Biofilm Formation by Escherichia coli Isolated from Urinary Tract Infections from Aguascalientes, Mexico
Abstract
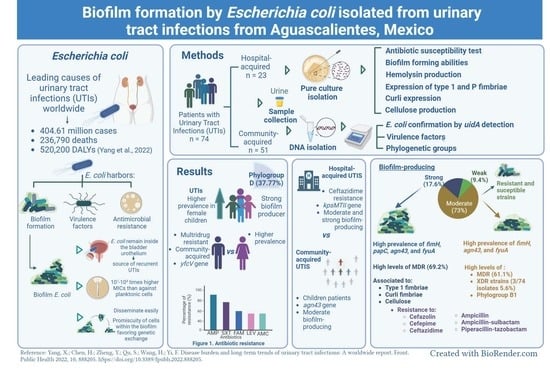
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Detection of Phylogroups and Virulence Genes
2.2. Antimicrobial Susceptibility Testing
2.3. Biofilm Formation Anlaysis
2.4. Statistical Analysis
3. Results
3.1. Demographic Characteristics of Patients with UTI Infection and the Relationship between Antimicrobial Susceptibility and Virulence Genes on E. coli Isolates Strains
3.2. Biofilm-Forming Abilities
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Chen, H.; Zheng, Y.; Qu, S.; Wang, H.; Yi, F. Disease burden and long-term trends of urinary tract infections: A worldwide report. Front. Public Health 2022, 10, 888205. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Sharma, M.; Chaudhary, U. Biofilm and multidrug resistance in uropathogenic Escherichia coli. Pathog. Glob. Health 2015, 109, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Ulett, G.C.; Totsika, M.; Schaale, K.; Carey, A.J.; Sweet, M.J.; Schembri, M.A. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr. Opin. Microbiol. 2013, 16, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. Uropathogenic Escherichia coli (UPEC) Infections: Virulence factors, bladder responses, antibiotic and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- Zagaglia, C.; Ammendolia, M.G.; Maurizi, K.; Nicoletti, M.; Longhi, C. Urinary tract infections caused by uropathogenic Escherichia coli strains—New strategies for an old pathogen. Microorganisms 2022, 10, 1425. [Google Scholar] [CrossRef]
- Kot, B. Antibiotic resistance among uropathogenic Escherichia coli. Pol. J. Microbiol. 2019, 68, 403–415. [Google Scholar] [CrossRef]
- Bartoletti, R.; Cai, T.; Wagenlehner, F.M.; Naber, K.; Bjerklund Johansen, T.E. Treatment of urinary tract infections and antibiotic stewardship. Eur. Urol. Suppl. 2016, 4, 81–87. [Google Scholar] [CrossRef]
- Sanchez, G.V.; Babiker, A.; Master, R.N.; Luu, T.; Mathur, A.; Bordon, J. Antibiotic resistance among urinary isolates from female outpatients in the United States in 2003 and 2012. Antimicro. Agents Chemother. 2016, 660, 2680–2883. [Google Scholar] [CrossRef]
- Adamus-Białek, W.; Baraniak, A.; Wawszczak, M.; Głuszek, S.; Gad, B.; Wróbel, K.; Bator, P.; Majchrzak, M.; Parniewski, P. The genetic background of antibiotic resistance among clinical uropathogenic Escherichia coli strains. Mol. Biol. Rep. 2018, 45, 1055–1065. [Google Scholar] [CrossRef]
- Amabile-Cuevas, C.F. Antibiotic usage and resistance in Mexico: An update after a decade of change. J. Infect. Dev. Ctries. 2021, 15, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.J.; McEwen, S.A. One Health-Its Importance in Helping to Better Control Antimicrobial Resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef]
- Abernethy, J.; Guy, R.; Sheridan, E.A.; Hopkins, S.; Kiernan, M.; Wilcox, M.H.; Johnson, A.P.; Hope, R.; E. coli bacteraemia sentinel surveillance group. Epidemiology of Escherichia coli bacteraemia in England: Results of an enhanced sentinel surveillance programme. J. Hosp. Infect. 2017, 95, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Naziri, Z.; Derakhshandeh, A.; Soltani Borchaloee, A.; Poormaleknia, M.; Azimzadeh, N. Treatment Failure in Urinary Tract Infections: A warning witness for virulent multi-drug resistant ESBL- Producing Escherichia coli. Infect. Drug Resist. 2000, 13, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.C.; Palermo, J.J.; Schilling, J.D.; Roth, R.; Heuser, J.; Hultren, S.J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003, 301, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, P.; Natarajan, V.; Sevanan, M. In vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac. J. Trop. Med. 2012, 5, 210–213. [Google Scholar] [CrossRef]
