HLA-DQ2/8 and COVID-19 in Celiac Disease: Boon or Bane
Abstract
:1. Introduction
1.1. Protective HLA Alleles in Celiac Disease
1.2. The Human Gastrointestinal Tract Is a Target Organ for SARS-CoV-2
1.3. COVID-19-Celiac Disease Interplay
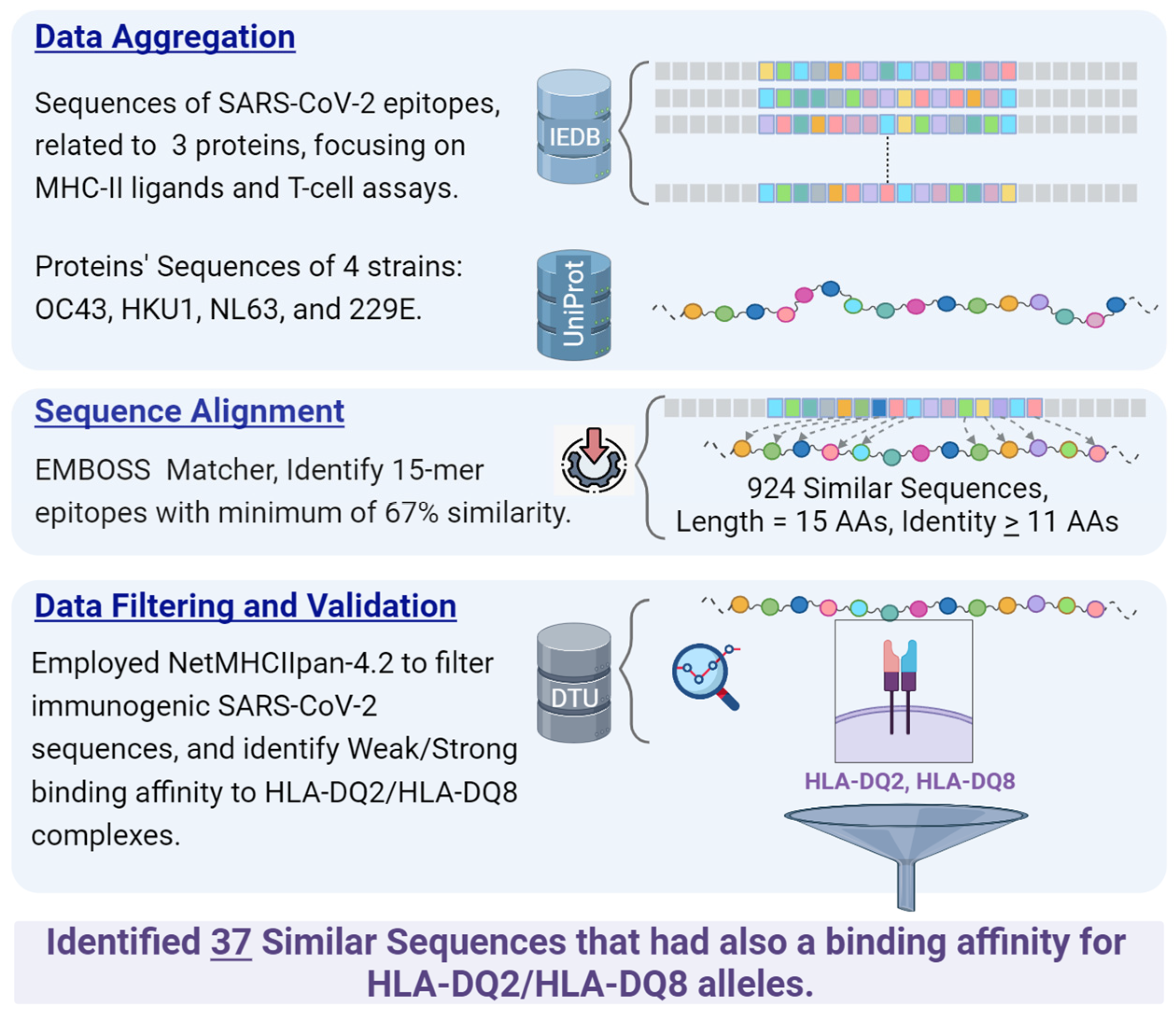
2. Materials and Methods
2.1. Data Sources
2.1.1. Protein Sequences Extraction of Common Cold Coronaviruses
2.1.2. Data Retrieval from IEDB
- Organism: SARS-CoV-2 (ID: 2697049).
