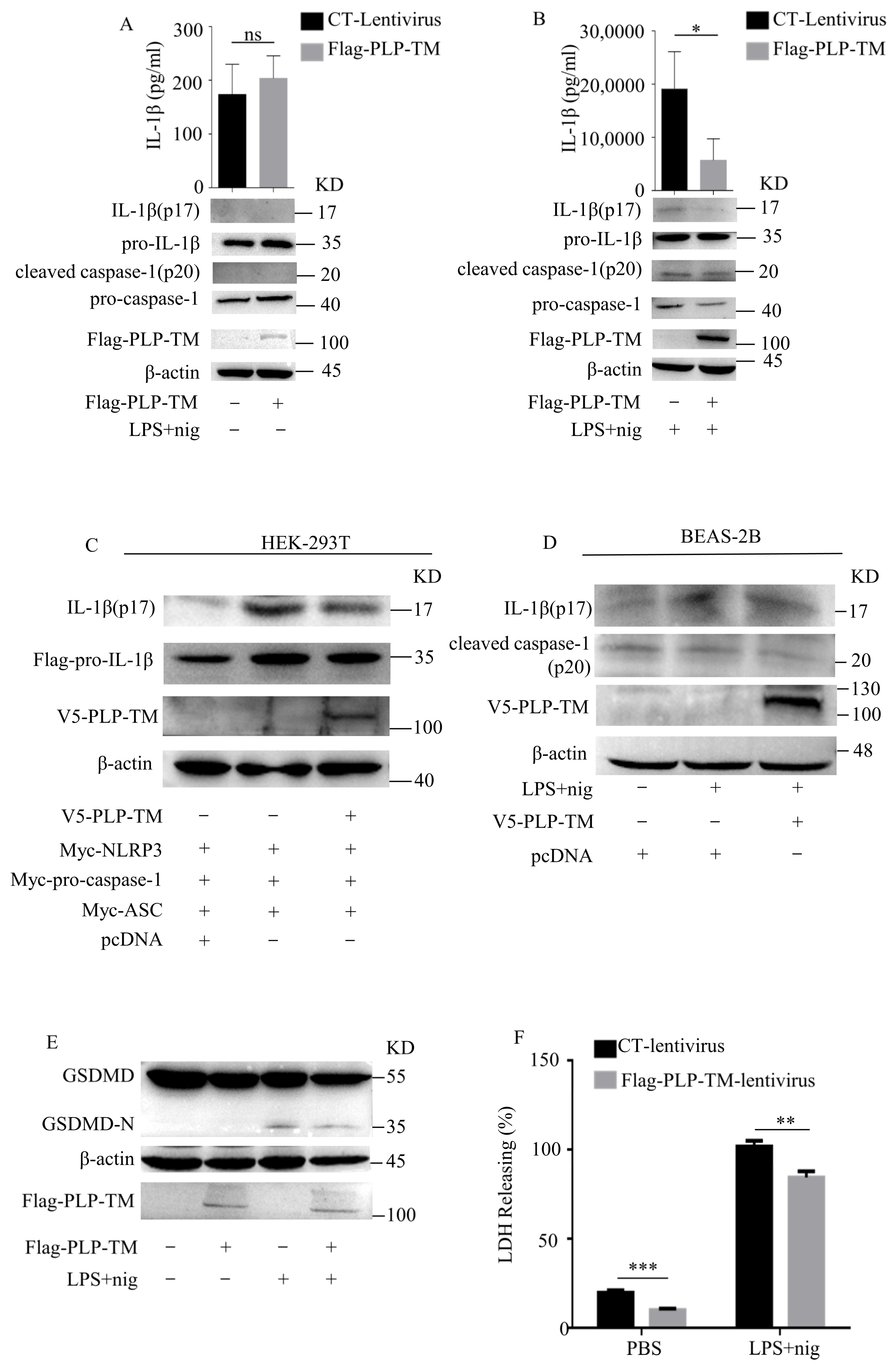
3.2. SARS-CoV-2 PLP Inhibits the Activation of NLRP3 Inflammasomes and Caspase-1-Mediated GSDMD Cleavage
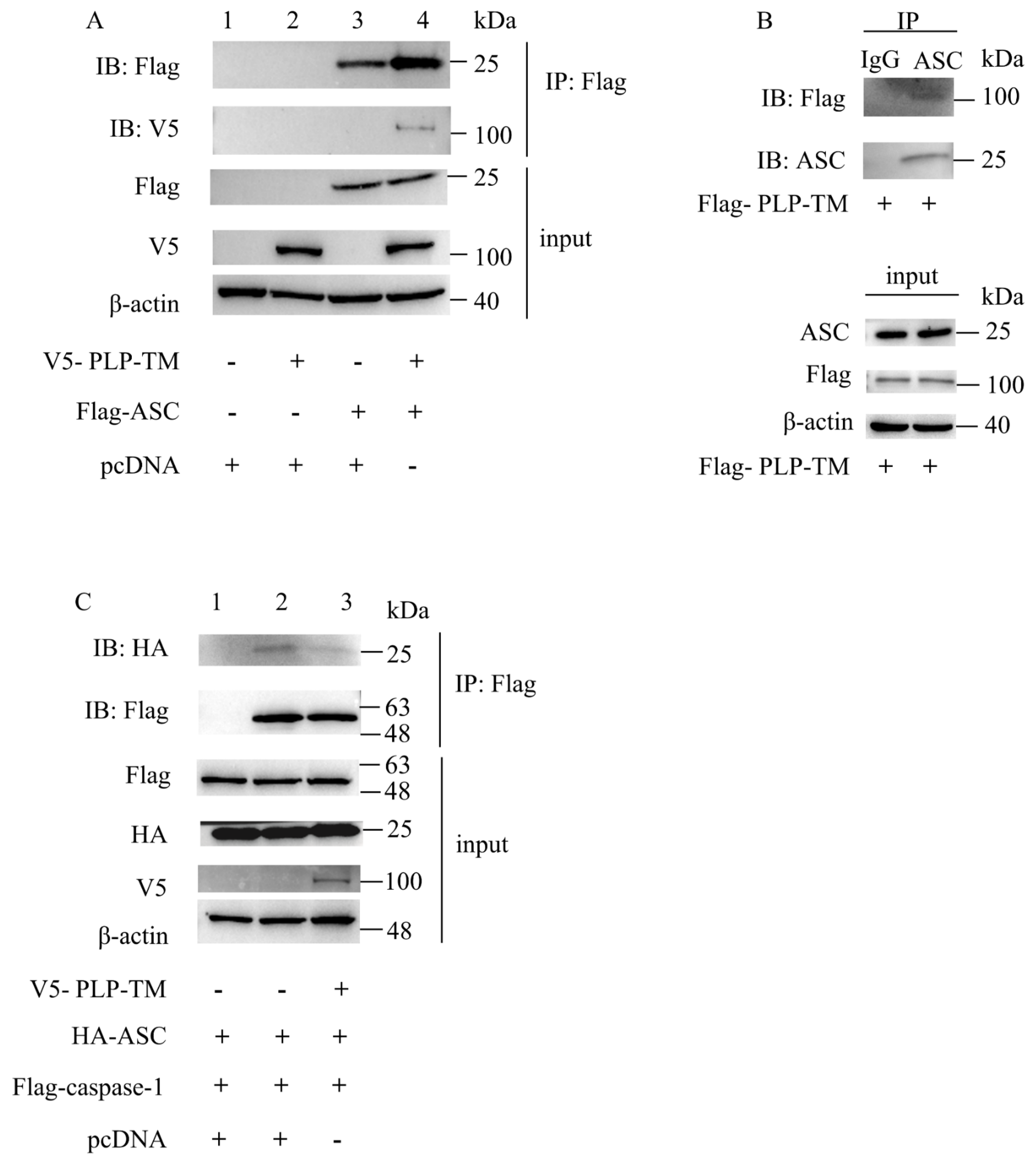
3.3. SARS-CoV-2 PLP-TM May Coimmunopreciptate with ASC and Blunt the ASC–Caspase-1 Complex
3.4. SARS-CoV-2 PLP Reduces the Oligomerization of ASC and ASC Speck Formation
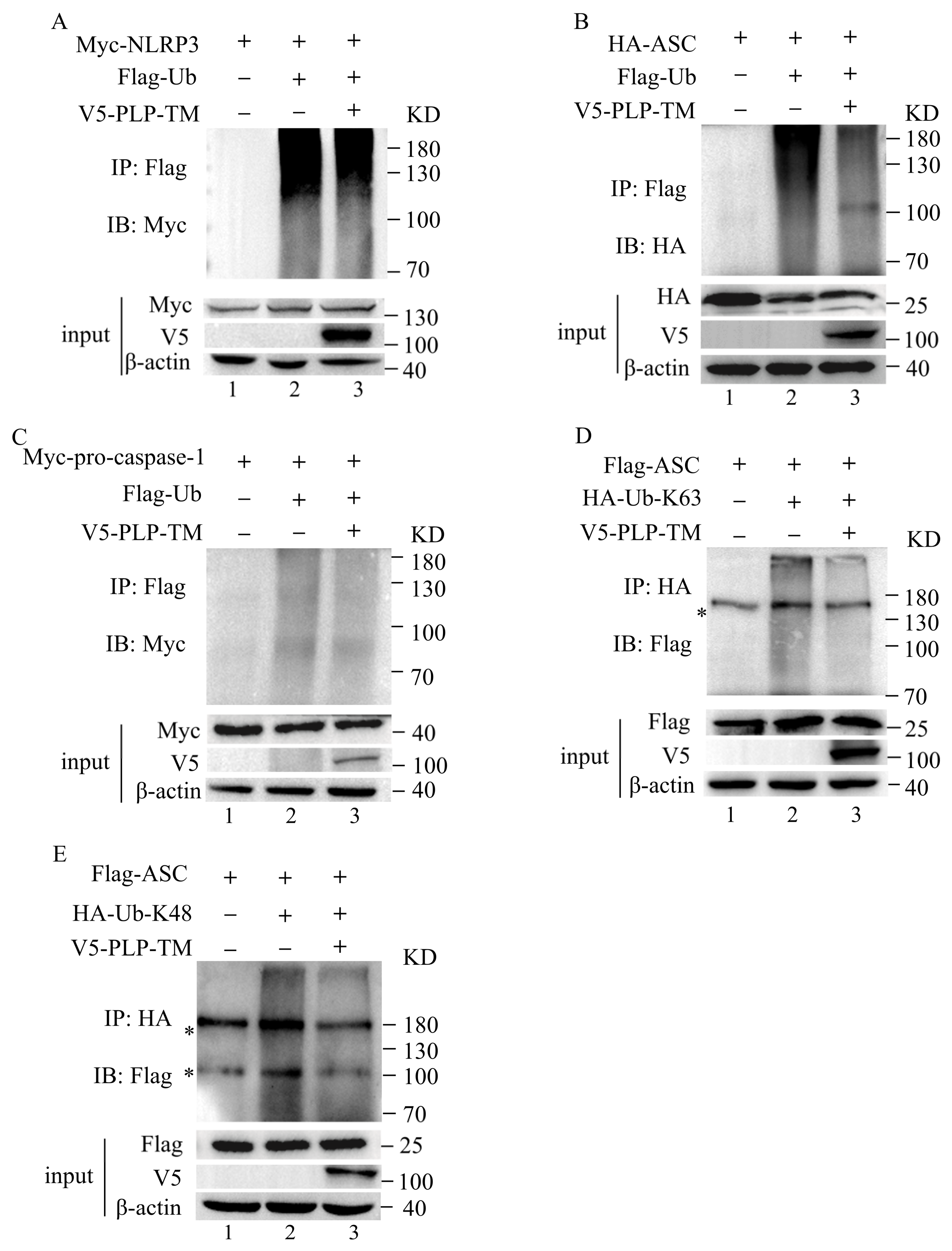
3.5. SARS-CoV-2 PLP Reduces the Ubiquitination of ASC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 genome-encoded proteins. Science China. Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef]
- Lei, J.; Kusov, Y.; Hilgenfeld, R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antivir. Res. 2018, 149, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. SARS-CoV-2 Papain-Like Protease: Structure, Function and Inhibition. Chembiochem. Eur. J. Chem. Biol. 2022, 23, e202200327. [Google Scholar] [CrossRef] [PubMed]
- Clementz, M.A.; Chen, Z.; Banach, B.S.; Wang, Y.; Sun, L.; Ratia, K.; Baez-Santos, Y.M.; Wang, J.; Takayama, J.; Ghosh, A.K.; et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010, 84, 4619–4629. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE 2012, 7, e30802. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, K.; Xing, Y.; Tu, J.; Yang, X.; Zhao, Q.; Li, K.; Chen, Z. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell 2014, 5, 912–927. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M.; et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B 2021, 11, 237–245. [Google Scholar] [CrossRef]