- Sharma, M.; Aparna; Yadav, S.; Chaudhary, U. Biofilm production in uropathogenic Escherichia coli. Indian J. Pathol. Microbiol. 2009, 52, 294. [Google Scholar] [CrossRef]
- Tapiainen, T.; Hanni, A.M.; Salo, J.; Ikäheimo, I.; Uhari, M. Escherichia coli biofilm formation and recurrences of urinary tract infections in children. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 111–115. [Google Scholar] [CrossRef]
- Dossouvi, K.M.; Ba, B.S.; Lo, G.; Ndiaye, I.; Tine, A.; Karam, F.; Diagne-Samb, H.; Ngom-Cisse, S.; Diop-Ndiaye, H.; Mamman, I.; et al. High prevalence of virulence genes and in-vitro biofilm production in clinical multidrug resistant Escherichia coli in Dakar Senegal. Res. Sq. 2023, 8, 69–81. [Google Scholar] [CrossRef]
- Rafaque, Z.; Abid, N.; Liaqat, N.; Afridi, P.; Siddique, S.; Masood, S.; Kanwal, S.; Dasti, J.I. In-vitro investigation of antibiotics efficacy against uropathogenic Escherichia coli biofilms and antibiotic induced biofilm formation at sub-minimum inhibitory concentration of ciprofloxacin. Infect. Drug Resist. 2023, 13, 2801–2810. [Google Scholar] [CrossRef] [PubMed]
- Blango, M.G.; Mulvey, M.A. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob. Agents Chemother. 2010, 54, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.C.; Han, X.M.; Shi, M.; Pang, Z.L. Persistence of uropathogenic Escherichia coli in the bladders of female patients with sterile urine after antibiotic therapies. J. Huazhong Univ. Sci. Technol. Med. Sci. 2016, 36, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Niveditha, S.; Pramodhini, S.; Umadevi, S.; Kumar, S.; Stephen, S. The Isolation and the Biofilm Formation of uropathogens in the patients with catheter associated urinary tract infections (UTIs). J. Clin. Diagn. Res. 2012, 6, 1478–1482. [Google Scholar] [CrossRef] [PubMed]
- Naves, P.; del Prado, G.; Huelves, L.; Gracia, M.; Ruiz, V.; Blanco, J.; Rodriguez-Cerrato, V.; Ponte, M.C.; Soriano, F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 2008, 45, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Zamani, H.; Salehzadeh, A. Biofilm formation in uropathogenic Escherichia coli: Association with adhesion factor genes. Turk. J. Med. Sci. 2018, 48, 162–167. [Google Scholar] [CrossRef]
- Ballén, V.; Gabasa, Y.; Ratia, C.; Sanchez, M.; Soto, S. Correlation between antimicrobial resistance, virulence determinants and biofilm formation ability among extraintestinal Pathogenic Escherichia coli strains isolated in Catalonia, Spain. Front. Microbiol. 2021, 12, 803862. [Google Scholar] [CrossRef]
- Fattahi, S.; Kafil, H.S.; Nahai, M.R.; Asgharzadeh, M.; Nori, R.; Aghazadeh, M. Relationship of biofilm formation and different virulence genes in uropathogenic Escherichia coli isolates from Northwest Iran. GMS Hyg. Infect. Control 2015, 10, Doc11. [Google Scholar] [CrossRef]
- Tajbakhsh, E.; Ahmadi, P.; Abedpour-Dehkordi, E.; Arbab-Soleimani, N.; Khamesipour, F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob. Resist. Infect. Control 2016, 5, 11. [Google Scholar] [CrossRef]
- Tarchouna, M.; Ferjani, A.; Ben-Selma, W.; Boukadida, J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int. J. Infect. Dis. 2013, 17, e450–e453. [Google Scholar] [CrossRef] [PubMed]
- Davari Abad, E.; Khameneh, A.; Vahedi, L. Identification phenotypic and genotypic characterization of biofilm formation in Escherichia coli isolated from urinary tract infections and their antibiotics resistance. BMC Res. Notes 2019, 12, 796. [Google Scholar] [CrossRef] [PubMed]
- M100; Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013.