- Antigens: Spike glycoprotein (P0DTC2), nucleoprotein (P0DTC9), replicase polyprotein 1ab (P0DTD1).
- Epitope structure: Linear sequence.
- Assay type: Only positive assays pertaining to T-cell epitopes and MHC ligands were considered. This refers to epitopes validated through laboratory experiments.
- MHC restriction: Class II.
- Host: Human.
- Diseases: No specific restrictions; all diseases were considered.
2.2. Sequence Similarity Identification
2.3. Binding Affinity Prediction to HLA-DQ2, DQ8
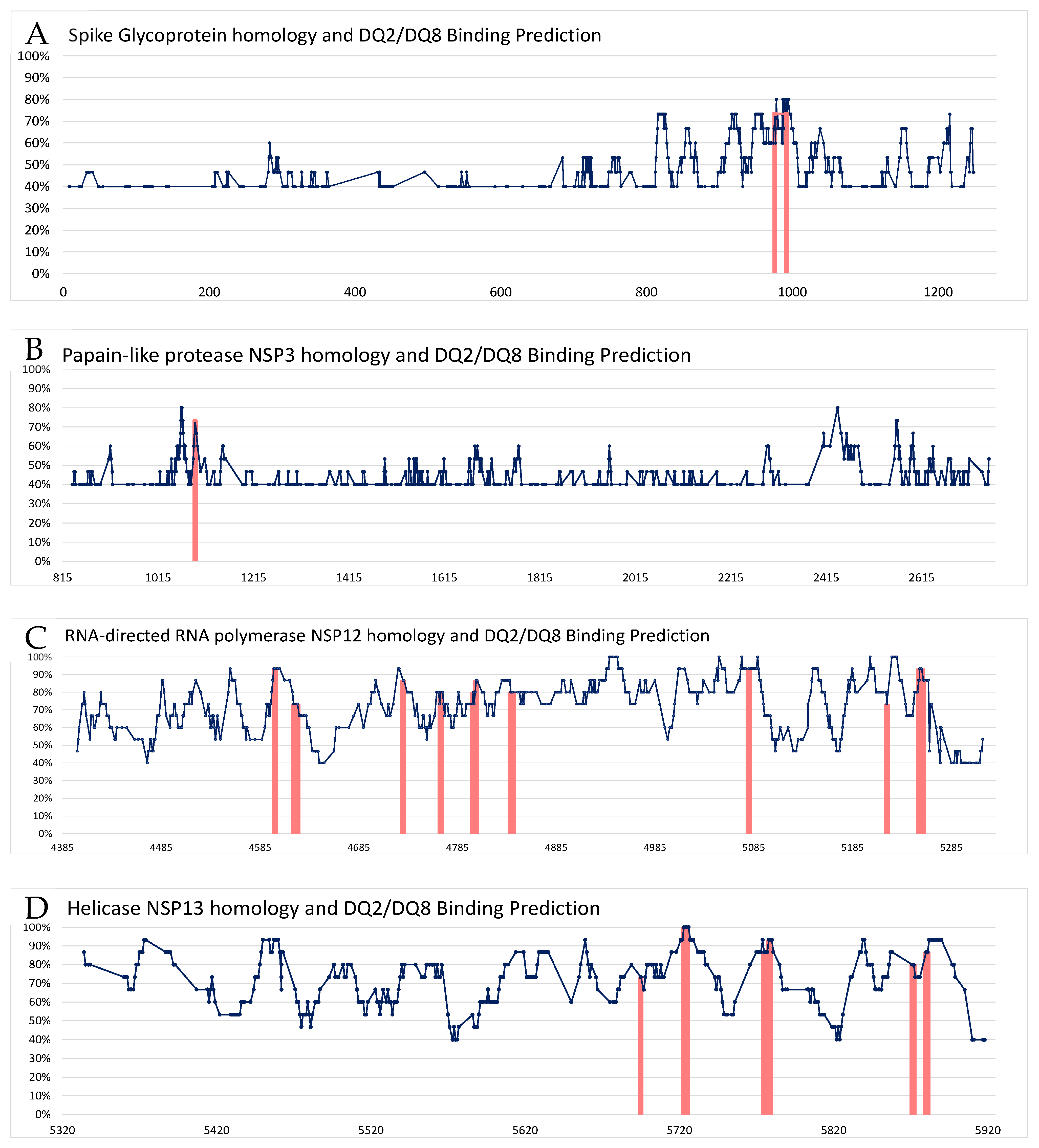
3. Results
4. Discussion
- High affinity between SARS-CoV-2 antigens and HLA-DQ immune presentation to the T cells enhances anti-SARS-CoV-2 immunity. In fact, the HLA allele most associated with COVID-19 deterioration is HLA-A*11. However, HLA II also plays a role in the disease severity, with HLA-DRB1*15:01 and HLA-DRB1*04 alleles being examples [122]. Unfortunately, the CD-associated HLA-DQs were not explored when the binding affinities of 438 HLA alleles were screened [122]. Nevertheless, asymptomatic and mildly/moderately affected patients likely develop an effective early immune response to clear the virus [123]. A reasonable explanation for the associations between CD, SARS-CoV-2, and HLA-DQ2/8 observed presently is that most of the strong HLA binders of coronavirus peptides are also strong binders of other sequences, and hence, are likely to be general strong binders that probably underwent selection in the past [122].
- SARS-CoV-2-naïve people might have a certain measure of HLA-dependent immune defense presented by antibodies cross-reactive to other CCC [124]. Most of those HLAs belong to HLA class I, hence, a minority of them are part of class II. Unfortunately, the CD-associated HLA-DQ were not explored [124]. The topic of a potential protective cross-reactivity against the COVID-19 virus in uninfected CD patients conferred by their HLA-DQ2/8 is a subject for further investigation.
- An individual HLA variant has its unique repertoire of peptides with a specific sequence structure to stick in the peptide-binding groove of HLA. It appears that certain HLA haplotypes have higher preferences to present peptides with specific molecular functions [125]. This HLA preferential presentation was extrapolated to explain the protective effect of certain HLA alleles in infectious