Abstract
Bacteria of the genus Cutibacterium are Gram-positive commensals and opportunistic pathogens that represent a major challenge in the diagnosis and treatment of implant-associated infections (IAIs). This study provides insight into the distribution of different sequence types (STs) of C. acnes, and the presence of virulence factors (VFs) in 64 Cutibacterium spp. isolates from suspected or confirmed IAIs obtained during routine microbiological diagnostics. Fifty-three C. acnes, six C. avidum, four C. granulosum, and one C. namnetense isolate, collected from different anatomical sites, were included in our study. Using whole-genome sequencing and a single-locus sequencing typing scheme, we successfully characterized all C. acnes strains and revealed the substantial diversity of STs, with the discovery of six previously unidentified STs. Phylotype IA1, previously associated with both healthy skin microbiome and infections, was the most prevalent, with ST A1 being the most common. Some minor differences in STs’ distribution were observed in correlation with anatomical location and association with infection. A genomic analysis of 40 investigated VFs among 64 selected strains showed no significant differences between different STs, anatomical sites, or infection-related and infection undetermined/unlikely groups of strains. Most differences in VF distribution were found between strains of different Cutibacterium spp., subspecies, and phylotypes, with CAMP factors, biofilm-related VFs, lipases, and heat shock proteins identified in all analyzed Cutibacterium spp.
1. Introduction
Representatives of the genus Cutibacterium are Gram-positive, commensal, lipophilic, facultative anaerobic, and slow-growing rod-shaped bacteria [1]. As members of normal skin microbiota, they are often considered laboratory contaminants [2]. However, they are increasingly recognized as important opportunistic pathogens that can cause serious infections such as implant-associated infections (IAIs) [3,4,5,6], especially in immunocompromised individuals or when introduced into sterile areas during surgical procedures [7].
Bacteria of the genus Cutibacterium are intrinsically resistant to metronidazole but generally susceptible to clindamycin, macrolides, β-lactams, fluoroquinolones, and tetracycline. However, resistance to clindamycin, tetracycline, and macrolides is emerging [4,7]. Together with biofilm development on implant surfaces [8], the extended incubation period required to detect Cutibacterium spp., the frequent risk of contamination [9], and the absence of local or systemic signs of the infection [7] contribute to the challenging diagnosis and treatment of IAIs caused by Cutibacterium spp.
In addition to the best-known species, Cutibacterium acnes, there are several other Cutibacterium species (e.g., C. avidum, C. granulosum, C. namnetense, and C. humerusii), whose detection rates in clinical samples have increased significantly over the last decade due to improved microbiological diagnostics such as the sonication of removed medical devices, incubation period up to 14 days, and use of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) identification [1,10,11,12].
Members of the genus Cutibacterium are generally considered to be opportunistic pathogens with low virulence, causing subacute or chronic infections. However, in 2008, Million et al. addressed the sepsis potential of Cutibacterium spp., particularly C. avidum [13], which differs from the other members of the genus in its specific tropism for a moist environment on the skin surface. On the contrary, C. acnes and C. granulosum require a lipid-rich skin surface [2,12,14,15,16] and are therefore less commonly associated with acne patients [16]. There is limited information on IAIs caused by C. granulosum and C. namnetense, with the latter being more similar to C. acnes than to other Cutibacterium species [4,6,14,17].
The members of Cutibacterium spp. (formerly known as Propionibacterium spp.) have been reclassified several times: in 1909, they were first recognized as members of the genus Bacillus, later as Corynebacterium spp., and only in 1946 as the genus Propionibacterium. The most recent reclassification occurred in 2016, when all cutaneous Propionibacterium spp. (C. acnes, C. avidum, C. granulosum, C. namnetense, and C. humerusii) were placed in a new genus called Cutibacterium spp. [4,14,16]. Currently, three subspecies and six phylotypes of C. acnes are known: C. acnes subsp. acnes (phylotypes IA1, IA2, IB, IC), C. acnes subsp. defendens (phylotype II), and C. acnes subsp. elongatum (phylotype III) [1,18,19]. Based on an analysis of the cell wall and the non-ribosomal housekeeping genes recA and tly, C. acnes strains were initially classified into two main types, I and II, followed by the additional type III, corresponding to strains with filamentous appendages [20,21,22]. To further increase the discriminatory power, multi-locus sequence typing (MLST) schemes have been developed, further classifying phylotypes into clonal complexes (CC) based on four (MLST4) [23], nine (MLST9—Aarhus scheme) [24], or eight housekeeping genes (MLST8—Belfast scheme) [25]. The development of a new SLST (single-locus sequencing type) scheme in 2014 divided C. acnes strains into clades IA-1, IA-2, IB-1, IB-2, IB-3, IC, II, III, which are further classified into sequence types (STs), of which 188 are currently known [26]. In 2023, a novel bi-locus sequence typing scheme CUTIS-SEQ was introduced, combining SLST and camp2 loci [19].
The first complete genome sequence of C. acnes published in 2004 revealed a single circular chromosome with a size of 2,560,265 base pairs, corresponding to 2333 potential genes [27]. The differences in the genome size between different Cutibacterium spp. are not substantial, except for C. granulosum, which has a slightly smaller genome size (2.18 Mbp) than others.
Several previous studies have indicated a possible association between different phylotypes and specific clinical manifestations or anatomical sites. Phylotypes IB and II were commonly associated with IAI, soft tissue infections, and bacteremia, and were considered commensals of healthy skin [7,25,28]. Phylotype IA was mostly associated with acne, but the latest reports also report its presence in prosthetic joint infections [1]. Phylotype III is more commonly found on the trunk [1]. In spinal instrument infections, phylotype IA, especially CC18 (MLST9) and CC28 (MLST9) or ST A and F (SLST), were the predominant types [28,29]. Thus, these results are contradictory and may indicate that there is no predominant disease-specific phylotype [7]. No classification is currently available for other species of the genus Cutibacterium, likely due to the fact that the proportion of infections caused by C. granulosum (2.4–4.8%) and C. avidum (4.2–10.7%) among all Cutibacterium infections is low, and C. avidum is present in a minor proportion in skin microbiota (0.4%) compared to C. acnes [30,31,32]. Infections with C. namnetense and C. humerusii have rarely been reported. Several recent studies identified putative virulence factors (VFs) and virulence-associated genes important for environmental adaptation, adherence to target cells, enzymatic degradation of host tissues, and especially bacterial biofilm [1]. Biofilm production was observed in vivo and in vitro in several studies [7,8]. Kuehnast et al. suggested a possible correlation between the C. acnes phylotype and biofilm production rather than with the anatomical site of infection [33]. However, data on putative VFs in other Cutibacterium spp. are limited [1]. Therefore, the aim of this study was to perform an in silico comparative genomic analysis of 64 Cutibacterium spp. isolates from confirmed or suspected IAIs showing different clinical manifestations and isolated from different anatomical sites, with an emphasis on the presence of VFs and virulence-associated genes among the different Cutibacterium spp.
