Abstract
Carotenoids are secondary metabolites that exhibit antioxidant properties and are characterized by a striking range of colorations from red to yellow. These natural pigments are synthesized by a wide range of eukaryotic and prokaryotic organisms. Among the latter, carotenoid-producing methanotrophic bacteria, which display fast growth on methane or natural gas, are of particular interest as potential producers of a feed protein enriched with carotenoids. Until recently, Methylomonas strain 16a and Methylomonas sp. ZR1 remained the only representatives of the genus for which detailed carotenoid profile was determined. In this study, we analyzed the genome sequences of five strains of Methylomonas species whose pigmentation varied from white and yellow to orange and red, and identified carotenoids produced by these bacteria. Carotenoids synthesized using four pigmented strains included C30 fraction, primarily composed of 4,4’-diaplycopene-4,4’-dioic acid and 4,4’-diaplycopenoic acid, as well as C40 fraction with the major compound represented by 1,1’-dihydroxy-3,4-didehydrolycopene. The genomes of studied Methylomonas strains varied in size between 4.59 and 5.45 Mb and contained 4201–4735 protein-coding genes. These genomes and 35 reference Methylomonas genomes available in the GenBank were examined for the presence of genes encoding carotenoid biosynthesis. Genomes of all pigmented Methylomonas strains harbored genes necessary for the synthesis of 4,4’-diaplycopene-4,4’-dioic acid. Non-pigmented “Methylomonas montana” MW1T lacked the crtN gene required for carotenoid production. Nearly all strains possessed phytoene desaturases, which explained their ability to naturally synthesize lycopene. Thus, members of the genus Methylomonas can potentially be considered as producers of C30 and C40 carotenoids from methane.
1. Introduction
Carotenoids are a class of abundant naturally occurring pigments synthesized by all photosynthetic organisms and some non-photosynthetic bacteria and fungi [1]. To date, the Carotenoids Database (http://carotenoiddb.jp, accessed on 20 November 2023) [2] provides information on the chemical structures of more than 1200 carotenoids found in >720 organisms from all domains of life. This diversity and ubiquity establish carotenoids as the second most prevalent natural compounds after chlorophyll [3]. Carotenoids are isoprenoid compounds with a polyene backbone that contains a variable number of conjugated double bounds (with a maximum absorbance of 400–500 nm), a feature that confers coloration in the yellow-to-red range [4].
Carotenoids can be categorized into two primary groups based on the oxygenation status: (i) oxygenated molecules or xanthophylls (e.g., astaxanthin, canthaxanthin, lutein, zeaxanthin) and (ii) non-oxygenated molecules or carotenes (e.g., lycopene, α-carotene, β-carotene) [5]. Carotenoids are highly sensitive to oxygen, heat, and light, as well as acidic or basic conditions [6,7]. The precursors for carotenoid biosynthesis, isopentenyl pyrophosphate (IPP), and its isomer dimethylallyl pyrophosphate (DMAPP) are generated through two distinct metabolic routes, i.e., the mevalonate pathway and non-mevalonate (MEP/DOXP) pathways [8,9]. Isoprenoid synthesis in prokaryotes predominantly relies on the MEP/DOXP pathway. The mevalonate pathway is rarely present and only a few prokaryotes have both [10]. Chain elongation from C10 to C20 occurs through the successive head-to-tail condensation of IPP to DMAPP, resulting in the synthesis of geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), and geranylgeranyl diphosphate (GGPP) [11]. Consequently, carotenoids originating from C30 and C40 chains follow distinct metabolic routes. C30 carotenoid synthesis proceeds with the addition of two FPP units, whereas C40 carotenoid synthesis is driven by the incorporation of two GGPP units. Carotenoids belonging to the C30 group are subject to early functionalization through oxidation processes, resulting in the production of aldehyde, ketone, and/or carboxyl groups. In contrast, carotenoids in the C40 group undergo dehydrogenation and cyclization processes prior to these oxidation reactions [7].
Currently, 179 representatives of the domain Bacteria have been described as a source of carotenoids. However, the number of pigmented bacteria is much bigger. Among them are aerobic methanotrophs, characterized by the unique ability to utilize methane (CH4) as their sole source of energy. Methanotrophs possess methane monooxygenase, the enzyme that can activate the stable C-H bond in the CH4 molecule (104 kcal/mol), which explains its unique ability to use methane for growth [12,13,14]. The currently described diversity of aerobic methanotrophs encompasses ~30 genera within Gamma- and Alphaproteobacteria and the Verrucomicrobia [15], many of which are represented by pigmented phenotypes. In particular, the presence of pigments is one of the genus-specific features of Methylomonas species [16]. The only exception is “M. montana”, which was described as a non-pigmented methanotroph [17]. Growing interest in the field of methanotrophic biocatalysis is driven by the availability and abundance of methane as a component of natural gas. Methanotrophs have been intensively exploited for the conversion of CH4 to single-cell protein [18,19] and various value-added products, including methanol [20], polyhydroxybutyrate [21], lipids [22], and organic acids [23]. High biotechnological potential has been shown for Methylomonas and Methylococcus genera, which accommodate fast-growing methanotrophs [24,25,26]. Thus, Methylococcus capsulatus was used for a large-scale commercial production of microbial proteins from natural gas [19,27]. Methylomonas species were also considered as potential producers of a single-cell protein, i.e., protein produced in microbial cells [28,29]. Notably, Methylomonas bacteria have a clear advantage over non-pigmented Methylococcus methanotrophs due to the presence of carotenoids in their biomass. Recently described Methylomonas rapida MP1T displayed a high growth rate in a bioreactor supplied with natural gas and produced both C30 (4,4’-diaplycopene-4,4’-dioic acid, 4,4’-diaplycopenoic acid) and C40 (1,1’-dihydroxy-3,4-didehydrolycopene) carotenoids [30]. Few studies described the construction of the metabolic route leading to C40 carotenoid, astaxanthin, in Methylomonas hosts [31,32]. This was implemented by blocking the production of C30 carotenoids via gene deletions and via the subsequent introduction of the C40 carotenoid biosynthesis genes (crtE, crtY, crtI, crtB, crtW, and crtZ).
The biosynthesis of C30 carotenoids in methanotrophic bacteria was studied on the basis of Methylomonas sp. 16a. Tao et al. [33] identified the crtNb gene involved in the conversion of 4,4’-diapolycopene to 4,4’-diapolycopene aldehyde, as well as the aldehyde dehydrogenase gene (ald) responsible for the subsequent oxidation of 4,4’-diapolycopene aldehyde to 4,4’-diapolycopene acid. The group of C30 4,4’-diapocarotenoids also encompasses compounds like 4,4’-diapocarotene-4-oic acid and fatty acid esters di-(β,D-glucosyl)-4,4’-diapocarotene-4,4’-dioate from Methylobacterium rhodinum [34]. It also includes compounds like 4,4’-diaponeurosporene and OH-diaponeurosporene found in Heliobacteria [35,36], as well as staphyloxanthin, 4,4’-diapophytoene, 4,4’-diapophytofluene, 4-4’-diapo-zeta-carotene, 4,4’-diaponeurosporene, and its derivatives from Staphylococcus aureus [37,38].
In this study, we obtained three novel isolates of Methylomonas bacteria and sequenced their genomes. The pool of studied bacteria was expanded by the recently described red-colored M. rapida [30] and non-pigmented “M. montana” [17]. The objective of this study was to identify carotenoid composition in these Methylomonas strains and to examine genomes of these bacteria for the presence of genes related to carotenoid biosynthesis.
2. Materials and Methods
2.1. Strains and Cultivation Procedures
Previously described representatives of the genus Methylomonas, M. rapida MP1T [30], and “M. montana” MW1T [17], as well as three newly isolated strains of Methylomonas species, MY1, MO1, and MV1, were used in this study. New isolates were obtained from surface (0–2 cm) sediment samples collected in 2021 and 2022 from various freshwater bodies. Aliquots of sediment samples were used as inoculum to obtain enrichment cultures of methanotrophic bacteria. The latter were obtained using dilute NMS medium (dNMS), containing (in grams per liter of distilled water) KNO3, 0.1; MgSO4 × 7H2O, 0.2; CaCl2 × 2H2O, 0.04; 100 mM phosphate buffer, pH 5.8, 1% (v/v); and a trace element solution 0.1% (v/v) containing the following (g/L): EDTA, 5; FeSO4 × 7H2O, 2; ZnSO4 × 7H2O, 0.1; MnCl2 × 4H2O, 0.03; CoCl2 × 6H2O, 0.2; CuSO4 × 5H2O, 0.1; NiCl2 × 6H2O, 0.02; and Na2MoO4, 0.03. One gram of sediment was added to a 500 mL bottle containing 100 mL of dNMS medium. The bottle was sealed with silicone rubber septa, and methane was added aseptically using a syringe equipped with a disposable filter (0.22 µm) to achieve a 10–20% mixing ratio in the headspace. The incubation was performed on a rotary shaker (120 r.p.m.) at 30–35 °C. After 10 days of incubation, the cultures enriched with methanotrophic bacteria were subjected to serial dilutions. After several serial dilution steps, cell suspensions were plated on agar-solidified dNMS medium. The plates were incubated at 30 °C in desiccators containing approximately 30% (v/v) methane in air. The colonies appearing on the plates were picked and re-streaked on the same agar medium. The set of finally selected colonies was subjected to several additional serial dilution steps in a liquid dNMS medium at 30–35 °C until isolates of methanotrophic bacteria were obtained. Culture purity was verified via examination using phase-contrast microscopy and by plating on 10-fold-diluted Luria–Bertani agar (1.0% tryptone, 0.5% yeast extract, 1.0% NaCl).