diseases, including COVID-19. Indeed, Karnaukhov V. et al. reported on HLA-A/HLA-B and HLA-A/HLA-C variants having a more distinct functional antigen preference presentation, but the HLA-DQ2/8 ones were not explored [125]. The authors reported on HLA differential presentation of SARS-CoV-2 antigens mainly by HLA type I alleles, hence, the CD-associated HLA-DQ haplotypes might play a protective or attenuative role in COVID-19 disease. Notably, several studies reported on HLA-DQ variants associated with a dominant T cell response against the SARS-CoV-2 virus, resulting in a milder disease [120,126,127], including a higher production of antibodies post mRNA-based vaccination [128].
- Cross-reactive antibodies shared between SARS-CoV-2 and gluten. If cross-reactivity exists between the virus and gluten, those reactive antibodies might attenuate the severity of COVID-19 and protect the untreated or the non-compliant CD patients. In fact, Vojdani A, et al. reported on such cross-reactive antibodies [129]. Screening 180 different food antigens and peptides, the authors showed that SARS-CoV-2 proteins share cross-reactive epitopes with various food antigens that had not been previously explored. Wheat and alpha-gliadin were shown to cross-react with SARS-CoV-2 spike protein and nucleoprotein [129]. More so, the authors reported on sequence similarity between SARS-CoV-2 proteins and alpha-gliadin toxic peptides and glutenin, thus, reinforcing a potential effect of the COVID-19–food axis relationships. It should be stressed that the potential protective effects of the above-mentioned cross-reactive antibodies and the sequence similarity were not substantiated and should be further evaluated.
- Increase in anti-inflammatory factors in COVID-19-infected celiac patients. Recently, Asri N et al. studied naïve CD patients for various inflammatory and anti-inflammatory markers [85]. The CD patients exposed an increased expression of anti-inflammatory molecules like CD4, CD25 (IL-2Rα), and FOXP3, compared to severe COVID-19 patients and controls. However, the HLA-DQs’ allelic status was not investigated. The increase in the anti-inflammatory profile might be beneficial to the CD patients by lowering COVID-19 severity and attenuating the disease course. The relationship of those markers to the HLA-DQs should farther be explored.