- Vollmerhausen, T.L.; Katouli, M. Molecular characterisation of Escherichia coli isolated from hospitalised children and adults with urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Rezatofighi, S.E.; Mirzarazi, M.; Salehi, M. Virulence genes and phylogenetic groups of uropathogenic Escherichia coli isolates from patients with urinary tract infection and uninfected control subjects: A case-control study. BMC Infect. Dis. 2021, 21, 361. [Google Scholar] [CrossRef]
- Soto, S.M.; Smithson, A.; Martinez, J.A.; Horcajada, J.P.; Mensa, J.; Vila, J. Biofilm formation in uropathogenic Escherichia coli strains: Relationship with prostatitis, urovirulence factors and antimicrobial resistance. J. Urol. 2007, 177, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yang, H.; Bi, D.; Khaledi, A.; Qiao, M. A systematic review and meta-analysis of antibiotic resistance patterns, and the correlation between biofilm formation with virulence factors in uropathogenic E. coli isolated from urinary tract infections. Microb. Pathog. 2020, 144, 104196. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P.; Urbán, E.; Gajdács, M. Association between biofilm-production and antibiotic resistance in uropathogenic Escherichia coli (UPEC): An in vitro study. Diseases 2020, 8, 17. [Google Scholar] [CrossRef]
- Whelan, S.; O’Grady, M.C.; Corcoran, D.; Finn, K.; Lucey, B. Uropathogenic Escherichia coli biofilm-forming capabilities are not predictable from clinical details or from colonial morphology. Diseases 2020, 8, 11. [Google Scholar] [CrossRef]
- Mao, B.H.; Chang, Y.F.; Scaria, J.; Chang, C.C.; Chou, L.W.; Tien, N.; Wu, J.J.; Tseng, C.C.; Wang, M.C.; Chang, C.C.; et al. Identification of Escherichia coli genes associated with urinary tract infections. J. Clin. Microbiol. 2012, 50, 449–456. [Google Scholar] [CrossRef]
- Toval, F.; Köhler, C.D.; Vogel, U.; Wagenlehner, F.; Mellmann, A.; Fruth, A.; Schmidt, M.A.; Karch, H.; Bielaszewska, M.; Dobrindt, U. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 2014, 52, 407–418. [Google Scholar] [CrossRef]
- Ramírez Castillo, F.Y.; Avelar Gonzalez, F.J.; Garneau, P.; Marquez Diaz, F.; Guerro Barrera, A.L.; Harel, J. Presence of multidrug resistant pathogenic Escherichia coli in the San Pedro River located in the State of Aguascalientes, Mexico. Front. Microbiol. 2013, 4, 147. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Dinh, P.C., Jr.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Sik Kim, K.; Jhonson, J.R.; Mobley, H.L.T. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Ramos, N.L.; Saayman, M.L.; Chapman, T.A.; Tucker, J.R.; Smith, H.V.; Faogali, J.; Chin, J.C.; Brauner, A.; Katouli, M. Genetic relatedness and virulence gene profiles of Escherichia coli strains isolated from septicaemic and uroseptic patients. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Terai, A.; Yuri, K.; Kurazono, H.; Takeda, Y.; Yoshida, O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol. Med. Microbiol. 1995, 12, 85–90. [Google Scholar] [CrossRef]
- Chen, C.; Liao, X.; Jiang, H.; Zhu, H.; Yue, L.; Li, S.; Fang, B.; Liu, Y. Characteristics of Escherichia coli biofilm production, genetic typing, drug resistance pattern and gene expression under aminoglycoside pressures. Environ. Toxicol. Pharmacol. 2010, 30, 5–10. [Google Scholar] [CrossRef]