2.2. Morphological Characterization and Growth Tests
Morphological observations and cell-size measurements were made with a Zeiss Axioplan 2 microscope and Axiovision 4.2 software (Zeiss, Oberkochen, Germany). Electron microscopy was conducted as described previously [30]. Briefly, exponentially grown cells were collected, pre-fixed with 2.5% glutaraldehyde in 0.05 M cacodylate buffer (pH 7.2), and fixed in OsO4 (1% OsO4 + 0.7% ruthenium red solution) in the same buffer. After fixation, samples were sequentially treated with a 3% uranyl acetate solution and 70% ethanol. Dehydration was performed in 96% ethanol and absolute acetone before embedding in Epon 812 epoxy resin. Ultrathin sections were cut, stained with 3% uranyl acetate and lead citrate, and examined using a JEM 100CXII transmission electron microscope (Jeol, Tokyo, Japan) at an 80 kV accelerating voltage. A comparative analysis of growth characteristics was carried out by observing the growth dynamics of the studied strains in a liquid mineral medium containing 20% methane in the headspace within the temperature range of 4–48 °C. All incubations were performed in triplicate.
2.3. Identification of Carotenoids
Centrifuged cell pellets (40–120 mg fresh weight) were extracted with ethanol. The extract was dried on a rotary evaporator. Then, the pigment film was dissolved in 50 µL of a mixture of acetone and methanol (7:2) and 20 µL of this mixture was used to identify carotenoids via HPLC with diode array detection. The HPLC device (Shimadzu, Kyoto, Japan) consisted of (1) a pump LC-10ADVP (Shimadzu, Kyoto, Japan) with a module FCV-10ALVP, (2) a detector with a diode matrix SPD-M20A, and (3) a thermostat CTO-20 AC. The separation of the carotenoids was performed on a 4.6 × 250 mm reversed phase column (Agilent Zorbax SB-C18, Agilent, Santa Clara, CA, USA) at 22 °C. The solvent gradient was described elsewhere [39]. The feed rate of all solvents was 1.0 mL/min. The obtained data were processed using the Origin8 program (OriginLab Corporation, Northampton, MA, USA). The carotenoids were identified by their retention time and absorption spectra [33,34,40,41,42]. The quantification of each carotenoid was performed by comparing its peak area in the region of 360–800 nm to the sum of all carotenoid peaks taken as 100% and was calculated with the LC-solution program (Shimadzu, Kyoto, Japan) using molar extinction coefficients [43].
2.4. DNA Extraction
Cultures of new isolates were grown in the liquid dNMS as described above. The cells were harvested after incubation at 30–35 °C on a rotary shaker at 150 rpm for 2 days. In total, 30 mL of cell suspensions were centrifuged at 10,000× g. Genomic DNA extraction was completed using the standard CTAB and phenol-chloroform protocol [44]. Briefly, the pellet was resuspended in 567 µL TE buffer. In total, 30 µL of 10% SDS and 3 µL of 20 mg/mL proteinase K were added, and the mixture was incubated at 37 °C for 1 h. Then, 100 µL of 5 M NaCl and 80 µL of CTAB/NaCl solution were sequentially added, with mixing after each addition. The resulting mixture was incubated at 65 °C for 10 min. An equal volume (0.7 to 0.8 mL) of chloroform/isoamyl alcohol was added, mixed, and centrifuged for 5 min at 10,000× g. The supernatant was transferred to a new tube and an equal volume of phenol/chloroform/isoamyl alcohol was added. After a 5 min centrifugation, the pellet was resuspended in 0.6 volume of isopropanol, centrifuged at 10,000× g for 5 min, and washed with 70% ethanol. After centrifugation, ethanol was discarded, and the pellet was redissolved in nuclease-free water.
2.5. Genome Sequencing and Annotation
The genomic paired-end (2 × 300) library was prepared with a NEBNext ultra II DNA Library kit (New England Biolabs, Ipswich, MA, USA) and sequenced using a MiSeq instrument (Illumina, San Diego, CA, USA). Adapter removal and quality filtering was completed using Trimmomatic [45]. Nanopore sequencing library was prepared using the 1D ligation sequencing kit (SQK-LSK108, Oxford Nanopore Technologies, Oxford, UK). Sequencing was performed on an R9.4 flow cell (FLO-MIN106, Oxford Nanopore Technologies, Oxford, UK) using a MinION device (Oxford Nanopore Technologies, Oxford, UK). Hybrid assembly of short and long reads was performed using Unicycler version 0.4.8 [46]. Assemblies were evaluated with Quast 5.0 [47] and Busco 5.1.2 [48]. Annotation was performed using PROKKA version 1.14.5 [49] and BlastKOALA version 3.0 [50].
2.6. Phylogenomic Analysis
The genome-based tree of the five Methylomonas strains and phylogenetically related members of the family Methylococcaceae was reconstructed using the Genome Taxonomy Database and GTDB-Tk version 2.3.2 (https://github.com/Ecogenomics/GtdbTk, accessed on 1 November 2023). The maximum likelihood tree was constructed using MEGA software version 10.2 [51]. The Pyani program version 0.2.12 was used to estimate average nucleotide identities (ANIs) across Methylomonas genomes [52].
2.7. Identification of Carotenoid Biosynthesis Genes
Reference amino-acid gene sequences of C30 and C40 carotenoid biosynthesis enzymes were retrieved from the “UniProtKB reference proteomes plus Swiss-prot” database [53] and from the KEGG database [54]. The analysis was primarily focused on gene categories related to the biosynthesis of C5, C15, and C20 building blocks as well as the genes involved in the biosynthesis of C30 and C40 carotenoids. The identification of carotenoid biosynthesis genes was conducted using BLASTP version 2.15.0 [55] against a local database that included genomes of five Methylomonas strains from this study and RefSeq Methylomonas genomes retrieved from GenBank [56]. A gene was considered homologous if a blast hit with E-value < 10−5 produced an alignment with a minimum of 50% sequence identity and 50% sequence similarity [57]. Conservation of function was verified by searching against Pfam database [58] and running HMMSCAN [59]. Binary data of presence/absence matrix were plotted using custom script in Python version 3.12.0 (http://www.python.org, accessed on 10 October 2023).
2.8. Sequence Accession Numbers
The 16S rRNA gene sequences of strains MO1, MY1, and MV1 have been deposited in the GenBank under the accession numbers OR234854-OR234856, respectively. The assembled genome sequences of strains MO1, MY1, and MV1 have been deposited in NCBI GenBank under the accession numbers JAVSMZ000000000.1, CP133985.1, and JAVSMY000000000.1, respectively.
3. Results
3.1. Identification and Characterization of Novel Isolates
The spectrum of examined methanotrophs included M. rapida MP1T, “M. montana” MW1T, and three new isolates obtained and identified in this study. Strain MO1 was isolated from a sediment of Meshchersky pond in the Moscow region (Russia) and was closely related to “M. denitrificans” FJG1 (99.67% 16S rRNA gene sequence similarity) and M. methanica MC09 (97.32% 16S rRNA gene sequence similarity). Strain MV1 was obtained from a sediment of an unnamed freshwater pond in the Krasnodar region, South Russia, and displayed 97.91% 16S rRNA gene sequence similarity to M. koyamae Fw12E-Y. Finally, strain MY1 was isolated from a sediment of an unnamed freshwater lake in the Krasnodar region and displayed 100% 16S rRNA gene sequence similarity to M. koyamae Fw12E-Y. Cells of strains MV1 and MO1 were represented by short rods, 0.7–0.9 μm in length, while cells of strain MY1 were up to 1.5 μm in length (Table 1).

Table 1.
Isolation sources and some characteristics of Methylomonas strains used in this study.