- HLA-DQ2/8 might be important in fighting human viruses. The mechanism of CD risk modification by HLA heterogeneity might involve differential presentation of autoantigenic sequences by HLA class II proteins. The HLA-DQ2 and DQ8 presentation of viral epitopes were reported concerning coxsackievirus-specific peptides [130]. The authors speculated that the phenomenon might represent a protective adaptive mechanism to maximize anti-enterovirus responses. The same can be speculated for the COVID-19 virus and the HLA-DQ2/8 epitopic presentation in CD, alluding to the potential protective role of those HLAs in fighting SARS-CoV-2 viruses.
- HLA class II: Evolutionary protective mechanisms for CD survival. The wide range of COVID-19 manifestations, morbidity, and mortality seen across various ethnicities and geographical distribution was suggested to be host genetic dependent [131]. This genetic adaptative diversity may apply to CD. Interestingly, selective advantage mechanisms for polymorphic genes were speculated to contribute to the evolutionary survival of the CD populations [38,39,132] (Table 1). In fact, the human HLAs’ genetic heterogeneity is a known major anti-infectious mechanism to fight microbes, parasites, and even viruses, SARS-CoV-2 included. Although the variants of class II HLA loci were less frequently analyzed, they can impact COVID-19 outcomes. Most recently, HLA class II DRB1*01:01, DRB1*04:01, and DRB1*03:01 were reported to reduce disease duration and attenuated COVID-19 course [133,134,135]. Unfortunately, the HLA-DQ repertoire was not screened in those studies. Of note, the topic is still controversial and some studies denied the association between HLA polymorphisms and COVID-19 outcomes [136,137].
5. Celiac Disease and Long COVID-19 Syndromes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wang, D.; Wu, X.; Li, C.; Han, J.; Yin, J.; Gan, J. The impact of geo-environmental factors on global COVID-19 transmission: A review of evidence and methodology. Sci. Total Environ. 2022, 826, 154182. [Google Scholar] [CrossRef]
- Begou, P.; Kassomenos, P. The ecosyndemic framework of the global environmental change and the COVID-19 pandemic. Sci. Total Environ. 2023, 857, 159327. [Google Scholar] [CrossRef]
- Zguro, K.; Fallerini, C.; Fava, F.; Furini, S.; Renieri, A. Host genetic basis of COVID-19: From methodologies to genes. Eur. J. Hum. Genet. 2022, 30, 899–907. [Google Scholar] [CrossRef]
- Ji, X.-S.; Chen, B.; Ze, B.; Zhou, W.-H. Human genetic basis of severe or critical illness in COVID-19. Front. Cell. Infect. Microbiol. 2022, 12, 963239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-J.; Dong, X.; Liu, G.-H.; Gao, Y.-D. Risk and Protective Factors for COVID-19 Morbidity, Severity, and Mortality. Clin. Rev. Allergy Immunol. 2023, 64, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Dobrijević, Z.; Robajac, D.; Gligorijević, N.; Šunderić, M.; Penezić, A.; Miljuš, G.; Nedić, O. The association of ACE1, ACE2, TMPRSS2, IFITM3 and VDR polymorphisms with COVID-19 severity: A systematic review and meta-analysis. EXCLI J. 2022, 21, 818–839. [Google Scholar] [CrossRef] [PubMed]
- Migliorini, F.; Torsiello, E.; Spiezia, F.; Oliva, F.; Tingart, M.; Maffulli, N. Association between HLA genotypes and COVID-19 susceptibility, severity and progression: A comprehensive review of the literature. Eur. J. Med Res. 2021, 26, 84. [Google Scholar] [CrossRef]