- Klein, R.D.; Hultgren, S.J. Urinary tract infections: Microbial pathogenesis, host-pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 2020, 18, 211–226. [Google Scholar] [CrossRef]
- Diaz, J.M.; Dozois, C.M.; Avelar-González, F.J. The vacuolating autotransporter toxin (Vat) of Escherichia coli causes cell cytoskeleton changes and produces non-lysosomal vacuole formation in bladder epithelial cells. Front. Cell. Infect. Microbiol. 2020, 20, 299. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Chakraborty, A.; Saralaya, V.; Adhikari, P.; Shenoy, S.; Baliga, S.; Hedge, A. Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in South Indian population. Ann. Med. Health Sci. Res. 2015, 5, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmetr, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2011, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, M.E.; Girard, V.; Nikolakakis, A.; Campos, M.; Berthiaume, F.; Dumas, F.; Lepine, F.; Mourez, M. O-linked glycosylation ensures the normal conformation of the autotransporter adhesin involved in diffuse adherence. J. Bacteriol. 2007, 189, 8880–8889. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, Y.D.; Vogeleer, P.; Jacques, M.; Harel, J. High-throughput microfluidic method to study biofilm formation and host-pathogen interactions in pathogenic Escherichia coli. Appl. Environ. Microbiol. 2015, 81, 2827–2840. [Google Scholar] [CrossRef]
- Novais, A.; Vuotto, C.; Pires, J.; Montenegro, C.; Donelli, G.; Coque, T.M. Diversity and biofilm-production ability among isolates of Escherichia coli phylogroup D belonging to ST69, ST393 and ST405 clonal groups. BMC Microbiol. 2013, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Crépin, S.; Houle, S.; Charbonneau, M.É.; Mourez, M.; Harel, J.; Dozois, C.M. Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infections. Infect. Immun. 2012, 80, 2802–2815. [Google Scholar] [CrossRef] [PubMed]
- Vagarali, M.A.; Karadesai, S.G.; Patil, C.S.; Metgud, S.C.; Mutnal, M.B. Haemagglutination and siderophore production as the urovirulence markers of uropathogenic Escherichia coli. Indian J. Med. Microbiol. 2008, 26, 68–70. [Google Scholar] [CrossRef]
- Vogeleer, P.; Tremblay, Y.D.N.; Jubelin, G.; Jacques, M.; Harel, J. Biofilm-forming abilities of Shiga toxin-producing Escherichia coli isolates associated with human infections. Appl. Environ. Microbiol. 2015, 82, 1448–1458. [Google Scholar] [CrossRef]
- Da Re, S.; Ghigo, J.M. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 2006, 188, 3073–3087. [Google Scholar] [CrossRef]
- Tabasi, M.; Asadi Karam, M.R.; Habibi, M.; Yekaninejad, M.S.; Bouzari, S. Phenotypic assays to determine virulence factors of uropathogenic Escherichia coli (UPEC) isolates and their correlation with antibiotic resistance pattern. Osong Public Health Res. Perspect. 2015, 6, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Cardiliya, A.P.; Chandrasekar, M.J.N.; Nanjan, M. Incidence of biofilms among the multidrug resistant E. coli, isolated from urinary tract infections in the Nilgiris districti, South India. Braz. J. Microbiol. 2023, 54, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Jombo, T.; Emanghe, U.E.; Amefule, E.N.; Damen, J.G. Urinary tract infections at a Nigerian university hospital: Causes, patterns and antimicrobial suceptibility profile. J. Microbiol. Antimicrob. 2011, 3, 153–159. Available online: http://www.academicjournals.org/JMA (accessed on 22 September 2023).