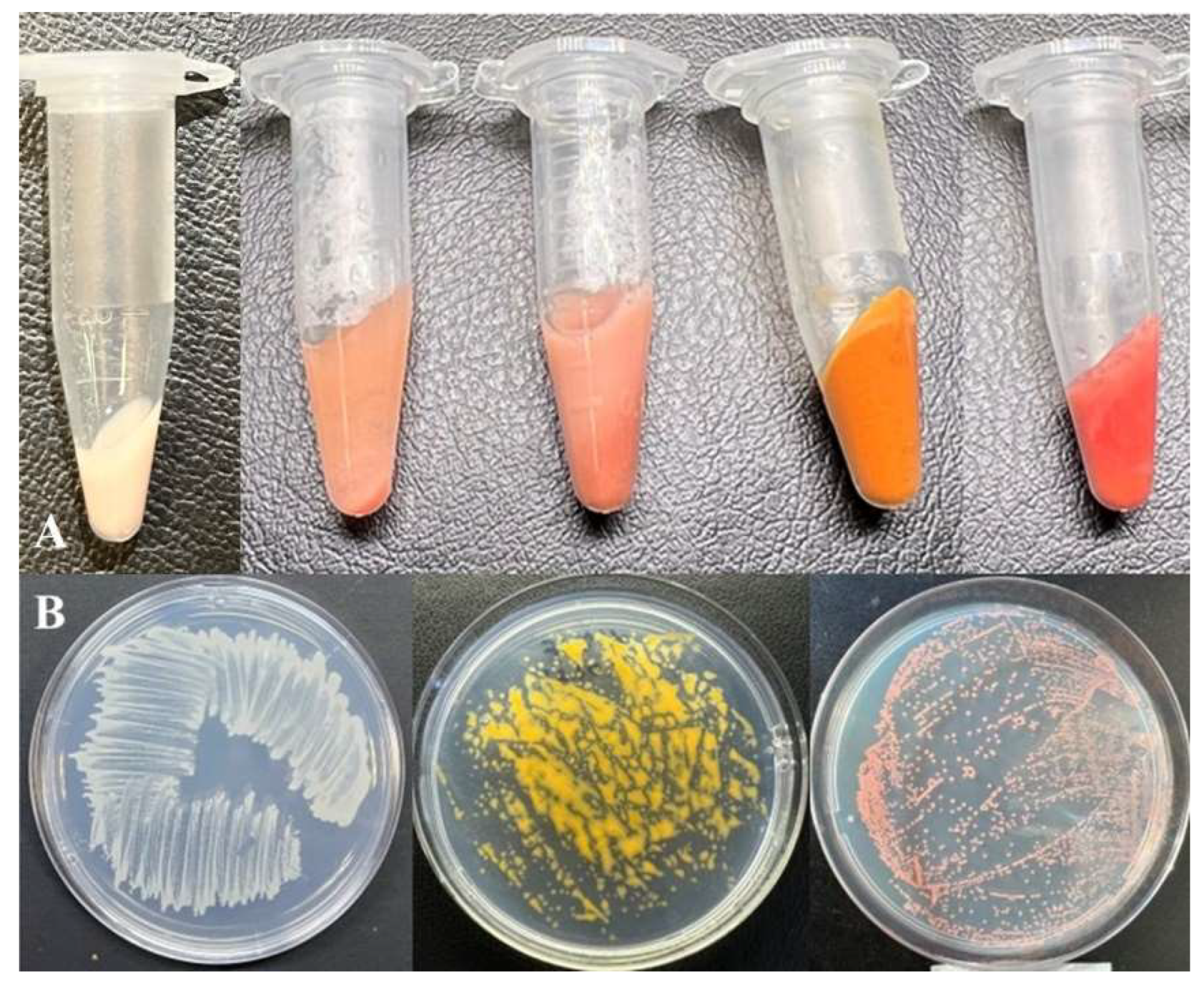
The slimy colonies formed by these isolates on agar dNMS medium after seven days of incubation with 20% (v/v) methane were 2–3 mm in diameter, round, and colored as orange (for strain MO1), yellow (for strain MY1), or pink to red (for strain MV1) (Figure 1).
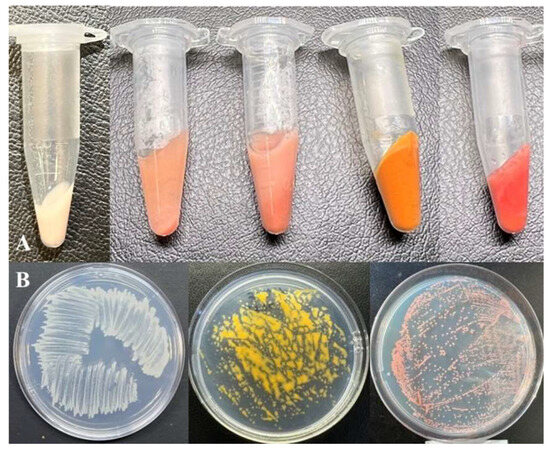
Figure 1.
Color spectrum of new Methylomonas strains. (A) Centrifuged biomass of strains, from left to right: MW1T, MV1, MO, MY1, and MP1T. Growth of strains on plates with agar medium, from left to right: strains MW1T, MY1, and MV1 (B).
Three novel strains and “M. montana” MW1T were capable of growth in a broad temperature range from below 10 °C to 37–38 °C, with an optimum temperature at 30–32 °C. M. rapida MP1T was capable of growth up to 45 °C, with an optimum temperature at 35 °C. Specific growth rates of new Methylomonas isolates ranged from 0.25 to 0.29 h−1, with the highest value (0.29 h−1) observed for M. rapida. Among all the studied Methylomonas representatives, the lowest growth rate (0.13 h−1) was observed for “M. montana” MW1T, while the highest growth rate (0.29 h−1) was recorded for M. rapida MP1T (Table 1).
3.2. Carotenoid Profiles
Carotenoids were detected in the pigment extracts of all strains except for strain MW1T. C30 carotenoids were predominant in the pigment fraction, with lesser amounts of C40 carotenoids also identified. The major carotenoid detected in colored isolates was 4,4’-diaplycopene-4,4’-dioic acid (C30). Strains MY1, MO1, and MP1 also contained 1,1’-dihydroxy-3,4-didehydrolycopene (C40) as the second major carotenoid (Table 2).

Table 2.
Carotenoid composition in studied strains of Methylomonas spp., mol %.
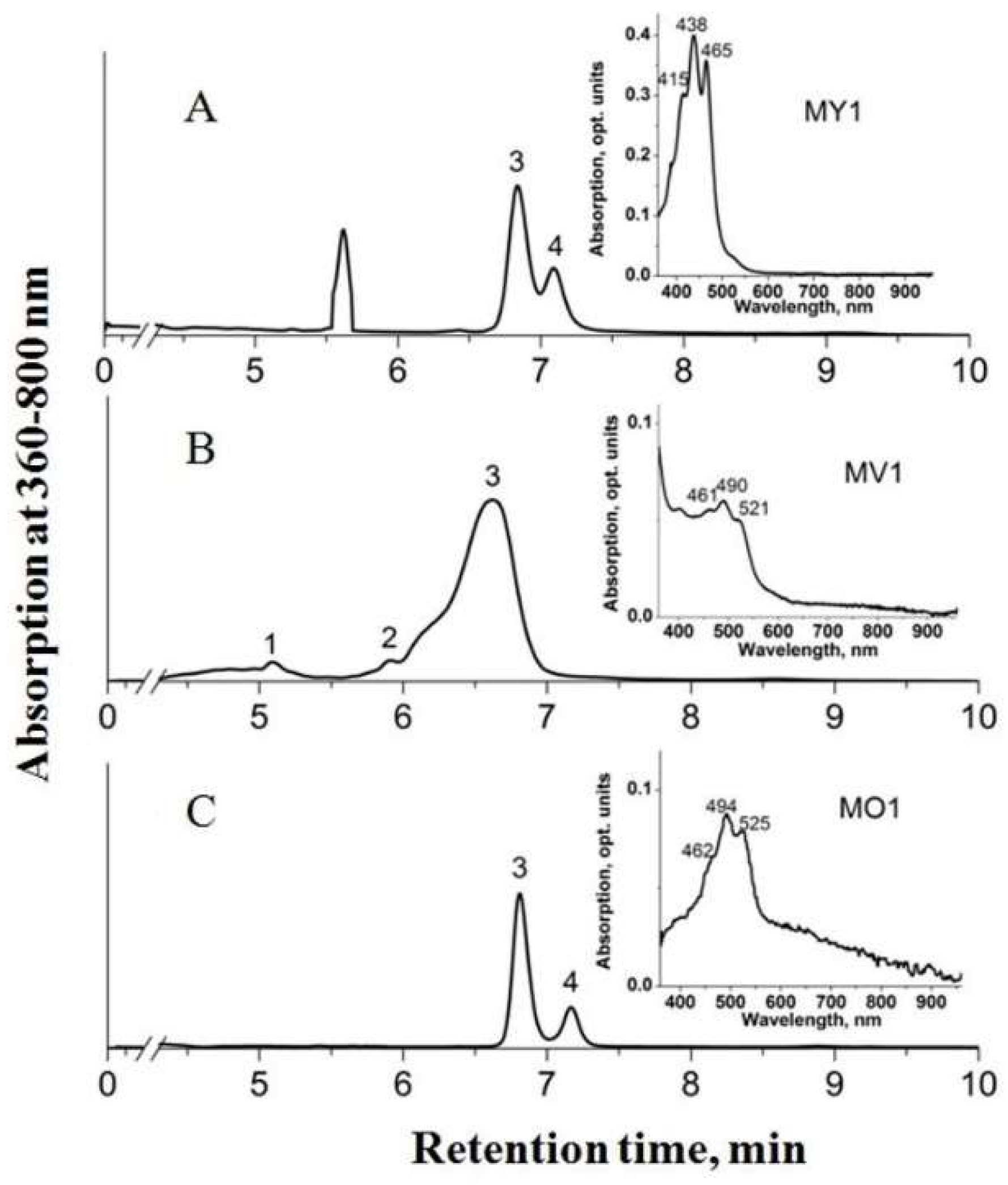
Minor amounts of 4,4’-diaplycopenoic acid (C30), 4’-apo-3,4-didehydrolycopene (C35), and tetradehydrolycopene (C40) were identified in strain MP1T. Strain MV1 lacked 1,1’-dihydroxy-3,4-didehydrolycopene, but contained minor amounts of another C40 carotenoid, 1,2-dihydro-3,4-dehydrolycopene, and the C35 carotenoid 4’-apo-3,4-didehydrolycopene. Thus, C35 carotenoids were exclusively present in pigment fraction of strains MV1 and MP1T. The HPLC chromatogram of pigment extracts from strains MY1, MV1, and MO1 is shown in Figure 2.