- Osipiuk, J.; Azizi, S.A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.; Wei, C.; Zheng, Z.; Song, T.; Wu, F.; Zhang, Y.; Cao, Y.; Ma, S.; Chen, W.; Xu, Q.; et al. MAVS Promotes Inflammasome Activation by Targeting ASC for K63-Linked Ubiquitination via the E3 Ligase TRAF3. J. Immunol. 2015, 194, 4880–4890. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Chan, J.F.; Wang, Y.; Yuen, T.T.; Chai, Y.; Hou, Y.; Shuai, H.; Yang, D.; Hu, B.; Huang, X.; et al. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study with Implications for the Pathogenesis of COVID-19. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Lau, C.C.Y.; Chan, K.H.; Li, C.P.Y.; Chen, H.; Jin, D.Y.; Chan, J.F.W.; Woo, P.C.Y.; Yuen, K.Y. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: Implications for pathogenesis and treatment. J. Gen. Virol. 2013, 94 Pt 12, 2679–2690. [Google Scholar] [CrossRef]
- Rivieccio, M.A.; John, G.R.; Song, X.; Suh, H.S.; Zhao, Y.; Lee, S.C.; Brosnan, C.F. The cytokine IL-1beta activates IFN response factor 3 in human fetal astrocytes in culture. J. Immunol. 2005, 174, 3719–3726. [Google Scholar] [CrossRef]
- Afonina, I.S.; Zhong, Z.; Karin, M.; Beyaert, R. Limiting inflammation-the negative regulation of NF-κB and the NLRP3 inflammasome. Nat. Immunol. 2017, 18, 861–869. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nature reviews. Immunology 2013, 13, 722–737. [Google Scholar] [CrossRef]
- Shi, J.; Zhou, J.; Zhang, X.; Hu, W.; Zhao, J.F.; Wang, S.; Wang, F.S.; Zhang, J.Y. Single-Cell Transcriptomic Profiling of MAIT Cells in Patients With COVID-19. Front. Immunol. 2021, 12, 700152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Wang, X.M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.W.; Fan, X.; Xia, P.; Fu, J.L.; Wang, S.Y.; et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Zhang, X.; Xiao, Y.; Zhang, W.; Wang, Y.; Ma, W.; Wang, X.; Song, P.; Lai, L.; Chen, H.; et al. ABRO1 promotes NLRP3 inflammasome activation through regulation of NLRP3 deubiquitination. EMBO J. 2019, 38, e100376. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, B.H.; Jukneliene, D.; Kanjanahaluethai, A.; Bechill, J.; Severson, K.M.; Smith, C.M.; Rota, P.A.; Baker, S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004, 78, 13600–13612. [Google Scholar] [CrossRef] [PubMed]
- Kanjanahaluethai, A.; Chen, Z.; Jukneliene, D.; Baker, S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 2007, 361, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Burdette, B.E.; Esparza, A.N.; Zhu, H.; Wang, S. Gasdermin D in pyroptosis. Acta Pharm. Sin. B 2021, 11, 2768–2782. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Bian, Y.; Zhang, Y.; Ren, G.; Li, G. Metformin activates AMPK/SIRT1/NF-κB pathway and induces mitochondrial dysfunction to drive caspase3/GSDME-mediated cancer cell pyroptosis. Cell Cycle 2020, 19, 1089–1104. [Google Scholar] [CrossRef]
- Dick, M.S.; Sborgi, L.; Rühl, S.; Hiller, S.; Broz, P. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 2016, 7, 11929. [Google Scholar] [CrossRef]
- Chung, I.C.; OuYang, C.N.; Yuan, S.N.; Li, H.P.; Chen, J.T.; Shieh, H.R.; Chen, Y.J.; Ojcius, D.M.; Chu, C.L.; Yu, J.S.; et al. Pyk2 activates the NLRP3 inflammasome by directly phosphorylating ASC and contributes to inflammasome-dependent peritonitis. Sci. Rep. 2016, 6, 36214. [Google Scholar] [CrossRef]
- Zangiabadi, S.; Akram, A.; Abdul-Sater, A.A. Detection of ASC Oligomerization by Western Blotting. Methods Mol. Biol. 2022, 2459, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ko, C.J.; Li, Y.; Jie, Z.; Zhu, L.; Zhou, X.; Xie, X.; Gao, T.; Liu, T.; Cheng, X.; et al. Peli1 facilitates NLRP3 inflammasome activation by mediating ASC ubiquitination. Cell Rep. 2021, 37, 109904. [Google Scholar] [CrossRef] [PubMed]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef] [PubMed]