2. Materials and Methods
2.1. Sample Selection
A genomic analysis of Cutibacterium spp. strains isolated from patients with confirmed or suspected IAIs and surgical site infection (SSI) was performed at the Institute of Microbiology and Immunology, Faculty of Medicine, Ljubljana, Slovenia. We retrospectively reviewed our archive collection which included strains from 2012 to 2022, and in total, 64 Cutibacterium spp. strains associated with infections at different anatomical sites were selected (53 C. acnes, 6 C. avidum, 4 C. granulosum from IAIs, and 1 C. namnetense strain isolated from SSI).
Among 64 strains, 51 strains were obtained from the sonicate fluid (SF) of removed implants and 13 strains from peri-implant tissue (PT) samples from different anatomical locations: hip (19 samples; 16 SF, 3 PT), knee (14 samples; 11 SF, 3 PT), shoulder (4 samples; 4 SF), spine (13 samples; 11 SF, 2 PT), ankle (2 samples; 1 SF, 1 PT), 8 samples from the cardiovascular system (1 prosthetic valve; 2 cardiovascular implantable electronic devices—CIED, 1 ventricular assist devices—VAD; 1 stent graft; 1 femoral popliteal bypass; 2 coronary stents—CST), 1 breast implant, 2 central nervous system devices (CNSD), and one SSI strain.
The strains were defined as infection-related or infection-undetermined/unlikely. Infection-related strains were defined as such if they met the following microbiological criteria for IAIs: at least two positive PT cultures or positive SF culture and one PT culture with the same microorganism identified, or if more than ≥50 CFU/mL was detected in the SF by conventional culture methods [34]. When detailed clinical data were not available, strains were characterized as infection undetermined/unlikely, since the criteria mentioned above may miss cases with clinical signs suggestive of infection but inconsistent with the microbiological results. The study protocol was approved by the Slovenian National Medical Ethics Committee of Slovenia (Ministry of Health, Republic of Slovenia) under identification number 0120-15/2022/6 (date of approval: 31 May 2022).
2.2. DNA Isolation
Isolates were collected from frozen stocks stored at −80 °C and incubated anaerobically on Schaedler agar plates at 35 °C for 72 h. Identification was confirmed using MALDI-TOF MS. Total DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen Ltd., West Sussex, United Kingdom) according to the manufacturer’s protocol for Gram-positive bacteria. The extracted DNA was stored at −80 °C until further use. A Qubit 3.0 fluorometer in combination with a Qubit 1× dsDNA HS assay kit (Thermo Fisher Scientific, Waltham, MA, USA) were used to determine the amount of DNA. In addition, DNA purity was assessed based on the absorbance ratio at A260/280 and A260/230 ratios using a NanoDrop 2000/2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA). Only DNA samples with a concentration higher than 1 ng/µL and an A260/280 ratio between 1.8 and 2.0 were selected for whole-genome sequencing (WGS) and included in further analysis.
2.3. Whole-Genome Sequencing
WGS of 64 Cutibacterium spp. strains was performed at the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana with Illumina NextSeq 550 (Illumina, San Diego, CA, USA) using a paired-end (2 × 150 bp) sequencing protocol. Strains were sequenced to a minimum coverage of 150×. A Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) was used for DNA library preparation. Raw reads were trimmed for adapter sequences and low-quality reads with Fastp v0.23.2 [35]. The quality of raw and trimmed reads was assessed with FastQC v0.11.9 [35]. Trimmed reads were assembled into contigs using SPAdes v3.15.3 [36] and default k-mer values and “--careful” parameters. Quast v5.2.0 [37] was used for a quality assessment of the assemblies. The minimum threshold for assembly quality was set at a value of 20,000 bp for N50 and the total number of contigs lower than 500. Prokka v1.14.6 [38] was used for annotation of the bacterial genomes, using the genome assembly of C. acnes strain HL096PA1 [39] (C. acnes) as the reference genome. The assembled genomes were further analyzed using Ridom SeqSphere v9.0.10 [40] and zDB software v1.1.1 [41].
2.4. SLST and Phylogenetic Analysis
For the C. acnes strains, the STs were determined in silico using the SLST database from Aarhus University (http://www.medbac.dk/slst_server_script.html, accessed on 27 July 2023).
A pan-genome analysis based on the accessory genome of 64 Cutibacterium spp. strains was performed using Roary v3.13.0 [42] and used for further phylogenetic analysis. A phylogenetic tree was constructed using FastTree v2.1.10 according to the varieties in groups of genes uniquely present/absent in accessory genome and visualized using iTOL v6.3 (https://itol.embl.de/, accessed on 24 July 2023) [43].
2.5. Analysis of Virulence-Associated Genes and Antimicrobial Resistance Genes
ABRicate v1.0.1 (https://github.com/tseemann/abricate, accessed on 10 May 2023) was used for the analysis of virulence factors by creating a custom database (Table S1) consisting of 40 VFs collected from the literature (Table 1). Additional species–specific sequences were analyzed in silico using blastn_ffa, tblastn, and blastp functions within the zDB tool. KEGG (Kyoto Encyclopedia of Genes and Genomes) Ortholog annotations, COG (Clusters of Orthologous Genes) annotations, or Pfam domains of VFs and their homologues were identified with the zDB software. The antimicrobial resistance genes present were identified with the AMRFinderPlus v3.11.2 database [44].

Table 1.
The list of putative VFs and their distribution among phylotypes of C. acnes and other Cutibacterium spp. Where VF-associated genes were analyzed, VFs are written in italics.
3. Results
3.1. Sequence Typing and Association with Clinical Relevance
The sequence typing results are summarized in Table 2 and in Figure 1 and Figure 2. All C. acnes strains were successfully assigned to specific STs using the SLST scheme. In addition to the already known STs, six new STs were identified, namely A60, A61, C9, H18, K36, and K37. In our cohort, the most common phylotype among C. acnes strains was IA1, which was identified in 62% (33/53) of cases, followed by phylotype II in 19% (10/53), and phylotype IB in 15% (8/53). Phylotypes IA2 and III were both represented by a single strain.

Table 2.
WGS results, sequence typing, and clinical relevance data.