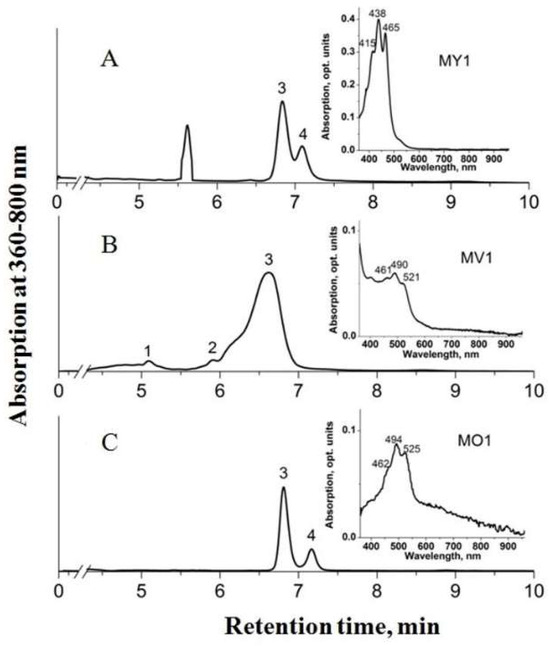
Figure 2.
HPLC chromatogram of pigment extracts from strains MY1 (A), MV1 (B), and MO1 (C). Peak identification: 1—1,2-dihydro-3,4-dehydrolycopene; 2—4’-apo-3,4-didehydrolycopene; 3—4,4’-diaplycopene-4,4’-dioic acid; 4—1,1’-dihydroxy-3,4-didehydrolycopene. Inserts: absorption spectra of pigment extracts dissolved in ethanol.
3.3. Genome Sequencing and Assembly
The genomes of three novel isolates were sequenced using a hybrid approach. Oxford Nanopore sequencing yielded 211,318–258,525 reads with a total length of 1.2–1.4 Gb (Table 3).

Table 3.
Sequencing statistics.
Sequencing on Illumina MiSeq platform generated a total of 211,318–258,525 paired-end reads (300 bp) with a total length of 0.6–1.1 Gb. Both short and long reads were combined to perform a hybrid assembly. The genome of strain MY1 was assembled into a circular contig. Genome assembly for strains MO1 and MV1 consisted of 2 (N50 = 4.93 Mbp) and 10 (N50 = 2.78 Mbp) contigs. The genome characteristics of studied strains are summarized in Table 4.

Table 4.
General genome characteristics of studied Methylomonas strains.
Genome sizes varied from 4.95 Mb in strain MY1 to 5.42 Mb in strain MV1. The DNA G + C content ranged from 51.44% to 56.16%. M. rapida MP1T possessed four copies of rRNA operon, while other Methylomonas genomes contained only three copies of rRNA operon. Each genome contained one copy of the pMMO-encoding gene cluster. The number of coding sequences varied between 4201 and 4735.
3.4. Genome-Based Phylogeny and Genome-to-Genome Comparison
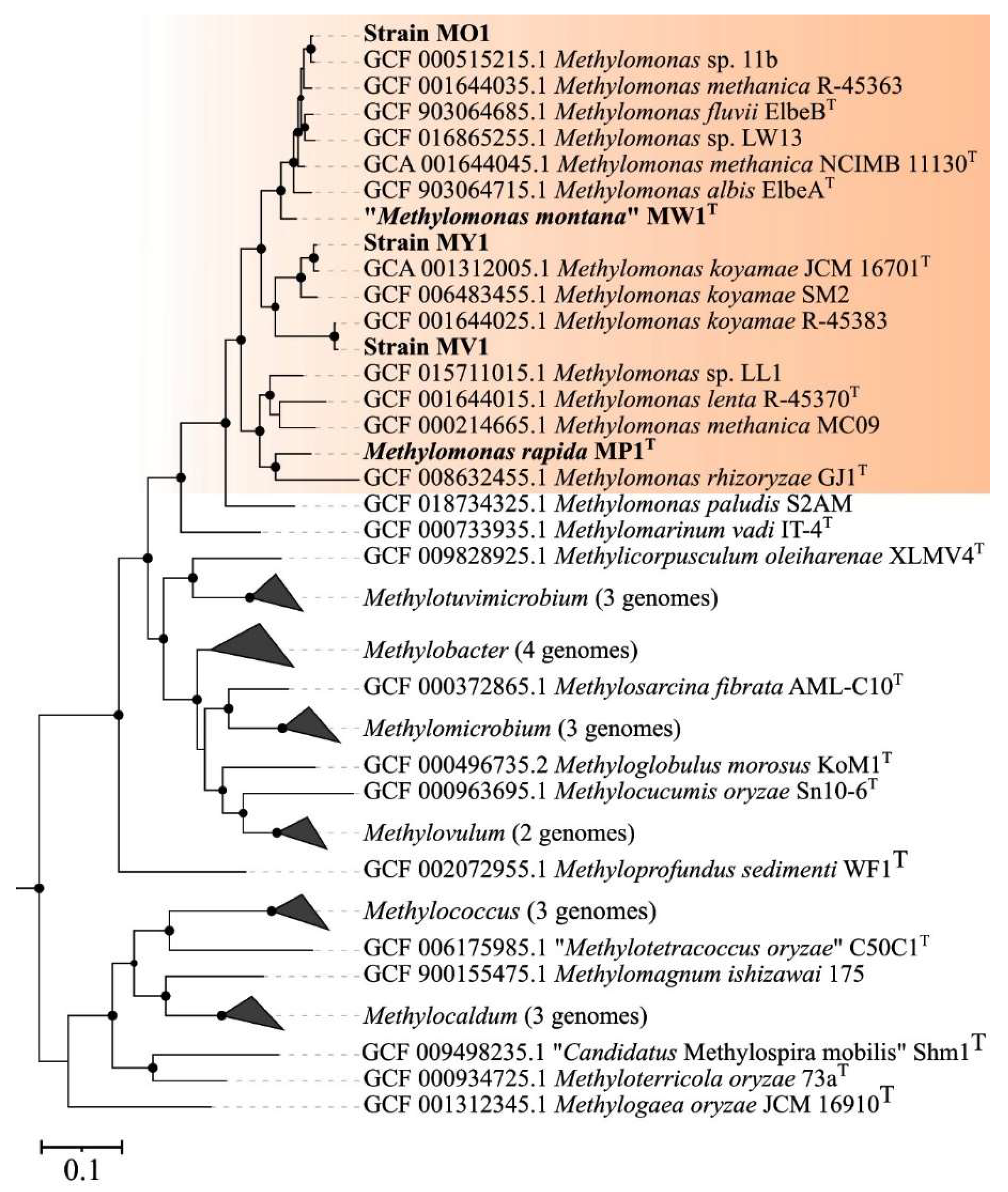
The genome-based phylogeny of new Methylomonas-affiliated isolates was elucidated using the comparative sequence analysis of 120 ubiquitous single-copy proteins (Figure 3).