- Srivastava, A.; Hollenbach, J.A. The immunogenetics of COVID-19. Immunogenetics 2023, 75, 309–320. [Google Scholar] [CrossRef]
- Ghazy, A.A.; Alrasheedi, A.N.; Elashri, M.; Moussa, H.H.; Rashwan, E.K.; Amer, I.; El Sharawy, S.; Elgamal, S.; Tawfik, S.; Abdelnasser, M.; et al. Relevance of HLA-DP/DQ and INF-λ4 Polymorphisms to COVID-19 Outcomes. Br. J. Biomed. Sci. 2023, 80, 11044. [Google Scholar] [CrossRef]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef]
- Arab, F.; Mollazadeh, S.; Ghayourbabaei, F.; Moghbeli, M.; Saburi, E. The role of HLA genotypes in understanding the pathogenesis of severe COVID-19. Egypt. J. Med Hum. Genet. 2023, 24, 14. [Google Scholar] [CrossRef]
- Lin, F.; Lin, X.; Fu, B.; Xiong, Y.; Zaky, M.Y.; Wu, H. Functional studies of HLA and its role in SARS-CoV-2: Stimulating T cell response and vaccine development. Life Sci. 2023, 315, 121374. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; Muller, S.; Kanduc, D.; David, P.; Halpert, G.; Shoenfeld, Y. The SARS-CoV-2 as an instrumental trigger of autoimmunity. Autoimmun. Rev. 2021, 20, 102792. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; Mahroum, N.; Bogdanos, D.P.; Shoenfeld, Y. COVID-19 as an infectome paradigm of autoimmunity. J. Allergy Clin. Immunol. 2022, 149, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Baiocchi, G.C.; Vojdani, A.; Rosenberg, A.Z.; Vojdani, E.; Halpert, G.; Ostrinski, Y.; Zyskind, I.; Filgueiras, I.S.; Schimke, L.F.; Marques, A.H.C.; et al. Cross-sectional analysis reveals autoantibody signatures associated with COVID-19 severity. J. Med Virol. 2023, 95, e28538. [Google Scholar] [CrossRef] [PubMed]
- Lavi, Y.; Vojdani, A.; Halpert, G.; Sharif, K.; Ostrinski, Y.; Zyskind, I.; Lattin, M.T.; Zimmerman, J.; Silverberg, J.I.; Rosenberg, A.Z.; et al. Dysregulated Levels of Circulating Autoantibodies against Neuronal and Nervous System Autoantigens in COVID-19 Patients. Diagnostics 2023, 13, 687. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Chen, T.Y.-T.; Wang, S.-I.; Hung, Y.-M.; Chen, H.-Y.; Wei, C.-C.J. Risk of autoimmune diseases in patients with COVID-19: A retrospective cohort study. EClinicalMedicine 2023, 56, 101783. [Google Scholar] [CrossRef] [PubMed]
- Miyadera, H.; Tokunaga, K. Associations of human leukocyte antigens with autoimmune diseases: Challenges in identifying the mechanism. J. Hum. Genet. 2015, 60, 697–702. [Google Scholar] [CrossRef]
- Jara, L.J.; Vera-Lastra, O.; Mahroum, N.; Pineda, C.; Shoenfeld, Y. Autoimmune post-COVID vaccine syndromes: Does the spectrum of autoimmune/inflammatory syndrome expand? Clin. Rheumatol. 2022, 41, 1603–1609. [Google Scholar] [CrossRef]
- Dotan, A.; David, P.; Arnheim, D.; Shoenfeld, Y. The autonomic aspects of the post-COVID19 syndrome. Autoimmun. Rev. 2022, 21, 103071. [Google Scholar] [CrossRef]
- Brown, N.K.; Guandalini, S.; Semrad, C.; Kupfer, S.S. A Clinician’s Guide to Celiac Disease HLA Genetics. Am. J. Gastroenterol. 2019, 114, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Del Pozzo, G.; Farina, F.; Picascia, S.; Laezza, M.; Vitale, S.; Gianfrani, C. HLA class II genes in precision-based care of childhood diseases: What we can learn from celiac disease. Pediatr. Res. 2021, 89, 307–312. [Google Scholar] [CrossRef]