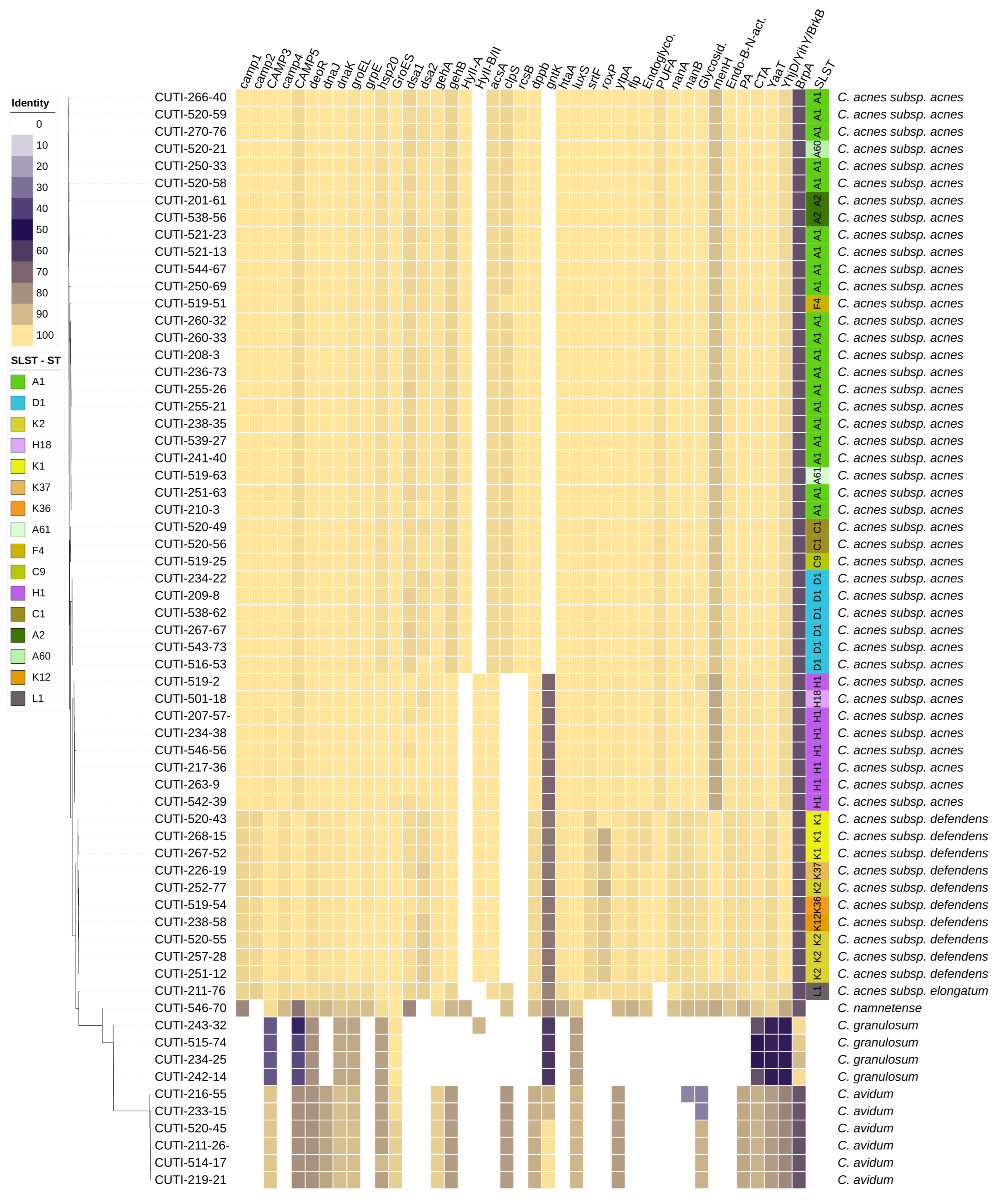
Figure 1.
Phylogenetic tree of the accessory genome of 64 strains of Cutibacterium spp. The distribution of VFs is represented as a heat map showing the percentage of homology of putative, mostly C. acnes-specific, VFs. The tree is based on groups of genes uniquely present/absent in the accessory genome.
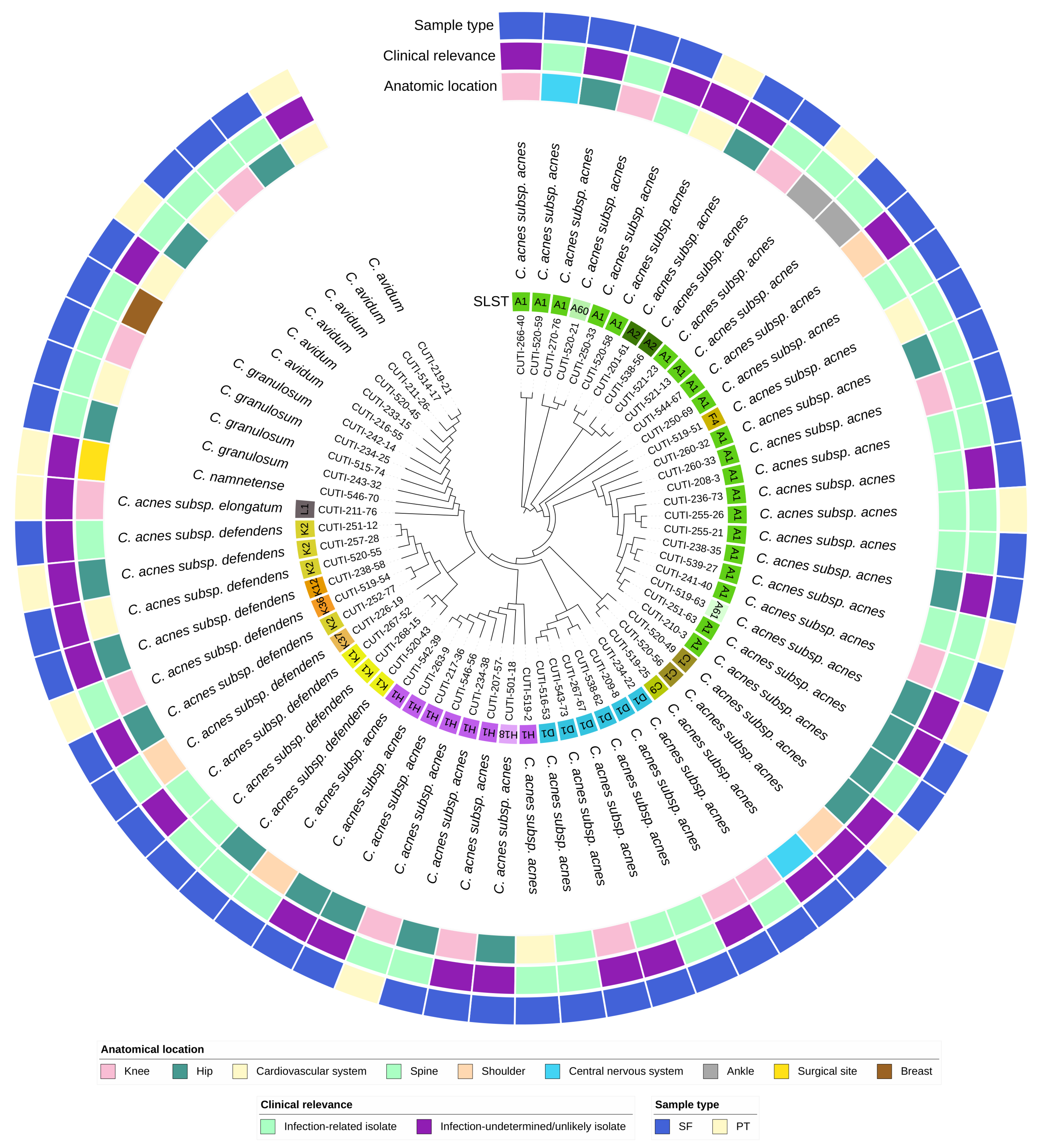
Figure 2.