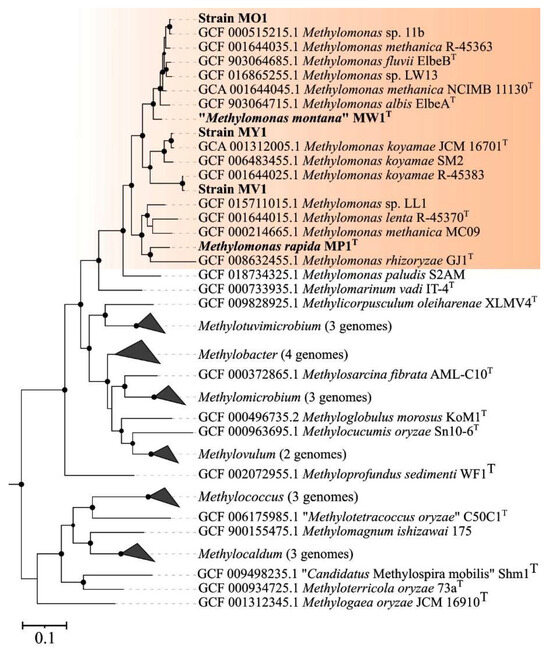
Figure 3.
Genome-based phylogeny showing the position of new isolates in relation to other gammaproteobacterial methanotrophs based on the comparative sequence analysis of 120 ubiquitous single-copy proteins. The clade of Methylomonas methanotrophs is highlighted in orange. The tree was constructed using the GTDB-Tk version 2.3.2 (https://github.com/Ecogenomics/GtdbTk, accessed on 1 November 2023). The significance levels of interior branch points obtained in maximum-likelihood analysis were determined by bootstrap analysis (100 data re-samplings). Bootstrap values of >70% are shown. The root (not shown) is composed of 52 genomes affiliated to type II methanotrophs Methylocella, Methylocapsa, Methyloferula, Methylocystis, and Methylosinus). Bar, 0.1 substitutions per amino acid position.
Strains MV1 and MY1 belonged to the phylogenetic clade defined by members of the species M. koyamae. Strain MO1 was positioned close to an uncharacterized Methylomonas species within a well-supported clade that also included M. fluvii ElbeB, M. methanica NCIMB 11130, and M. albis ElbeAT. “M. montana” MW1T constituted a separate branch located near the aforementioned clade. The average nucleotide identity (ANI) values were estimated for each pair within a set comprising publicly available Methylomonas genomes and genomes of studied strains. Below are the data on the pairwise comparison of ANI values between the new isolates and closely related taxonomically characterized methanotrophs. ANI values calculated for strains MY1 and M. koyamae JCM 16701T and for strain MV1 in comparison to M. koyamae JCM 16701T constituted 96.8% and 77.9%, respectively. The determined ANI value for strain MO1 and M. fluvii EbB was 89.0%. The fact that ANI values between the genomes of strains MV1, MO1, and closely related methanotrophs were estimated below a 95% threshold indicates that these novel isolates may potentially represent new species within the genus Methylomonas, according to current standards for using genome data in the taxonomy of prokaryotes [60,61,62].
3.5. Analysis of Carotenoid Biosynthesis Genes
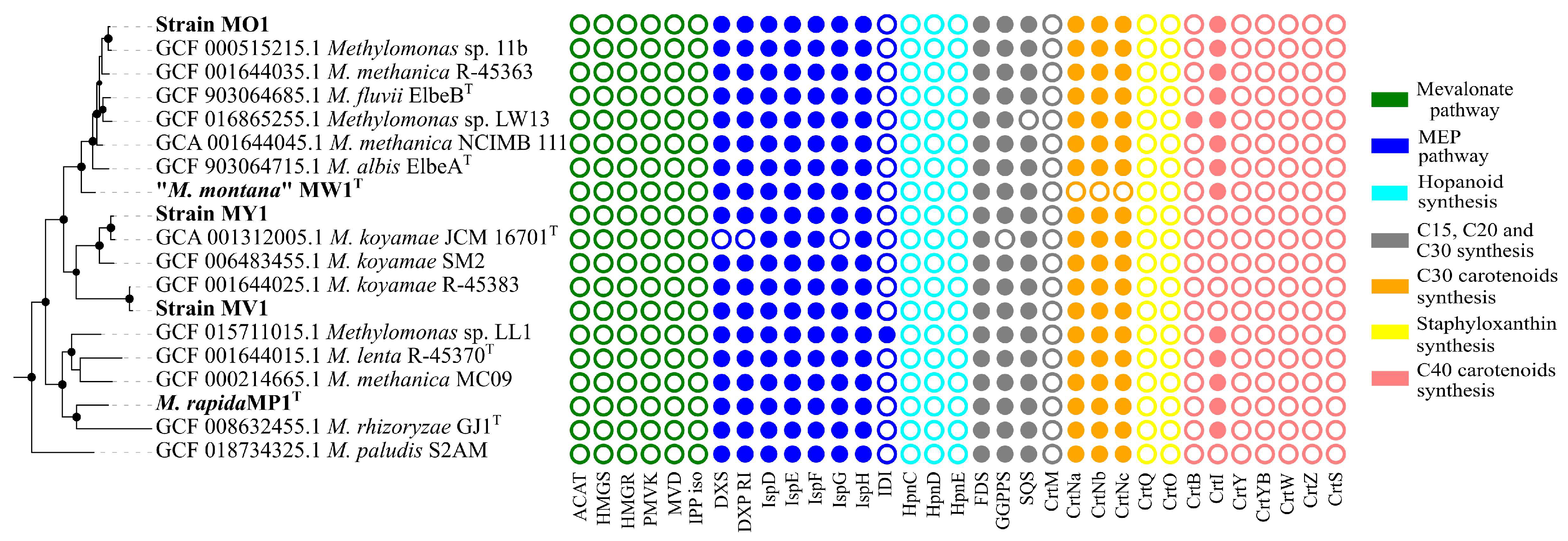
All studied Methylomonas genomes possessed the entire set of genes encoding enzymes of methyl-D-erythritol phosphate (MEP) pathway for the biosynthesis of precursor molecules, isopentenyl diphosphate (IPP), and dimethylallyl diphosphate (DMAPP) (Figure 4).
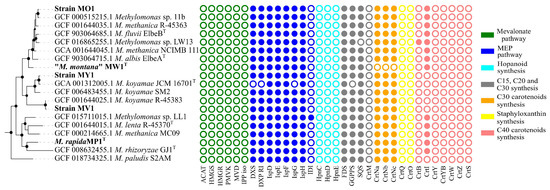
Figure 4.
Distribution of genes related to carotenoid biosynthesis in Methylomonas genomes. Abbreviations for genes: ACAT, acetyl-CoA C-acetyltransferase; HMGS, hydroxymethylglutaryl-CoA synthase; HMGR, hydroxymethylglutaryl-CoA reductase (NADPH); PMVK, phosphomevalonate kinase; MVD, diphosphomevalonate decarboxylase; IPP iso, isopentenyl-diphosphate delta-isomerase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXS RI, 1-deoxy-D-xylulose 5-phosphate reductoisomeras; ispD, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; ispE, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; ispF, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; IspG, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; IspH, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase; IDI, isopentenyl-diphosphate Delta-isomerase; HpnC, hydroxysqualene_synthase; HpnD, presqualene_diphosphate_synthase; HpnE, hydroxysqualene_dehydroxylase; FDS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; SQS, squalene synthase; CrtM, diapophytoene synthase; CrtNa, diapolycopene oxygenase; CrtNc, 4,4’-diapolycopenoate synthase; CrtQ, 4,4’-diaponeurosporenoate glycosyltransferase; CrtO, Glycosyl-4,4’-diaponeurosporenoate acyltransferase; CrtB, phytoene synthase; CrtI, phytoene desaturase; CrtY, lycopene beta-cyclase; CrtYB, bifunctional lycopene cyclase/phytoene synthase; CrtW, beta-carotene ketolase; CrtZ, beta-carotene hydroxylase; and CrtS, astaxanthin synthase.