- Greco, N.; Meacci, A.; Mora, B.; Vestri, A.; Picarelli, A. Coeliac disease in the COVID-19 pandemic: Does HLA have a protective effect? Ann. Med. 2022, 54, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, H.; Oka, S.; Tsuchiya, N.; Shimada, K.; Hashimoto, A.; Tohma, S.; Kawasaki, A. The role of common protective alleles HLA-DRB1*13 among systemic autoimmune diseases. Genes Immun. 2017, 18, 1–7. [Google Scholar] [CrossRef]
- Bettencourt, A.; Carvalho, C.; Leal, B.; Brás, S.; Lopes, D.; da Silva, A.M.; Santos, E.; Torres, T.; Almeida, I.; Farinha, F.; et al. The Protective Role of HLA-DRB113 in Autoimmune Diseases. J. Immunol. Res. 2015, 2015, 948723. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al Mutair, A.; Aljeldah, M.; Al Shammari, B.R.; Sulaiman, T.; Alshukairi, A.N.; Alfaresi, M.; Al-Jishi, J.M.; Al Bati, N.A.; Al-Mozaini, M.A.; et al. Genetic Variants and Protective Immunity against SARS-CoV-2. Genes 2022, 13, 2355. [Google Scholar] [CrossRef] [PubMed]
- Lani, R.; Senin, N.A.; AbuBakar, S.; Hassandarvish, P. Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines. Vaccines 2022, 10, 1606. [Google Scholar] [CrossRef]
- Troshina, E.; Yukina, M.; Nuralieva, N.; Vasilyev, E.; Rebrova, O.; Akhmatova, R.; Ikonnikova, A.; Savvateeva, E.; Gryadunov, D.; Melnichenko, G.; et al. Association of Alleles of Human Leukocyte Antigen Class II Genes and Severity of COVID-19 in Patients of the ‘Red Zone’ of the Endocrinology Research Center, Moscow, Russia. Diseases 2022, 10, 99. [Google Scholar] [CrossRef]
- Hernández-Doño, S.; Sánchez-González, R.A.; Trujillo-Vizuet, M.G.; Zamudio-Castellanos, F.Y.; García-Silva, R.; Bulos-Rodríguez, P.; Vazquez-Guzmán, C.A.; Cárdenas-Ramos, X.; Rodríguez, D.d.L.; Elías, F.; et al. Protective HLA alleles against severe COVID-19: HLA-A*68 as an ancestral protection allele in Tapachula-Chiapas, Mexico. Clin. Immunol. 2022, 238, 108990. [Google Scholar] [CrossRef]
- Mocci, S.; Littera, R.; Tranquilli, S.; Provenzano, A.; Mascia, A.; Cannas, F.; Lai, S.; Giuressi, E.; Chessa, L.; Angioni, G.; et al. A Protective HLA Extended Haplotype Outweighs the Major COVID-19 Risk Factor Inherited From Neanderthals in the Sardinian Population. Front. Immunol. 2022, 13, 891147. [Google Scholar] [CrossRef]
- Suslova, T.A.; Vavilov, M.N.; Belyaeva, S.V.; Evdokimov, A.V.; Stashkevich, D.S.; Galkin, A.; Kofiadi, I.A. Distribution of HLA-A, -B, -C, -DRB1, -DQB1, -DPB1 allele frequencies in patients with COVID-19 bilateral pneumonia in Russians, living in the Chelyabinsk region (Russia). Hum. Immunol. 2022, 83, 547–550. [Google Scholar] [CrossRef]
- Deb, P.; Zannat, K.E.; Talukder, S.; Bhuiyan, A.H.; Alam Jilani, S.; Saif-Ur-Rahman, K.M. Association of HLA gene polymorphism with susceptibility, severity, and mortality of COVID-19: A systematic review. HLA 2022, 99, 281–312. [Google Scholar] [CrossRef]
- Bubnova, L.; Pavlova, I.; Terentieva, M.; Glazanova, T.; Belyaeva, E.; Sidorkevich, S.; Bashketova, N.; Chkhingeria, I.; Kozhemyakina, M.; Azarov, D.; et al. HLA Genotypes in Patients with Infection Caused by Different Strains of SARS-CoV-2. Int. J. Environ. Res. Public Health 2022, 19, 14024. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.H.L.; Lau, K.; Li, L.; Cheng, S.; Chan, W.Y.; Hui, P.K.; Zee, B.; Leung, C.; Sung, J.J.Y. Association of Human-Leukocyte-Antigen Class I (B*0703) and Class II (DRB1*0301) Genotypes with Susceptibility and Resistance to the Development of Severe Acute Respiratory Syndrome. J. Infect. Dis. 2004, 190, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tavasolian, F.; Rashidi, M.; Hatam, G.R.; Jeddi, M.; Hosseini, A.Z.; Mosawi, S.H.; Abdollahi, E.; Inman, R.D. HLA, Immune Response, and Susceptibility to COVID-19. Front. Immunol. 2021, 11, 601886. [Google Scholar] [CrossRef]