Circular phylogenetic tree of the accessory genome of 64 isolates of Cutibacterium spp. with ignored branch lengths. The tree is based on groups of genes uniquely present/absent in the accessory genome. The tree is annotated with the following strain metadata: SLST ST (for C. acnes strains), classification of strains as infection-related or infection-undetermined/unlikely, sample type (SF—sonication fluid; PT—peri-implant tissue), and anatomical location.
We observed no significant difference in the distribution of phylotypes among the infection-related and infection-undetermined/unlikely strains (Table 2). Interestingly, C. acnes ST A2, A61, C1, C9, and K2 were found exclusively in the infection-undetermined/unlikely group of strains.
3.2. Phylogenetic Analysis
The WGS results are summarized in Table S2 and run accession numbers for each strain in Table S4. The initial genome screening of the 53 C. acnes strains revealed no significant overall differences in genome size (2.49 ± 0.09 Mbp; mean ± SD) or GC content (60%). Phylotype III (2.57 Mbp) had a slightly larger genome than the phylotypes IB (2.55 Mbp ± 0.02) and IA1 (2.52 Mbp ± 0.03). Among the other Cutibacterium spp., C. avidum (2.5 Mbp ± 0.02) and C. namnetense (2.41 Mbp) had a similar genome size, whereas C. granulosum (2.17 Mbp ± 0.05) had the smallest genome size. C. granulosum had the highest GC content with 64%, followed by C. avidum with 63% and C. namnetense with 61%. The number of putative protein coding sequences in the genomes varied from 1762 (C. granulosum) to 2464 (C. acnes subsp. elongatum), with an average of 2335.36 ± 33.81 in C. acnes, 2266.17 ± 26.49 in C. avidum, 2218 in C. namnetense, and 1805.5 ± 30.12 in C. granulosum. To assess the genomic relatedness of strains belonging to different Cutibacterium species, the presence and absence of genes according to a pan-genome analysis were determined. A total of 11,770 coding sequences were identified.
3.3. Antimicrobial Resistance Genes
Cutibacterium spp. isolates are intrinsically resistant to metronidazole. Phenotypically, we detected resistance to clindamycin using the gradient diffusion method in two isolates (CUTI-242-14 and CUTI-260-33) during routine microbiological diagnostics. Both strains contained the erm(X) gene associated with clindamycin resistance. In addition, we found erm(X) in one strain (CUTI-260-32) which was phenotypically susceptible to clindamycin. No other antimicrobial resistance genes were identified, which was in perfect agreement with the phenotypical testing.
3.4. Analysis of Putative Virulence Factors
We investigated the presence of 40 putative VFs in 64 Cutibacterium spp. strains with the aim of determining their distribution in different Cutibacterium species, subspecies, phylotypes, STs, anatomical sites, and association with infection. Most of the selected VFs (Table 1 and Table S1) and their associated genes that were analyzed in this study were previously identified and described in C. acnes strains. This could potentially lead to the misidentification of homologs that were identified based on whether they belonged to the same KEGG ortholog annotation, COG annotation, or Pfam domain (Table S2). We characterized homologs into four subgroups according to the percentage of identity: homologs with very high identity (>95%), homologs with high identity (>80% and <95%), homologs with low identity (>60% and <80%), and homologs with very low identity (<60%) (Table S2). The VFs’ distribution in different Cutibacterium spp. strains is summarized in Figure 1. C. acnes and C. namnetense had the highest number of identified VFs and most similar distribution of VFs, whereas C. granulosum had the lowest number of identified putative VFs and the most diverse distribution compared with other Cutibacterium spp. The present results did not show the specific distribution of VFs in association with particular anatomic sites or clinical manifestations. The main differences in the presence of VFs were observed primarily at the level of the Cutibacterium species, subspecies, and phylotypes.
4. Discussion
We successfully characterized all selected C. acnes strains and identified six new STs in our study cohort: A60, A61, C9, H18, K36, and K37. Consistent with previous studies, phylotype IA1 was found to be the most prevalent, with ST A1 being the most common [1,49,50].
Phylotype IA1 has been considered a common commensal in a healthy skin microbiota and associated with severe acne [24,29,51,52,53], but is becoming increasingly recognized as the predominant phylotype in IAI infections [7,23,25,29,54]. In the past, phylotype IB was the one most commonly associated with IAI. Among the C. acnes strains, the group of strains characterized as phylotype IB, had with 63% (5/8) the highest percentage of infection-related strains in our cohort. Certain STs were found exclusively in the infection-indeterminate/unlikely group of strains. Although some minor differences in specific STs were seen in association with the anatomic location and association with infection among the different STs, any correlation between STs, anatomic location, or association with infection was inconclusive due to limited clinical data and sample sizes.