No genes were identified for the alternative mevalonate pathway, which is common in archaea, fungi, and animals, and can also be found in some bacteria [11]. All genomes harbored the gene for farnesyl diphosphate (FPP) synthase that catalyzes the sequential addition of the hydrocarbon moieties of dimethylallyl diphosphate (C5) and geranyl diphosphate (C10) to IPP (C5) to give FPP (C15) [63]. The crtM gene, encoding a homologue of the diapophytoene synthase in Staphylococcus, was not found in any of the genomes. In contrast, a single-copy gene for squalene/phytoene synthase family protein, containing SQS_PSY domain (PF00494, Pfam database), was identified in a cluster with the gene encoding squalene-hopene cyclase in all examined Methylomonas genomes. SQS catalyzes a two-step conversion of two FPP molecules into squalene. The hpnCDE genes, which are also responsible for the synthesis of squalene, were not detected in Methylomonas genomes. Strain MW1 had no genes for the synthesis of C30 carotenoids. In all pigmented strains, genes involved in the subsequent steps of carotenoid biosynthesis were identified as a clustered unit, which included crtN, crtNb, and crtNc. Consequently, CrtN serves as the first amino oxidase enzyme in the pathway, catalyzing the first desaturation of the C30 backbone with the formation of 4,4’-diapolycopene. Diapolycopene oxygenase (CrtNb) catalyzes the oxidation of 4,4’-diapolycopene to yield 4,4’-diapolycopene-4,4’-dial. Then, the oxidative activity of 4,4’-diapolycopenoate synthase (CtrNc) leads to the formation of 4,4’-diapolycopene-4,4’-dioate. As expected, the non-pigmented strain MW1T had no genes for C30 carotenoid biosynthesis. To examine the pattern of carotenoid-encoding genes in other Methylomonas representatives, we included all available Methylomonas genomes from Genbank’s Refseq Database in the analysis. The metabolic route leading to the production of 4,4’-diapolycopene-4,4’-dioate, encoded by crtN, crtP, and crtNc genes, was identified in all analyzed genomes except that of strain MW1T. We also examined the genomes for the presence of genes related to C40 carotenoid pathways. The gene encoding the squalene/phytoene synthase family protein is the sole identified candidate for the synthesis of the C40 molecule. Genomes of strains MP1 and MO1 also harbor genes encoding phytoene desaturases, which are responsible for the conversion of colorless phytoene into lycopene.
4. Discussion
In this study, we identified carotenoids produced by five strains of Methylomonas species which displayed a spectrum of pigmentation ranging from white and yellow to orange and red. We also sequenced genomes and identified genome-encoded metabolic routes of carotenoid biosynthesis. The biosynthesis of C30 carotenoids in prokaryotes was originally elucidated on a Staphylococcus aureus model where it starts with the enzyme CrtM, 4,4’-diapophytoene synthase, which catalyzes the head-to-head condensation of two molecules of farnesyl diphosphate (FPP) into the C30 carotenoid 4,4’-diapophytoene [64,65]. However, CrtM is specific to Firmicutes, meaning that an alternative biochemical route should exist in other carotenoid-producing bacteria. Recently, squalene was proposed as a precursor for biosynthetic pathway of C30 carotenoids [66]. Squalene is an intermediate in the biosynthesis of sterols, hopanoids, and related pentacyclic triterpenes in bacteria [67]. The presence of the squalene/phytoene synthase-encoding gene in all Methylomonas genomes may suggest that they also utilize the C30 backbone of squalene for carotenoid biosynthesis. Previously, squalene synthases from yeast, humans, and bacteria subjected to directed evolution were shown to produce the C30 carotenoid backbone, dehydrosqualene [68]. Dehydrosqualene, or 4,4’-diapophytoene, is converted to 4,4’-diapolycopene through the enzymatic activity of 4,4’-diapophytoene desaturase. Notably, among publicly available Methylomonas genomes, strain MW1T stands out as the sole example lacking genes for carotenoid biosynthesis. This distinct feature suggests that strain MW1T may represent the only non-pigmented phenotype among currently known Methylomonas bacteria. The closest phylogenetic relative to strain MW1T, M. albis, was characterized as pink-pigmented methanotroph [69]. All other studied strains harbor genes encoding CrtN, CrtNb, and CrtNc necessary for the biosynthesis of 4,4’-diapolycopene, 4,4’-diapolycopene-4,4’-dial, and 4,4’-diapolycopene-4,4’-dioate [33]. CrtN is involved in the synthesis of the yellow carotenoid 4,4’-diaponeurosporene or the red carotenoid 4,4’-diapolycopene. Notably, the introduction of crtM and crtN genes from S. aureus into B. subtilis led to the accumulation of two C30 carotenoids, 4,4’-diapolycopene and 4,4’-diaponeurosporene, which resulted in the yellow pigmentation of the engineered strain [70]. From a genomic point of view, the accumulation of these compounds in different proportions may explain the yellow, orange, and red color of studied Methylomonas phenotypes.
According to the HPLC analysis of pigment extracts, carotenoids were produced by all strains except for strain MW1T. This strongly correlates with the absence of carotenoid pathway genes in the complete genome of strain MW1 and is consistent with observations that the culture of strain MW1 remained colorless regardless of cultivation conditions. C30 carotenoids were predominant in the pigment extracts of all other studied Methylomonas strains, which was expected from the genome analysis due to the presence of genome-encoded metabolic pathway leading to the production of 4,4’-diapolycopene-4,4’-dioic acid (C30). In a previous study, another Methylomonas bacterium, strain 16a (ATCC PTA-2402), produced esters of both 4,4-diapolycopene-4-oic acid and 4,4-diapolycopene-4,4-dioic acid [33]. The nonsaponified carotenoids from this strain migrated slightly differently compared to the 4,4’-diapolycopene-4,4’-dioic acid and 4,4’-diapolycopene-4-oic acid found in Methylobacterium rhodinum by using HPLC analysis. C35 carotenoids were exclusively present in pigment fractions of strains MV1 and MP1T. Previously, the co-expression of S. aureus CrtM and Erwinia geranylgeranyldiphosphate (GGDP) synthase in Escherichia coli led to the production of carotenoids with the asymmetrical C35 backbone as the result of the uptake of GGDP by CrtM when FDP was depleted [40]. Considering enzyme promiscuity, the C35 carotenoids in our study could have been synthesized from GGDP and FDP by SQS, followed by further desaturation by crtN. Remarkably, C40 carotenoids were detected in pigment extracts from strains MP1T, MY1, and MV1. Previously, another Methylomonas bacterium, strain ZR1, was shown to synthesize lycopene [25]. As far as we know, the genetic determinants of C40 carotenoid pathways have never been reported for Methylomonas bacteria. In this study, genome analysis did not definitively confirm whether Methylomonas strains can produce the C40 carotenoid phytoene. One of the possible explanations is that all studied genomes harbor the squalene/phytoene synthase family protein with the SQS_PSY domain (PF00494, Pfam database). Squalene synthase and phytoene synthase share a number of functional similarities [71,72]. It is tempting to speculate that squalene synthase from Methylomonas may also accept GGPP molecules as a substrate to produce phytoene. Notably, some Methylomonas strains, in addition to having 4,4’-diapophytoene desaturases, also possess phytoene desaturases, enabling them to synthesize the C40 carotenoid lycopene. The growing data on the production of various C35 and C40 carotenoid compounds by Methylomonas species offer avenues for investigating the diversity of carotenoid synthesis pathways in these bacteria.
The capability for fast growth on methane is a necessary requirement in certain biotechnological applications, such as single-cell protein production. All five studied strains were capable of robust growth on methane. However, the highest growth rates (0.25–0.33 h−1) were observed for strains MV1, MY1, and MP1T, which are comparable to those of thermotolerant Methylococcus bacteria [24]. Notably, the growth rates of some Methylomonas methanotrophs were reported to be even higher than those of Methylococcus strains [24,25]. Pigmented methanotrophs of the genus Methylomonas offer an alternative to commonly used non-pigmented Methylococcus capulatus for the production of single-cell protein enriched with carotenoids. A number of carotenoids are recognized for their ability to influence the antioxidative state and immune system, which leads to improved disease resistance, growth performance, survival, and egg quality in farmed fish without causing any cytotoxicity or side effects [73]. Many fish species accumulate carotenoids in their integuments and muscles, contributing quality criteria to meet consumer demands of aquaculture products [74]. Thus, the biomass of carotenoid-producing Methylomonas bacteria may represent a promising source of fish feed in modern aquaculture.