- Oran, D.P.; Topol, E.J. The Proportion of SARS-CoV-2 Infections That Are Asymptomatic: A Systematic Review. Ann. Intern. Med. 2021, 174, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.E.; Kim, D.K.; Song, Y.J. SARS-CoV-2 Nonstructural Proteins 1 and 13 Suppress Caspase-1 and the NLRP3 Inflammasome Activation. Microorganisms 2021, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chang, W.; Zhang, L.; Zhang, Y. Pyroptotic cell death in SARS-CoV-2 infection: Revealing its roles during the immunopathogenesis of COVID-19. Int. J. Biol. Sci. 2022, 18, 5827–5848. [Google Scholar] [CrossRef]
- Anantharaj, A.; Gujjar, S.; Verma, N.; Khan, N.A.; Shaman, H.; Sharanabasava, P.; Das, A.; Pandey, R.; Pandey, A.K.; Medigeshi, G.R. Resolution of viral load in mild COVID-19 patients is associated with both innate and adaptive immune responses. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2022, 146, 105060. [Google Scholar] [CrossRef]
- Chen, N.; Xia, P.; Li, S.; Zhang, T.; Wang, T.T.; Zhu, J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 2017, 69, 297–304. [Google Scholar] [CrossRef]
- Zhen, Y.; Zhang, H. NLRP3 Inflammasome and Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Alnemri, T.; Wu, J.; Yu, J.W.; Datta, P.; Miller, B.; Jankowski, W.; Rosenberg, S.; Zhang, J.; Alnemri, E.S. The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007, 14, 1590–1604. [Google Scholar] [CrossRef]