No significant differences in the presence of VFs and their associated genes were observed between the different STs of C. acnes, different anatomic sites, or between the infection-related and infection indeterminate/unlikely group of strains. However, differences in the presence of VFs were detected at the level of bacterial species and, in the case of C. acnes, also at the level of subspecies and phylotypes. The most prominent differences in the gene presence were observed for hyaluronate lyase (HYL) genes, which encode enzymes responsible for the degradation of hyaluronic acid in the host extracellular matrix, thereby facilitating bacterial invasion and tissue colonization, particularly HYL-IB/II and HYL-IA. HYL-IB/II was only detected in phylotypes IB and II, whereas HYL-IA was found exclusively in phylotype IA. This result is in contrast to previous reports showing the presence of HYL-IA in phylotypes IA and IB and HYL-IB/II in phylotypes IA, IB, and II [1]. Another interesting observation is that HYL-IA was detected in one C. namnetense strain and HYL-IB/II in one C. granulosum strain (CUTI-243-32) isolated from a hip prosthesis infection.
The clpS gene, which encodes the Clp protease adaptor protein essential for the control of intracellular protein degradation, has been observed in a previous study mainly in IA phylotypes [46], which is in agreement with the present study where it was identified in phylotypes IA, phylotype III, and in C. avidum strains. Remarkably, we did not find it in phylotypes II, IB, C. granulosum, and C. namnetense. The clpS gene was also detected in the non-coding region in other C. acnes strains, characterized by the insertion of nucleotide T at position 129. We hypothesize that this insertion could potentially cause a frameshift mutation that disrupts the reading frame and results in the absence of the protein product.
Heat shock proteins (HSPs) play a critical role in various prokaryotes, performing functions in stress response, protein folding, intracellular survival, potential evasion of host immune responses, and more. All investigated HSPs and genes related to HSPs’ production (e.g., dnaK, groEL, dnaJ, grpE, GroES, and hsp20) were detected in all 64 strains [55,56]. In cases where the HSPs were not initially recognized as homologs, a further analysis confirmed the presence of species-specific HSPs (accession numbers in Table S1).
All five CAMP factors previously known as Christie-Atkins-Munch-Petersen factors, responsible for triggering tissue damage by membrane pore formation, were found in all C. acnes strains analyzed, which is consistent with the previous findings [7,23,57]. C. acnes-specific CAMP factors 1, 3, 4, and 5 were additionally confirmed in the C. namnetense strain. CAMP factors 3 and 5 were identified in all species, but as homologs with a very low identity in C. granulosum. Other CAMP factors were absent in both C. granulosum and C. avidum. Instead, two previously described species-specific CAMP factors were identified in C. granulosum and C. avidum (accession numbers in Table S1). Both CAMP genes in C. avidum showed a fairly high identity with the CAMP 3 and CAMP 5 genes of C. acnes, consistent with the conclusions of Mak et al. [45].
Numerous putative VFs may be associated with the process of biofilm formation, including cell envelope-related transcriptional attenuator, rcsB, ytpA, flp, luxS, YhjD/YihY/BrkB family envelope integrity protein, cell fate regulator YaaT, and, indirectly, probably the proteins GroES and CAMP 1. Biofilm-regulating protein A (BrpA), C. granulosum-specific VF, previously described in Streptococcus species, was detected in all our C. granulosum strains identified as the putative cell wall biosynthetic protein LcpB, as well as homologs with lower identity in other species (Tables S1 and S3). In contrast, homologs of the cell fate regulator YaaT, which controls sporulation, competence, and biofilm development, were found with high sequence identity in all Cutibacterium species from this study cohort, except for homologs in C. granulosum, which had a lower identity (Table S2). The YhjD/YihY/BrkB family envelope integrity protein, originally found in Bordetella pertussis, was identified as VF BrkB in all Cutibacterium spp. strains, but the homologs in C. granulosum showed very low identity (Table S2). A hypothetical protein common to all strains of phylotype IA was identified as a response regulator (rcsB), confirming the results of Cavallo et al. [46]. Putative adhesion proteins may also play a critical role in biofilm formation, as the previously described putative adhesin in the reference genome of C. acnes (HL096PA1) was identified in all strains, which may also play a critical role in biofilm formation. Putative adhesive protein homologs were not identified in C. granulosum, while in C. avidum and C. namnetense they were present as homologs with high identity. Genes for dermatan sulfate adhesion proteins dsA1 and dsA2 were absent in C. avidum and C. granulosum, in our study as well as in Mak et al., but we confirmed the presence of a low identity dsA1 homolog in the strain of C. namnetense [45].
We identified the presence of the genes-encoding enzymes phospholipase ytpA and luxS and adhesive flp pili (TadE/G) in all Cutibacterium strains. Previous reports indicated the exclusive presence of luxS, which is involved in quorum sensing, in phylotypes IA1, IB, and II [46]; adhesive flp pili (TadE/G) in phylotype II [1]; and ytpA in phylotype IA1 [46].
The genus Cutibacterium has lipolytic properties; it possesses several lipases that can hydrolyze triglycerides into fatty acids. They were one of the first putative VFs identified because lipid degradation can promote inflammation. One of them, triacylglycerol lipase, occurs in two variants as gehA and gehB, with a 42% identity at the protein level between them [1], and the products, free fatty acids, are thought to contribute to the pathogenesis of acne. In this study, we confirmed the presence of gehA in all phylotypes as well as in C. avidum and C. namnetense as homologs with very high identity and gehB in all phylotypes and also as homologs with high identity in C. avidum and C. namnetense. We also identified three triacylglycerol lipases as potential homologs in C. granulosum (Table S1).
Polyunsaturated fatty acid (PUFA) isomerase was absent in the strain of C. acnes subsp. elongatum and other Cutibacterium species [1]. The presence of this enzyme remains relatively poorly understood, and its role in Cutibacterium spp. virulence has not been extensively studied. This enzyme is involved in the production of short-chain fatty acids (SCFAs) produced by C. acnes during fermentative growth. These SCFAs include propionate, acetate, butyrate, and valerate, and they may be associated with the suppression of S. aureus growth [58], inhibition of S. epidermidis biofilm formation [59], and possible adverse effects on skin barrier functions [1,60].
In addition to lipid hydrolysis, C. acnes has several enzymes that can process glycolipids. Sialidase A and B (nanA, nanB) were present in all C. acnes strains and in C. namnetense, while absent in other Cutibacterium spp., except in one C. avidum strain (CUTI-216-55), characterized as infection undetermined/unlikely, where sialidase B was present as a homolog with a very low identity. Glycosidase was present only in two strains C. granulosum (CUTI-243-32 and CUTI-515-74). In the other species, it was present in all other Cutibacterium species, in two C. avidum strains as a homolog with a very low identity (CUTI-233-15 and CUTI-216-55), and in others as a homolog with a high identity. Lipashydrolase (menH) and endo-β-N-acetylglucosaminidase were identified in both C. acnes and C. namnetense. In addition, endoglycoceramidase, which has been previously described as a potential VF because of its presence in the infundibulum of sebaceous follicles [1,55], was found in C. acnes, C. namnetense, and C. avidum.