- Gu, J.; Chen, X.; Yang, Z.; Bai, Y.; Zhang, X. Gender differences in the microbial spectrum and antibiotic sensitivity of uropathogens isolated from patients with urinary stones. J. Clin. Lab. Anal. 2022, 36, e24155. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.; Marijam, A.; Mitrani-Gold, F.S.; Terry, L.; Taylor-Stokes, G.; Joshi, A. Unmet needs in uncomplicated urinary tract infection in the United States and Germany: A physician survey. BMC Infect. Dis. 2023, 23, 281. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, N.; Ağuş, N.; Bayram, A.; Şamlıoğlu, P.; Şirin, M.C.; Derici, Y.K.; Hancı, S.Y. Antimicrobial susceptibilities of Escherichia coli isolates as agents of community-acquired urinary tract infection (2008–2014). Turk. J. Urol. 2016, 42, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Khamari, B.; Kumar, P.; Pradeep, B.E. Resistance to nitrofurantoin is an indicator of extensive drug-resistant (XDR) Enterobacteriaceae. J. Med. Microbiol. 2021, 70. [Google Scholar] [CrossRef]
- Iranpour, D.; Hassanpour, M.; Ansari, H.; Tajbakhsh, S.; Khamisipour, G. Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. Biomed. Res. Int. 2015, 2015, 846219. [Google Scholar] [CrossRef]
- Mirzahosseini, H.K.; Najmeddin, F.; Najafi, A.; Ahmadi, A.; Sharifnia, H.; Khaleidi, A.; Mojtahedzadeh, M. Correlation of biofilm formation, virulence factors, and phylogenetic groups among Escherichia coli strains causing urinary tract infections: A global systematic review and meta-analysis. J. Res. Med. Sci. 2023, 28, 66. [Google Scholar] [CrossRef]
- Atray, D.; Atray, M. Correlation between biofilm production and antibiotic resistance pattern in uropathogenic Escherichia coli in tertiary care hospital in Southern Rajasthan, India. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 640–646. Available online: https://www.ijcmas.com/vol-4-7/Deepika%20Atray%20and%20Meena%20Atray.pdf (accessed on 2 September 2023).
- Poursina, F.; Sepehrpour, S.; Mobasherizadeh, S. Biofilm Formation in Nonmultidrug-resistant Escherichia coli Isolated from Patients with Urinary Tract Infection in Isfahan, Iran. Adv. Biomed. Res. 2018, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, J.; Mishra, B.; Srivastava, S.; Srivastava, R. Genotypic characteristics and biofilm formation among Escherichia coli isolates from Indian women with acute cystitis. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Neupane, S.; Pant, N.D.; Khatiwada, S.; Chaudhary, R.; Banjara, M.R. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrob. Resist. Infect. Control 2016, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Naves, P.; del Prado, G.; Huelves, L.; Gracia, M.; Ruiz, V.; Blanco, J.; Rodriguez-Cerrato, V.; Ponte, M.C.; Soriano, F. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J. Appl. Microbiol. 2008, 105, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Sharifi, A.; Amin-Ghatei, M.; Sadrinasab, M. Evaluation of biofilm formation and virulence genes and association with antibiotic resistance patterns of uropathogenic Escherichia coli strains in Southwestern Iran. Judishapur. J. Microbiol. 2021, 14, e117785. [Google Scholar] [CrossRef]