5. Conclusions
Overall, this study provided insights into the carotenoid biosynthesis in Methylomonas bacteria. The major carotenoids identified in the pigmented strains were 4,4’-diaplycopene-4,4’-dioic acid (C30) and 1,1’-dihydroxy-3,4-didehydrolycopene (C40). Genome analysis revealed the presence of C30 carotenoid biosynthesis pathways leading to 4,4’-diaplycopene-4,4’-dioic acid. Notably, strain MW1T lacked carotenoid biosynthesis genes and may represent the only colorless phenotype among Methylomonas bacteria. Several strains also possess phytoene desaturases, which explains the presence of lycopene compounds in the pigment extracts. Thus, Methylomonas bacteria possess the ability to naturally synthesize both C30 and C40 carotenoids. Novel isolates display robust growth on methane and represent promising candidates for certain industrial applications, such as single-cell protein production enriched with carotenoids. Further research into the metabolic pathways and genetic manipulation of Methylomonas bacteria may unlock their full potential in biotechnology.
Author Contributions
Conceptualization, S.N.D. and I.Y.O.; Data curation, I.Y.O., E.N.T., R.Z.S., A.A.A. and A.A.I.; Formal analysis, I.Y.O.; Funding acquisition, N.V.P. and S.N.D.; Investigation, I.Y.O., R.Z.S. and A.A.A.; Methodology, I.Y.O. and A.A.A.; Software, I.Y.O.; Supervision, S.N.D.; Visualization, I.Y.O., E.N.T. and A.A.A.; Writing—original draft, I.Y.O. and S.N.D.; Writing—review and editing, S.N.D. and N.V.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant for the development of genomic editing technologies for innovation in industrial biotechnology (grant 075-15-2021-1071) from the Ministry of Science and Higher Education of the Russian Federation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Polyakov, N.E.; Focsan, A.L.; Gao, Y.; Kispert, L.D. The Endless World of Carotenoids—Structural, Chemical and Biological Aspects of Some Rare Carotenoids. Int. J. Mol. Sci. 2023, 24, 9885. [Google Scholar] [CrossRef]
- Yabuzaki, J. Carotenoids Database: Structures, Chemical Fingerprints and Distribution among Organisms. Database 2017, 2017, bax004. [Google Scholar] [CrossRef]
- Zia-Ul-Haq, M.; Dewanjee, S.; Riaz, M. Carotenoids: Structure and Function in the Human Body; Springer International Publishing: New York, NY, USA, 2021; ISBN 3-030-46458-8. [Google Scholar]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A Global Perspective on Carotenoids: Metabolism, Biotechnology, and Benefits for Nutrition and Health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cámara, S.; Ibañez, A.; Rubio, S.; Barreiro, C.; Barredo, J.-L. Main Carotenoids Produced by Microorganisms. Encyclopedia 2021, 1, 1223–1245. [Google Scholar] [CrossRef]
- Siziya, I.N.; Hwang, C.Y.; Seo, M.J. Antioxidant Potential and Capacity of Microorganism-Sourced C30 Carotenoids—A Review. Antioxidants 2022, 11, 1963. [Google Scholar] [CrossRef]
- López, G.D.; Álvarez-Rivera, G.; Carazzone, C.; Ibáñez, E.; Leidy, C.; Cifuentes, A. Bacterial Carotenoids: Extraction, Characterization, and Applications. Crit. Rev. Anal. Chem. 2023, 53, 1239–1262. [Google Scholar] [CrossRef]
- Goldstein, J.L.; Brown, M.S. Regulation of the Mevalonate Pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Non-Mevalonate Isoprenoid Biosynthesis: Enzymes, Genes and Inhibitors. Biochem. Soc. Trans. 2000, 28, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Sangari, F.J.; Pérez-Gil, J.; Carretero-Paulet, L.; García-Lobo, J.M.; Rodríguez-Concepción, M. A New Family of Enzymes Catalyzing the First Committed Step of the Methylerythritol 4-Phosphate (MEP) Pathway for Isoprenoid Biosynthesis in Bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 14081–14086. [Google Scholar] [CrossRef]
- Paniagua-Michel, J.; Olmos-Soto, J.; Ruiz, M.A. Pathways of Carotenoid Biosynthesis in Bacteria and Microalgae. Methods Mol. Biol. 2012, 892, 1–12. [Google Scholar] [CrossRef]
- Lieberman, R.L.; Rosenzweig, A.C. Crystal Structure of a Membrane-Bound Metalloenzyme That Catalyses the Biological Oxidation of Methane. Nature 2005, 434, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hakemian, A.S.; Rosenzweig, A.C. The Biochemistry of Methane Oxidation. Annu. Rev. Biochem. 2007, 76, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.C. Biochemistry: Breaking Methane. Nature 2015, 518, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Knief, C. Diversity and Phylogeny of Described Aerobic Methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M., Xing, X., Eds.; Springer: Cham, Switzerland, 2018; pp. 17–42. ISBN 9783319748665. [Google Scholar]
- Bowman, J.P. Methylomonas. Bergey’s Man. Syst. Archaea Bact. 2016, 8, 748. [Google Scholar] [CrossRef]
- Suleimanov, R.Z.; 3Tikhonova, E.N.; Oshkin, I.Y.; Danilova, O.V.; Dedysh, S.N. Methylomonas montana sp. nov., the First Nonpigmented Methanotroph of the Genus Methylomonas, Isolated from Mountain River Sediments. Microbiology 2023, 92, 766–774. [Google Scholar]
- Yazdian, F.; Hajizadeh, S.; Shojaosadati, S.A.; Jahanshahi, M.; Nosrati, M. Production of Single Cell Protein from Natural Gas: Parameter Optimization and RNA Evaluation. Iran. J. Biotechnol. 2005, 3, 235–242. [Google Scholar]
- Skrede, A.; Berge, G.M.; Storebakken, T.; Herstad, O.; Aarstad, K.G.; Sundstøl, F. Digestibility of Bacterial Protein Grown on Natural Gas in Mink, Pigs, Chicken and Atlantic Salmon. Anim. Feed Sci. Technol. 1998, 76, 103–116. [Google Scholar] [CrossRef]
- Bjorck, C.E.; Dobson, P.D.; Pandhal, J. Biotechnological Conversion of Methane to Methanol: Evaluation of Progress and Potential. AIMS Bioeng. 2018, 5, 1–38. [Google Scholar] [CrossRef]
- Safaeian, P.; Yazdian, F.; Khosravi-Darani, K.; Rashedi, H.; Lackner, M. P3HB from CH4 Using Methanotrophs: Aspects of Bioreactor, Fermentation Process and Modelling for Cost-Effective Biopolymer Production. Front. Bioeng. Biotechnol. 2023, 11, 1137749. [Google Scholar] [CrossRef]
- Fei, Q.; Puri, A.W.; Smith, H.; Dowe, N.; Pienkos, P.T. Enhanced Biological Fixation of Methane for Microbial Lipid Production by Recombinant Methylomicrobium buryatense. Biotechnol. Biofuels 2018, 11, 129. [Google Scholar] [CrossRef]
- Henard, C.A.; Smith, H.; Dowe, N.; Kalyuzhnaya, M.G.; Pienkos, P.T.; Guarnieri, M.T. Bioconversion of Methane to Lactate by an Obligate Methanotrophic Bacterium. Sci. Rep. 2016, 6, 21585. [Google Scholar] [CrossRef]
- Oshkin, I.Y.; Danilova, O.V.; But, S.Y.; Miroshnikov, K.K.; Suleimanov, R.Z.; Belova, S.E.; Tikhonova, E.N.; Kuznetsov, N.N.; Khmelenina, V.N.; Pimenov, N.V.; et al. Expanding Characterized Diversity and the Pool of Complete Genome Sequences of Methylococcus Species, the Bacteria of High Environmental and Biotechnological Relevance. Front. Microbiol. 2021, 12, 756830. [Google Scholar] [CrossRef]
- Guo, W.; Li, D.; He, R.; Wu, M.; Chen, W.; Gao, F.; Zhang, Z.; Yao, Y.; Yu, L.; Chen, S. Synthesizing Value-Added Products from Methane by a New Methylomonas. J. Appl. Microbiol. 2017, 123, 1214–1227. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Kim, D.; Lee, E.Y. A Comparative Transcriptome Analysis of the Novel Obligate Methanotroph methylomonas sp. DH-1 Reveals Key Differences in Transcriptional Responses in C1 and Secondary Metabolite Pathways during Growth on Methane and Methanol. BMC Genom. 2019, 20, 130. [Google Scholar] [CrossRef]