Previously, acetyl-CoA synthetase (acsA), which is important for lipid transport and metabolism, had been identified mainly in the IA1 phylotype. However, our study has now confirmed that very high identity homologs exist in all phylotypes of C. acnes and in C. namnetense. There are limited data available on several other VFs that were analyzed in this study. The shikimate kinase, encoded by gntK, has been identified in phylotypes IB, II, and III, and was previously reported only in the phylotypes IB and II [46]. In other Cutibacterium species, it was found as a homolog. RoxP, short for “resistance to oxidative stress protein P”, known for its role in protecting the bacterium from oxidative stress by reactive oxygen species produced by the host immune system, was detected only in strains of C. acnes and C. namnetnese. The surface protein transpeptidase—sortase F (srtF) was previously identified in all phylotypes, which was confirmed in our study. Its homologs as hypothetical proteins were found in C. avidum and in C. granulosum (Table S1). Iron acquisition protein (htaA), a polyunsaturated fatty acid isomerase, was present in all C. acnes phylotypes and in C. namnetense. Additionally, it was identified as a species-specific homolog in C. avidum. The dipeptide transport system permease protein dppB, previously described in the IA1 phylotype [46], has been identified as dppB_1 in C. acnes, C. namnetense, and C. avidum and as homolog dppB_2 in C. granulosum and is important for amino acid transport and inorganic ion transport in metabolism. The repressor gene of porphyrin synthesis, deoR, was confirmed in all C. acnes phylotypes and other Cutibacterium spp. with a high identity.
5. Conclusions
While C. acnes is increasingly recognized as an opportunistic pathogen in IAIs, causing mostly low-grade and chronic infections, the role of other Cutibacterium species remains poorly understood. This study provides additional information on the genomic diversity of C. acnes strains and shows the distribution of VFs among 64 Cutibacterium spp. strains from IAIs. While some of the present results differ from the previously reported distributions of VFs in C. acnes, previous studies have mainly focused on strains from patients with acne, in contrast to our study, which focuses on implant-associated infections. While our study found some diversity in the prevalence of virulence factors (VFs) within the genus Cutibacterium, no significant differences in the presence of VFs were observed between different STs, different anatomical sites, or association with infection. Overall, the present results improve the understanding of the genetic diversity and virulence potential of C. acnes and other Cutibacterium species and emphasize the need for further studies to fully understand the specific relationships between different virulence factors, bacterial genotypes, and disease pathogenesis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11122971/s1, Table S1: Accession numbers of the specific virulence factors of C. acnes used in our study, with additional information on other identified virulence factors of C. namnetense, C. avidum, and C. granulosum; Table S2: Percentage of identity for each of the virulence factors searched with additional information on subspecies, phylotype, sequence type, COG annotation, Pfam domain, and Kegg ortholog annotations. Where these data were not available, the orthogroup number was given; Table S3: This table shows the presence and absence of all searched virulence factors specific for C. acnes, C. avidum, C. namnetense, and C. granulosum. The gray shading indicates the presence of the virulence factor, while the white shading indicates its absence in the specific strains; Table S4: The generated raw data were submitted to the Nucleotide Archive under the study accession number PRJEB67661. Additional information on the study, sample, experiment, and run accession numbers can be found in the table for each strain.
Author Contributions
Conceptualization, A.E., P.M.V., K.S.S. and T.T.; methodology, software and validation, A.E., A.C.Š. and K.S.S.; data curation, A.E. and A.C.Š.; writing—original draft preparation, A.E., P.M.V. and A.C.Š.; writing—review and editing, A.E., P.M.V., A.C.Š., K.S.S. and T.T.; visualization, A.E.; supervision, project administration, and resources, P.M.V., T.T. and K.S.S. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge financial support from the Slovenian Research and Innovation Agency (research core funding No. P3-0083).
Data Availability Statement
Generated raw reads were submitted to the Nucleotide Archive under the study accession number PRJEB67661.
Acknowledgments
The authors would like to acknowledge the technical support of research colleagues and laboratory technicians Petra Čamernik, Urša Kolenc, Tinka Lampe, Jure Keber, Danijela Petrović, Aldina Bajramovič, Vesna Cvitković Špik, Petra Kmetič, Tina Mikuletič, Irena Šest, and Marko Kolenc. Special thanks to Bojan Papić for comments that greatly improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mayslich, C.; Grange, P.A.; Dupin, N. Cutibacterium acnes as an Opportunistic Pathogen: An Update of Its Virulence-Associated Factors. Microorganisms 2021, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Boman, J.; Nilson, B.; Sunnerhagen, T.; Rasmussen, M. True infection or contamination in patients with positive Cutibacterium blood cultures-a retrospective cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.E.; Corvec, S.; Borens, O.; Trampuz, A. Propionibacterium acnes: An Underestimated Pathogen in Implant-Associated Infections. BioMed Res. Int. 2013, 2013, 804391. [Google Scholar] [CrossRef]
- Achermann, Y.; Goldstein, E.J.C.; Coenye, T.; Shirtliff, M.E. Propionibacterium acnes: From Commensal to Opportunistic Biofilm-Associated Implant Pathogen. Clin. Microbiol. Rev. 2014, 27, 419–440. [Google Scholar] [CrossRef] [PubMed]