- Kulkarni, S.R.; Peerapur, B.; Kulkarni, A. Biofilm formation in uropathogenic Escherichia coli strains; relationship with virulence factors and antimicrobial resistance. Int. J. Infect. Dis. 2020, 101, 137. [Google Scholar] [CrossRef]
- Katongole, P.; Nalubega, F.; Florence, N.C.; Asiimwe, B.; Andia, I. Biofilm formation, antimicrobial susceptibility and virulence genes of Uropathogenic Escherichia coli isolated from clinical isolates in Uganda. BMC Infect. Dis. 2020, 20, 453. [Google Scholar] [CrossRef]
- Qian, W.; Li, X.; Yang, M.; Liu, C.; Kong, Y.; Li, Y.; Wang, T.; Zhang, Q. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Escherichia coli Isolates from Ningbo, China. Infect. Drug Resist. 2022, 15, 2865–2878. [Google Scholar] [CrossRef]
- Ebrahimi, M.T.; Hedayati, M.A.; Pirlar, R.F.; Mortzavi, N.; Nazari, M.; Ahmadi, A.; Hemmati, J.; Erfani, Y. Investigation of the biofilm formation in extra-intestinal pathogenic Escherichia coli ST131 strains and its correlation with the presence of fimH, afa, and kpsMSTII genes. J. Appl. Genet. 2023, 64, 367–373. [Google Scholar] [CrossRef]
- Oskouie, A.N.; Hasani, A.; Rezaee, M.A.; Hasani, A.; Saleh, P.; Soltani, E. Phylogenetic characterization of UPEC and its relation with serotyping, distribution of virulence factors, antimicrobial resistance pattern and biofilm formation ability: An apparent elucidation of the bacterial nature. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Reisner, A.; Maierl, M.; Jörger, M.; Krause, R.; Berger, D.; Haid, A.; Tesic, D.; Zechner, E.L. Type 1 fimbriae contribute to catheter-associated urinary tract infections caused by Escherichia coli. J. Bacteriol. 2014, 196, 931–939. [Google Scholar] [CrossRef]
- Hancock, V.; Ferrieres, L.; Klemm, P. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiol. Lett. 2007, 267, 30–37. [Google Scholar] [CrossRef]
- Wright, K.J.; Seed, P.C.; Hultgren, S.J. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007, 9, 2230–2241. [Google Scholar] [CrossRef] [PubMed]
- Kai-Larsen, Y.; Lüthje, P.; Chromek, M.; Peters, V.; Wang, X.; Holm, A.; Kádas, L.; Hedlund, K.-O.; Johansson, J.; Chapman, M.R.; et al. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010, 6, e1001010. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, D.A.; Depas, W.H.; Chapman, M.R. The Biology of the Escherichia coli extracellular matrix. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Samet, M.; Ghaemi, E.; Nejad, M.H.; Jamali, A. Prevalence of different virulence factors and biofilm production ability of urinary Escherichia coli isolates. Int. J. Biol. Med. Res. 2014, 5, 4546–4549. [Google Scholar]
- Abdelhalim, K.A.; Uzel, A.; Unal, N.G. The role of major virulence factors and pathogenicity of adherent-invasive Escherichia coli in patients with Crohn’s disease. Prz. Gastroenterol. 2020, 15, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M.; Smithson, A.; Horcajada, J.P.; Martinez, J.A.; Mensa, J.P.; Vila, J. Implication of biofilm formation in the persistence of urinary tract infection caused by uropathogenic Escherichia coli. Clin. Microbiol. Infect. 2006, 12, 1034–1036. [Google Scholar] [CrossRef]