- Bothe, H.; Møller Jensen, K.; Mergel, A.; Larsen, J.; Jørgensen, C.; Bothe, H.; Jørgensen, L. Heterotrophic Bacteria Growing in Association with Methylococcus Capsulatus (Bath) in a Single Cell Protein Production Process. Appl. Microbiol. Biotechnol. 2002, 59, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Koffas, M.; Odom, J.; Schenzle, A. High Growth Methanotrophic Bacterial Strain. EP1320579A2, 25 June 2003. [Google Scholar]
- Gayazov, R.R.; Shishkina, V.N.; Mshenskii, Y.N.; Trotsenko, Y.A.; Ivanov, M.V. Effect of Temperature on Growth and Metabolism of Methylomonas methanica. Dokl. Acad. Sci. 1985, 284, 746–748. [Google Scholar]
- Tikhonova, E.N.; Suleimanov, R.Z.; Miroshnikov, K.K.; Oshkin, I.Y.; Belova, S.E.; Danilova, O.V.; Ashikhmin, A.A.; Konopkin, A.A.; But, S.Y.; Khmelenina, V.N.; et al. Methylomonas rapida sp. nov., a Novel Species of Fast-Growing, Carotenoid-Producing Obligate Methanotrophs with High Biotechnological Potential. Syst. Appl. Microbiol. 2023, 46, 126398. [Google Scholar] [CrossRef]
- Ye, R.W.; Yao, H.; Stead, K.; Wang, T.; Tao, L.; Cheng, Q.; Sharpe, P.L.; Suh, W.; Nagel, E.; Arcilla, D.; et al. Construction of the Astaxanthin Biosynthetic Pathway in a Methanotrophic Bacterium Methylomonas sp. Strain 16a. J. Ind. Microbiol. Biotechnol. 2007, 289–299. [Google Scholar] [CrossRef]
- Sharpe, P.L.; Dicosimo, D.; Bosak, M.D.; Knoke, K.; Tao, L.; Cheng, Q.; Ye, R.W. Use of Transposon Promoter-Probe Vectors in the Metabolic Engineering of the Obligate Methanotroph Methylomonas sp. Strain 16a for Enhanced C40 Carotenoid Synthesis. Appl. Environ. Microbiol. 2007, 73, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Schenzle, A.; Odom, J.M.; Cheng, Q. Novel Carotenoid Oxidase Involved in Biosynthesis of 4,4′- Diapolycopene Dialdehyde. Appl. Environ. Microbiol. 2005, 71, 3294–3301. [Google Scholar] [CrossRef]
- Kleinig, H.; Schmitt, R. On the Biosynthesis of C30 Carotenoic Acid Glucosyl Esters in Pseudomonas rhodos. Analysis of Car-Mutants. Z. Naturforsch.-Sect. C J. Biosci. 1982, 37, 758–760. [Google Scholar] [CrossRef]
- Takaichi, S.; Inoue, K.; Akaike, M.; Kobayashi, M.; Oh-oka, H.; Madigan, M.T. The Major Carotenoid in All Known Species of Heliobacteria Is the C30 Carotenoid 4,4′-Diaponeurosporene, Not Neurosporene. Arch. Microbiol. 1997, 168, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Takaichi, S.; Oh-oka, H.; Maoka, T.; Jung, D.O.; Madigan, M.T. Novel Carotenoid Glucoside Esters from Alkaliphilic Heliobacteria. Arch. Microbiol. 2003, 179, 95–100. [Google Scholar] [CrossRef]
- Marshall, J.H.; Wilmoth, G.J. Proposed Pathway of Triterpenoid Carotenoid Biosynthesis in Staphylococcus aureus: Evidence from a Study of Mutants. J. Bacteriol. 1981, 147, 914–919. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.H.; Wilmoth, G.J. Pigments of Staphylococcus aureus, a Series of Triterpenoid Carotenoids. J. Bacteriol. 1981, 147, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Ashikhmin, A.; Makhneva, Z.; Bolshakov, M.; Moskalenko, A. Incorporation of Spheroidene and Spheroidenone into Light-Harvesting Complexes from Purple Sulfur Bacteria. J. Photochem. Photobiol. B. 2017, 170, 99–107. [Google Scholar] [CrossRef]
- Umeno, D.; Tobias, A.V.; Arnold, F.H. Evolution of the C30 Carotenoid Synthase CrtM for Function in a C40 Pathway. J. Bacteriol. 2002, 184, 6690–6699. [Google Scholar] [CrossRef]
- Steiger, S.; Takaichi, S.; Sandmann, G. Heterologous Production of Two Unusual Acyclic Carotenoids, 1,1-Dihydroxy-3,4-Didehydrolycopene and 1-Hydroxy-3,4,3,4-Tetradehydrolycopene by Combination of the CrtC and CrtD Genes from Rhodobacter and Rubrivivax. J. Biotechnol. 2002, 97, 51–58. [Google Scholar] [CrossRef]
- Enzell, C.R.; Francis, G.W.; Liaaen-Jensen, S. Mass Spectrometric Studies of Carotenoids. 2. A Survey of Fragmentation Reactions. Acta Chem. Scand. 1969, 23, 727–750. [Google Scholar] [CrossRef][Green Version]
- Britton, G. UV/Visible Spectroscopy. In Carotenoids, V. 1B., Spectroscopy; Britton, G., Liaaen-Jensen, S.H.P., Eds.; Birkhäuser Verlag: Basel, Switzerland, 1995. [Google Scholar]
- Wilson, K. Preparation of Genomic DNA from Bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M. Data, Information, Knowledge and Principle: Back to Metabolism in KEGG. Nucleic Acids Res. 2014, 42, 199–205. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus agalactiae: Implications for the Microbial “Pan-Genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic Insights That Advance the Species Definition for Prokaryotes Konstantinos. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Thulasiram, H.V.; Poulter, C.D. Farnesyl Diphosphate Synthase: The Art of Compromise between Substrate Selectivity and Stereoselectivity. J. Am. Chem. Soc. 2006, 128, 15819–15823. [Google Scholar] [CrossRef]
- Wieland, B.; Feil, C.; Gloria-Maercker, E.; Thumm, G.; Lechner, M.; Bravo, J.M.; Poralla, K.; Gotz, F. Genetic and Biochemical Analyses of the Biosynthesis of the Yellow Carotenoid 4,4′-Diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 1994, 176, 7719–7726. [Google Scholar] [CrossRef]
- Pelz, A.; Wieland, K.P.; Putzbach, K.; Hentschel, P.; Albert, K.; Götz, F. Structure and Biosynthesis of Staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 2005, 280, 32493–32498. [Google Scholar] [CrossRef]
- Santana-Molina, C.; Henriques, V.; Hornero-Méndez, D.; Devos, D.P.; Rivas-Marin, E. The Squalene Route to C30 Carotenoid Biosynthesis and the Origins of Carotenoid Biosynthetic Pathways. Proc. Natl. Acad. Sci. USA 2022, 119, e2210081119. [Google Scholar] [CrossRef]
- Pan, J.J.; Solbiati, J.O.; Ramamoorthy, G.; Hillerich, B.S.; Seidel, R.D.; Cronan, J.E.; Almo, S.C.; Poulter, C.D. Biosynthesis of Squalene from Farnesyl Diphosphate in Bacteria: Three Steps Catalyzed by Three Enzymes. ACS Cent. Sci. 2015, 1, 77–82. [Google Scholar] [CrossRef]
- Furubayashi, M.; Umeno, D. Directed Evolution of Carotenoid Synthases for the Production of Unnatural Carotenoids; Humana Press: Totowa, NJ, USA, 2012; Volume 892, ISBN 9781617798788. [Google Scholar]
- Bussmann, I.; Horn, F.; Hoppert, M.; Klings, K.W.; Saborowski, A.; Warnstedt, J.; Liebner, S. Methylomonas albis sp. nov. and Methylomonas fluvii sp. nov.: Two Cold-Adapted Methanotrophs from the River Elbe and Emended Description of the Species Methylovulum psychrotolerans. Syst. Appl. Microbiol. 2021, 44, 126248. [Google Scholar] [CrossRef]
- Maeda, I. Genetic Modification in Bacillus subtilis for Production of C30 Carotenoids. Methods Mol. Biol. 2012, 892, 197–205. [Google Scholar] [CrossRef]
- Summers, C.; Karst, F.; Charles, A.D. Cloning, Expression and Characterisation of the CDNA Encoding Human Hepatic Squalene Synthase, and Its Relationship to Phytoene Synthase. Gene 1993, 136, 185–192. [Google Scholar] [CrossRef]
- Robinson, G.W.; Tsay, Y.H.; Kienzle, B.K.; Smith-Monroy, C.A.; Bishop, R.W. Conservation between Human and Fungal Squalene Synthetases: Similarities in Structure, Function, and Regulation. Mol. Cell. Biol. 1993, 13, 2706–2717. [Google Scholar] [CrossRef]
- Nakano, T.; Wiegertjes, G. Properties of Carotenoids in Fish Fitness: A Review. Mar. Drugs 2020, 18, 568. [Google Scholar] [CrossRef]
- de Carvalho, C.C.; Caramujo, M.J. Carotenoids in Aquatic Ecosystems and Aquaculture: A Colorful Business with Implications for Human Health. Front. Mar. Sci. 2017, 4, 93. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).