- Aubin, G.G.; Portillo, M.E.; Trampuz, A.; Corvec, S. Propionibacterium acnes, an Emerging Pathogen: From Acne to Implant-Infections, from Phylotype to Resistance. Méd. Mal. Infect. 2014, 44, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Renz, N.; Mudrovcic, S.; Perka, C.; Trampuz, A. Orthopedic Implant-Associated Infections Caused by Cutibacterium spp.—A Remaining Diagnostic Challenge. PLoS ONE 2018, 13, e0202639. [Google Scholar] [CrossRef]
- Brüggemann, H.; Salar-Vidal, L.; Gollnick HP, M.; Lood, R. A Janus-Faced Bacterium: Host-Beneficial and -Detrimental Roles of Cutibacterium acnes. Front. Microbiol. 2021, 31, 673845. [Google Scholar] [CrossRef]
- Coenye, T.; Spittaels, K.-J.; Achermann, Y. The role of biofilm formation in the pathogenesis and antimicrobial susceptibility of Cutibacterium acnes. Biofilm 2022, 9, 100063. [Google Scholar] [CrossRef]
- Zaid, M.; Chavez, M.R.; Carrasco, A.E.; Zimel, M.N.; Zhang, A.L.; Horvai, A.E.; Link, T.M.; O’Donnell, R.J. Cutibacterium (formerly Propionibacterium) acnes clavicular infection. J. Bone Jt. Infect. 2019, 4, 40–49. [Google Scholar] [CrossRef]
- Trampuz, A.; Piper, K.E.; Jacobson, M.J.; Hanssen, A.D.; Unni, K.K.; Osmon, D.R.; Mandrekar, J.N.; Cockerill, F.R.; Steckelberg, J.M.; Greenleaf, J.F.; et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 2007, 357, 654–663. [Google Scholar] [CrossRef]
- Bossard, D.A.; Ledergerber, B.; Zingg, P.O.; Gerber, C.; Zinkernagel, A.S.; Zbinden, R.; Achermann, Y. Optimal Length of Cultivation Time for Isolation of Propionibacterium acnes in Suspected Bone and Joint Infections Is More than 7 Days. J. Clin. Microbiol. 2016, 54, 3043–3049. [Google Scholar] [CrossRef] [PubMed]
- Scholz, C.F.P.; Kilian, M. The Natural History of Cutaneous Propionibacteria, and Reclassification of Selected Species within the Genus Propionibacterium to the Proposed Novel Genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Roux, F.; Solal, J.C.; Bréville, P.; Desplaces, N.; Barthas, J.; Van, J.-C.N.; Rajzbaum, G. Septic arthritis of the hip with Propionibacterium avidum bacteremia after intraarticular treatment for hip osteoarthritis. Jt. Bone Spine 2008, 75, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Aubin, G.G.; Bémer, P.; Kambalev, S.; Patel, N.B.; Lemenand, O.; Caillon, J.; Lawson, P.A.; Corvec, S. Propionibacterium namnetense sp. nov., isolated from a human bone infection. Int. J. Syst. Evol. Microbiol. 2016, 66, 3393–3399. [Google Scholar] [CrossRef] [PubMed]
- Butler-Wu, S.M.; Sengupta, D.J.; Kittichotirat, W.; Matsen, F.A.I.I.I.; Bumgarner, R.E. Genome sequence of a novel species, Propionibacterium humerusii. J. Bacteriol. 2011, 193, 3678. [Google Scholar] [CrossRef]
- Corvec, S. Clinical and Biological Features of Cutibacterium (Formerly Propionibacterium) avidum, an Underrecognized Microorganism. Clin. Microbiol. Rev. 2018, 31, e00064-17. [Google Scholar] [CrossRef]
- Benediktsdóttir, E.; Hambraeus, A. Dispersal of non-sporeforming anaerobic bacteria from the skin. J. Hyg. 1982, 88, 487–500. [Google Scholar] [CrossRef]
- Dagnelie, M.-A.; Khammari, A.; Dréno, B.; Corvec, S. Cutibacterium acnes Molecular Typing: Time to Standardize the Method. Clin. Microbiol. Infect. 2018, 24, 1149–1155. [Google Scholar] [CrossRef]
- McLaughlin, J.; Nagy, I.; Miliotis, G.; McDowell, A. CUTIS-SEQ, a flexible bilocus sequence typing scheme that provides high resolution of Cutibacterium acnes strains across all subspecies. Anaerobe 2023, 79, 102671. [Google Scholar] [CrossRef]
- Johnson, J.L.; Cummins, C.S. Cell wall composition and deoxyribonucleic acid similarities among the anaerobic coryneforms, classical propionibacteria, and strains of Arachnia propionica. J. Bacteriol. 1972, 109, 1047–1066. [Google Scholar] [CrossRef]
- McDowell, A.; Perry, A.L.; Lambert, P.A.; Patrick, S. A new phylogenetic group of Propionibacterium acnes. J. Med. Microbiol. 2008, 57, 218–224. [Google Scholar] [CrossRef] [PubMed]
- McDowell, A.; Valanne, S.; Ramage, G.; Tunney, M.M.; Glenn, J.V.; McLorinan, G.C.; Bhatia, A.; Maisonneuve, J.-F.; Lodes, M.; Persing, D.H.; et al. Propionibacterium acnes Types I and II Represent Phylogenetically Distinct Groups. J. Clin. Microbiol. 2005, 43, 326–334. [Google Scholar] [CrossRef] [PubMed]
- McDowell, A.; Nagy, I.; Magyari, M.; Barnard, E.; Patrick, S. The opportunistic pathogen Propionibacterium acnes: Insights into typing, human disease, clonal diversification and CAMP factor evolution. PLoS ONE 2013, 8, e70897. [Google Scholar] [CrossRef] [PubMed]
- Lomholt, H.B.; Kilian, M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS ONE 2010, 5, e12277. [Google Scholar] [CrossRef] [PubMed]
- McDowell, A.; Barnard, E.; Nagy, I.; Gao, A.; Tomida, S.; Li, H.; Eady, A.; Cove, J.; Nord, C.E.; Patrick, S. An Expanded Multilocus Sequence Typing Scheme for Propionibacterium Acnes: Investigation of ‘Pathogenic’, ‘Commensal’ and Antibiotic Resistant Strains. PLoS ONE 2012, 7, e41480. [Google Scholar] [CrossRef]
- Scholz, C.F.P.; Jensen, A.; Lomholt, H.B.; Brüggemann, H.; Kilian, M. A Novel High-Resolution Single Locus Sequence Typing Scheme for Mixed Populations of Propionibacterium acnes in Vivo. PLoS ONE 2014, 9, e104199. [Google Scholar] [CrossRef]
- Bruggemann, H. The Complete Genome Sequence of Propionibacterium acnes, a Commensal of Human Skin. Science 2004, 305, 671–673. [Google Scholar] [CrossRef] [PubMed]
- Aubin, G.G.; Lavigne, J.-P.; Foucher, Y.; Dellière, S.; Lepelletier, D.; Gouin, F.; Corvec, S. Tropism and virulence of Cutibacterium (formerly Propionibacterium) acnes involved in implant-associated infection. Anaerobe 2017, 47, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Pécastaings, S.; Corvec, S.; Veraldi, S.; Khammari, A.; Roques, C. Cutibacterium acnes (Propionibacterium acnes) and Acne Vulgaris: A Brief Look at the Latest Updates. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 5–14. [Google Scholar] [CrossRef]