- Hu, H.; Sun, S.C. Ubiquitin signaling in immune responses. Cell Res. 2016, 26, 457–483. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Liu, G.; Thulasi Raman, S.N.; Landreth, S.L.; Liu, Q.; Zhou, Y. NS1 Protein of 2009 Pandemic Influenza A Virus Inhibits Porcine NLRP3 Inflammasome-Mediated Interleukin-1 Beta Production by Suppressing ASC Ubiquitination. J. Virol. 2018, 92, e00022-18. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wen, J.X.; Gao, S.J.; He, X.; Li, P.Y.; Yang, Y.X.; Wei, S.Z.; Zhao, Y.L.; Xiao, X.H. Paeonia lactiflora Pall. regulates the NF-κB-NLRP3 inflammasome pathway to alleviate cholestasis in rats. J. Pharm. Pharmacol. 2018, 70, 1675–1687. [Google Scholar] [CrossRef]
- An, Y.; Zhang, H.; Wang, C.; Jiao, F.; Xu, H.; Wang, X.; Luan, W.; Ma, F.; Ni, L.; Tang, X.; et al. Activation of ROS/MAPKs/NF-κB/NLRP3 and inhibition of efferocytosis in osteoclast-mediated diabetic osteoporosis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 12515–12527. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, H.; Zhou, J.; Wang, M.; Zheng, M.; Xing, Y.; Wang, Y. SARS-CoV-2 Papain-like Protease Negatively Regulates the NLRP3 Inflammasome Pathway and Pyroptosis by Reducing the Oligomerization and Ubiquitination of ASC. Microorganisms 2023, 11, 2799. https://doi.org/10.3390/microorganisms11112799
Meng H, Zhou J, Wang M, Zheng M, Xing Y, Wang Y. SARS-CoV-2 Papain-like Protease Negatively Regulates the NLRP3 Inflammasome Pathway and Pyroptosis by Reducing the Oligomerization and Ubiquitination of ASC. Microorganisms. 2023; 11(11):2799. https://doi.org/10.3390/microorganisms11112799
Chicago/Turabian StyleMeng, Huan, Jianglin Zhou, Mingyu Wang, Mei Zheng, Yaling Xing, and Yajie Wang. 2023. "SARS-CoV-2 Papain-like Protease Negatively Regulates the NLRP3 Inflammasome Pathway and Pyroptosis by Reducing the Oligomerization and Ubiquitination of ASC" Microorganisms 11, no. 11: 2799. https://doi.org/10.3390/microorganisms11112799