- Reisner, A.; Krogfelt, K.A.; Klein, B.M.; Zechner, E.L.; Molin, S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: Impact of environmental and genetic factors. J. Bacteriol. 2006, 188, 3572–3581. [Google Scholar] [CrossRef]





| Category | Total n = 74 (%) | Hospital-Acquired n = 23 (%) | Community-Acquired n = 51 (%) | * p-Value | |
|---|---|---|---|---|---|
| Gender | Female | 56 (75.7) | 15 (65.2) | 41 (80.4) | |
| Male | 18 (24.3) | 8 (34.8) | 10 (19.6) | ||
| Age | Children | 45 (60.8) | 10 (43.5) | 35 (68.3) | 0.0402 |
| Adults | 29 (39.2) | 13 (56.5) | 16 (31.4) | ||
| Antimicrobial resistance | Ceftazidime resistance | 19 (25.7) | 10 (43.5) | 9 (17.6) | 0.0355 |
| Susceptible | 7 (9.5) | 2 (8.7) | 5 (9.8) | ||
| Resistant | 67 (90.5) | 21 (91.3) | 46 (90.2) | ||
| Multidrug-resistant | 47 (63.5) | 14 (60.9) | 33 (64.7) | ||
| Virulence gene | fimH | 65 (87.8) | 20 (87.9) | 45 (88.2) | |
| papC | 40 (54.1) | 14 (16.9) | 26 (51.0) | ||
| sfaS | 10 (13.5) | 2 (8.7) | 8 (15.7) | ||
| afa/Dr | 9 (12.2) | 2 (8.7) | 7 (13.7) | ||
| yfcV | 21 (28.4) | 8 (34.8) | 13 (24.5) | ||
| agn43 | 53 (71.6) | 15 (65.2) | 38 (74.5) | 0.0220 | |
| vat | 21 (28.4) | 5 (21.7) | 16 (31.4) | ||
| cnf1 | 3 (4.1) | 1 (4.3) | 2 (3.9) | ||
| hlyA | 9 (12.2) | 3 (13.0) | 6 (11.8) | ||
| fyuA | 59 (79.7) | 19 (82.6) | 40 (78.4) | ||
| chuA | 49 (66.2) | 14 (60.9) | 35 (68.6) | ||
| kpsMTII | 38 (51.4) | 16 (69.6) | 22 (43.1) | 0.0341 | |
| Virulence score | 5.09 ± 1.58 | 5.17 ± 1.66 | 5.06 ± 1.55 | ||
| Phylogenetic group | A | 4 (5.4) | 1 (4.5) | 3 (5.9) | |
| B1 | 9 (12.2) | 5 (21.7) | 4 (7.8) | ||
| B2 | 12 (16.2) | 3 (13.0) | 9 (17.6) | ||
| C | 9 (12.2) | 3 (13.0) | 6 (11.8) | ||
| D | 22 (29.7) | 4 (17.4) | 18 (35.3) | ||
| E | 1 (1.4) | 1 (4.3) | 0 (0.0) | ||
| F | 11 (14.9) | 4 (17.4) | 7 (13.7) | ||
| Clades | 6 (8.1) | 2 (8.7) | 4 (7.8) | ||
| Category | Total n = 74 (%) | Female Patients n = 56 (%) | Male Patients n = 18 (%) | * p-Value | |
|---|---|---|---|---|---|
| Age | Children | 45 (60.8) | 37 (66.07) | 8 (44.44) | |
| Adults | 29 (39.2) | 8 (14.28) | 10 (55.55) | ||
| Antimicrobial resistance | XDR and MDR | 47 (63.5) | 30 (53.6) | 17 (94.4) | 0.0017 |
| XDR | 3 (5.1) | 1 (1.8) | 2 (11.1) | ||
| MDR | 44 (59.5) | 29 (51.8) | 15 (83.3) | 0.0261 | |
| R | 20 (27) | 19 (33.9) | 1 (56.6) | 0.0297 | |
| Susceptible | 7 (9.5) | 7 (12.5) | 0 (0.0) | ||
| Virulence gene | fimH | 65 (87.8) | 47 (83.9) | 18 (100) | |
| papC | 40 (54.1) | 29 (51.8) | 11 (61.1) | ||
| sfaS | 10 (13.5) | 8 (14.3) | 2 (11.1) | ||
| afa/Dr | 9 (12.2) | 9 (16.1) | 0 (0.0) | ||
| yfcV | 21 (28.4) | 12 (21.4) | 9 (50.0) | 0.0193 | |
| agn43 | 53 (71.6) | 40 (71.4) | 13 (72.2) | ||
| vat | 21 (28.4) | 14 (25) | 7 (38.9) | ||
| cnf1 | 3 (4.1) | 2 (3.6) | 1 (5.6) | ||
| hlyA | 9 (12.2) | 6 (10.7) | 3 (16.7) | ||
| fyuA | 59 (79.7) | 47 (83.9) | 12 (66.7) | ||
| chuA | 49 (66.2) | 39 (69.6) | 10 (55.6) | ||
| kpsMTII | 38 (51.4) | 31 (55.4) | 7 (38.9) | ||
| Virulence score | 5.09 ± 1.58 | 5.07 ± 1.52 | 5.17 ± 1.79 | ||
| Phylogenetic group | A | 4 (5.4) | 2 (3.6) | 2 (11.1) | |
| B1 | 9 (12.2) | 6 (10.7) | 3 (16.7) | ||
| B2 | 12 (16.2) | 7 (12.5) | 5 (27.8) | ||
| C | 9 (12.2) | 6 (10.7) | 3 (16.7) | ||