- Prinz, J.; Schmid, B.; Zbinden, R.; Zingg, P.O.; Uçkay, I.; Achermann, Y.; Bosshard, P.P. Fast and sensitive multiplex PCR to detect Cutibacterium periprosthetic joint infections. J. Mol. Diagn. 2022, 24, 666–673. [Google Scholar] [CrossRef]
- Broly, M.; Ruffier d’Epenoux, L.; Guillouzouic, A.; Le Gargasson, G.; Juvin, M.-E.; Leroy, A.G.; Bémer, P.; Corvec, S. Propionibacterium/Cutibacterium Species–Related Positive Samples, Identification, Clinical and Resistance Features: A 10-Year Survey in a French Hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1357–1364. [Google Scholar] [CrossRef]
- Kuehnast, T.; Cakar, F.; Weinhäupl, T.; Pilz, A.; Selak, S.; Schmidt, M.A.; Rüter, C.; Schild, S. Multidrug-resistant Cutibacterium avidum isolated from patients with acne vulgaris and other infections. J. Glob. Antimicrob. Resist. 2022, 28, 151–157. [Google Scholar] [CrossRef]
- Kuehnast, T.; Cakar, F.; Weinhäupl, T.; Pilz, A.; Selak, S.; Schmidt, M.A.; Rüter, C.; Schild, S. Comparative Analyses of Biofilm Formation among Different Cutibacterium acnes Isolates. Int. J. Med. Microbiol. 2018, 308, 1027–1035. [Google Scholar] [CrossRef]
- McNally, M.; Sousa, R.; Wouthuyzen-Bakker, M.; Chen, A.F.; Soriano, A.; Vogely, H.C.; Clauss, M.; Higuera, C.A.; Trebše, R. The EBJIS definition of periprosthetic joint infection. Bone Jt. J. 2021, 103-B, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef]
- Marquis, B.; Pillonel, T.; Carrara, A.; Bertelli, C. zDB: Bacterial comparative genomics made easy. bioRxiv 2023, 2023-05, 543076. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Mak, T.N.; Schmid, M.; Brzuszkiewicz, E.; Zeng, G.; Meyer, R.; Sfanos, K.S.; Brinkmann, V.; Meyer, T.F.; Brüggemann, H. Comparative Genomics Reveals Distinct Host-Interacting Traits of Three Major Human-Associated Propionibacteria. BMC Genom. 2013, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Cavallo, I.; Sivori, F.; Truglio, M.; De Maio, F.; Lucantoni, F.; Cardinali, G.; Pontone, M.; Bernardi, T.; Sanguinetti, M.; Capitanio, B.; et al. Skin dysbiosis and Cutibacterium acnes biofilm in inflammatory acne lesions of adolescents. Sci. Rep. 2022, 12, 21104. [Google Scholar] [CrossRef] [PubMed]
- Cobian, N.; Garlet, A.; Hidalgo-Cantabrana, C.; Barrangou, R. Comparative Genomic Analyses and CRISPR-Cas Characterization of Cutibacterium acnes Provide Insights Into Genetic Diversity and Typing Applications. Front. Microbiol. 2021, 12, 758749. [Google Scholar] [CrossRef]
- Barnard, E.; Johnson, T.; Ngo, T.; Arora, U.; Leuterio, G.; McDowell, A.; Li, H. Porphyrin Production and Regulation in Cutaneous Propionibacteria. mSphere 2020, 5, e00793-19. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, F.; Roux, A.L.; Sapriel, G.; Salomon, E.; Bauer, T.; Gaillard, J.L.; Rottman, M. Molecular Typing of Multiple Isolates Is Essential to Diagnose Cutibacterium acnes Orthopedic Device-related Infection. Clin. Infect. Dis. 2019, 94, 534–538. [Google Scholar] [CrossRef]
- Liew-Littorin, C.; Brüggemann, H.; Davidsson, S.; Nilsdotter-Augustinsson, Å.; Hellmark, B.; Söderquist, B. Clonal Diversity of Cutibacterium acnes (Formerly Propionibacterium acnes) in Prosthetic Joint Infections. Anaerobe 2019, 59, 54–60. [Google Scholar] [CrossRef]
- Lomholt, H.; Kilian, M. Clonality and anatomic distribution on the skin of antibiotic resistant and sensitive Propionibacterium acnes. Acta Derm. Venereol. 2014, 94, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Scholz, C.F.P.; Lomholt, H.B. Multilocus Sequence Typing and Phylogenetic Analysis of Propionibacterium acnes. J. Clin. Microbiol. 2012, 50, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Lomholt, H.; Scholz, C.; Brüggemann, H.; Tettelin, H.; Kilian, M. A comparative study of Cutibacterium (Propionibacterium) acnes clones from acne patients and healthy controls. Anaerobe 2017, 47, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kasimatis, G.; Fitz-Gibbon, S.; Tomida, S.; Wong, M.; Li, H. Analysis of Complete Genomes of Propionibacterium acnes Reveals a Novel Plasmid and Increased Pseudogenes in an Acne Associated Strain. BioMed Res. Int. 2013, 2013, 918320. [Google Scholar] [CrossRef] [PubMed]
- Brzuszkiewicz, E.; Weiner, J.; Wollherr, A.; Thürmer, A.; Hüpeden, J.; Lomholt, H.B.; Kilian, M.; Gottschalk, G.; Daniel, R.; Mollenkopf, H.-J.; et al. Comparative Genomics and Transcriptomics of Propionibacterium acnes. PLoS ONE 2011, 6, e21581. [Google Scholar] [CrossRef]
- Farrar, M.D.; Ingham, E.; Holland, K.T. Heat Shock Proteins and Inflammatory Acne Vulgaris: Molecular Cloning, Overexpression and Purification of a Propionibacterium acnes GroEL and DnaK Homologue. FEMS Microbiol. Lett. 2000, 191, 183–186. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Holland, C.; Mak, T.N.; Zimny-Arndt, U.; Schmid, M.; Meyer, T.F.; Jungblut, P.R.; Brüggemann, H. Proteomic Identification of Secreted Proteins of Propionibacterium acnes. BMC Microbiol. 2010, 10, 230. [Google Scholar] [CrossRef]
- Shu, M.; Kuo, S.; Wang, Y.; Jiang, Y.; Liu, Y.-T.; Gallo, R.L.; Huang, C.-M. Porphyrin Metabolisms in Human Skin Commensal Propionibacterium acnes Bacteria: Potential Application to Monitor Human Radiation Risk. Curr. Med. Chem. 2013, 20, 562–568. [Google Scholar]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short Chain Fatty Acids Produced by Cutibacterium acnes Inhibit Biofilm Formation by Staphylococcus epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef]
- Sanford, J.A.; O’neill, A.M.; Zouboulis, C.C.; Gallo, R.L. Short-Chain Fatty Acids from Cutibacterium acnes Activate Both a Canonical and Epigenetic Inflammatory Response in Human Sebocytes. J. Immunol. 2019, 202, 1767–1776. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).