| D | 22 (29.7) | 21 (37.5) | 1 (5.6) | 0.0087 | |
| E | 1 (1.4) | 1 (1.8) | 0 (0.0) | ||
| F | 11 (14.9) | 9 (16.1) | 2 (11.1) | ||
| Clades | 6 (8.1) | 4 (7.1) | 2 (11.1) | ||
| Virulence Genes and Phylogroups | Biofilm-Producing Abilities | ||||
|---|---|---|---|---|---|
| Total n = 74 (%) | Strong n = 13 (%) | Moderate n = 54 (%) | Weak n = 7 (%) | ||
| Virulence gene | fimH | 65 (87.8) | 11 (84.6) | 47 (87.0) | 7 (100) |
| papC | 40 (54.1) | 10 (76.9) | 26 (48.1) | 4 (57.1) | |
| sfaS | 10 (13.5) | 2 (15.4) | 7 (12.9) | 1 (14.3) | |
| afa/Dr | 9 (12.2) | 2 (15.4) | 6 (11.1) | 1 (14.3) | |
| yfcV | 21 (28.4) | 5 (38.5) | 14 (25.9) | 2 (28.6) | |
| agn43 | 53 (71.6) | 9 (69.2) | 39 (72.2) | 5 (71.4) | |
| vat | 21 (28.4) | 3 (23.1) | 16 (29.6) | 2 (28.6) | |
| cnf1 | 3 (4.1) | 1 (7.7) | 1 (1.9) | 1 (14.3) | |
| hlyA | 9 (12.2) | 2 (15.4) | 7 (12.9) | 0 (0.0) | |
| fyuA | 59 (79.7) | 9 (69.2) | 44 (81.5) | 6 (85.7) | |
| chuA | 49 (66.2) | 9 (69.2) | 34 (62.9) | 6 (85.7) | |
| kpsMTII | 38 (51.4) | 5 (35.5) | 29 (53.7) | 4 (57.1) | |
| Virulence score | 5.09 ± 1.58 | 5.23 ± 1.79 | 5.00 ± 1.57 | 5.57 ± 1.40 | |
| Phylogenetic group | A | 4 (5.4) | 1 (7.7) | 3 (5.6) | 0 (0.0) |
| B1 | 9 (12.2) | 1 (7.7) | 8 (14.8) | 0 (0.0) | |
| B2 | 12 (16.2) | 1 (7.7) | 9 (16.7) | 2 (28.6) | |
| C | 9 (12.2) | 1 (7.7) | 7 (12.9) | 1 (14.3) | |
| D | 22 (1.4) | 4 (30.8) | 14 (25.7) | 4 (57.1) | |
| E | 1 (1.4) | 0 (0.0) | 1 (1.9) | 0 (0.0) | |
| F | 11 (14.9) | 3 (23.1) | 8 (14.8) | 0 (0.0) | |
| Clades | 6 (8.2) | 2 (15.4) | 4 (7.4) | 0 (0.0) | |
| Phenotypically Expressed Virulence Factors | Biofilm-Producing Abilities | ||||
|---|---|---|---|---|---|
| Total n = 74 (%) | Strong n =13 (%) | Moderate n = 54 (%) | Weak n = 7 (%) | * p-Value | |
| Type 1 fimbriae (MSHA) a | 55 (74.3) | 11 (84.6) | 40 (74.1) | 4 (57.1) | 0.0349 |
| P fimbriae (MRHA) b | 19 (25.7) | 2 (15.4) | 14 (25.9) | 3 (42.9) | |
| α-Hemolysis | 53 (71.6) | 9 (69.2) | 39 (72.2) | 5 (71.4) | |
| Curli fimbriae | 48 (64.9) | 8 (61.5) | 37 (68.5) | 3 (42.9) | 0.0477 |
| Cellulose | 41 (55.4) | 9 (69.2) | 30 (55.5) | 2 (25.7) | 0.0253 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez Castillo, F.Y.; Guerrero Barrera, A.L.; Harel, J.; Avelar González, F.J.; Vogeleer, P.; Arreola Guerra, J.M.; González Gámez, M. Biofilm Formation by Escherichia coli Isolated from Urinary Tract Infections from Aguascalientes, Mexico. Microorganisms 2023, 11, 2858. https://doi.org/10.3390/microorganisms11122858
Ramírez Castillo FY, Guerrero Barrera AL, Harel J, Avelar González FJ, Vogeleer P, Arreola Guerra JM, González Gámez M. Biofilm Formation by Escherichia coli Isolated from Urinary Tract Infections from Aguascalientes, Mexico. Microorganisms. 2023; 11(12):2858. https://doi.org/10.3390/microorganisms11122858
Chicago/Turabian StyleRamírez Castillo, Flor Yazmín, Alma Lilian Guerrero Barrera, Josée Harel, Francisco Javier Avelar González, Philippe Vogeleer, José Manuel Arreola Guerra, and Mario González Gámez. 2023. "Biofilm Formation by Escherichia coli Isolated from Urinary Tract Infections from Aguascalientes, Mexico" Microorganisms 11, no. 12: 2858. https://doi.org/10.3390/microorganisms11122858