- Ben Shachar, S.; Barda, N.; Manor, S.; Israeli, S.; Dagan, N.; Carmi, S.; Balicer, R.; Zisser, B.; Louzoun, Y. MHC Haplotyping of SARS-CoV-2 Patients: HLA Subtypes Are Not Associated with the Presence and Severity of COVID-19 in the Israeli Population. J. Clin. Immunol. 2021, 41, 1154–1161. [Google Scholar] [CrossRef]
- Aaron, L. The last two millennias echo-catastrophes are the driving forces for the potential genetic advantage mechanisms in celiac disease. Med. Hypotheses 2011, 77, 773–776. [Google Scholar] [CrossRef]
- Lerner, A. Balanced polymorphism: A survival advantage in celiac disease. Med. Hypotheses 2011, 77, 1–2. [Google Scholar] [CrossRef]
- King, J.A.; Jeong, J.; Underwood, F.E.; Quan, J.; Panaccione, N.; Windsor, J.W.; Coward, S.; Debruyn, J.; Ronksley, P.E.; Shaheen, A.-A.; et al. Incidence of Celiac Disease Is Increasing Over Time: A Systematic Review and Meta-analysis. Am. J. Gastroenterol. 2020, 115, 507–525. [Google Scholar] [CrossRef]
- Lerner, A.; Jeremias, P.; Matthias, T. The World Incidence of Celiac Disease Is Increasing: A Review. Int. J. Recent Sci. Res. 2015, 6, 5491–5496. [Google Scholar]
- Hadley, D.; Hagopian, W.; Liu, E.; She, J.-X.; Simell, O.; Akolkar, B.; Ziegler, A.-G.; Rewers, M.; Krischer, J.P.; Chen, W.-M.; et al. HLA-DPB1*04:01 Protects Genetically Susceptible Children from Celiac Disease Autoimmunity in the TEDDY Study. Am. J. Gastroenterol. 2015, 110, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Balamtekin, N.; Baysoy, G.; Tan, Ç.; Kızılkan, N.U.; Demir, H.; Temizel, N.S.; Özen, H.; Yüce, A.; Tezcan, I.; Gürakan, F. The hla groups and their relationship with clinical features in turkish children and adolescents with celiac disease. Turk. J. Pediatr. 2021, 63, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Del Prado, M.Y.; Olivares López, J.L.; Lázaro Almarza, A.; Lasierra Díaz, M.P. HLA system. Phenotypic and gene frequencies in celiac and healthy subjects from the same geographical area. Rev. Esp. Enfermedades Dig. 2001, 93, 110–113. [Google Scholar]
- Silva, E.M.B.T.; Fernandes, M.I.M.; Galvão, L.C.; Sawamura, R.; Donadi, E.A. Human Leukocyte Antigen Class II Alleles in White Brazilian Patients With Celiac Disease. J. Pediatr. Gastroenterol. Nutr. 2000, 31, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Laadhar, L.; Toumi, A.; Kallel-Sellami, M.; Zitouni, M.; Bouraoui, S.; Maherzi, A.; Makni, S.; Ben Hariz, M. HLA class II polymorphism in children with coeliac disease in Tunisia: Is there any influence on clinical manifestation? Eur. J. Gastroenterol. Hepatol. 2009, 21, 1286–1290. [Google Scholar] [CrossRef]
- Boy, M.F.; La Nasa, G.; Balestrieri, A.; Cherchi, M.V.; Usai, P. Distribution of HLA-DPBl, -DQBI-DQAl Alleles among Sardinian Celiac Patients. Dis. Markers 1994, 12, 199–204. [Google Scholar] [CrossRef]
- Lopez-Vazquez, A. MHC class I chain related gene A (MICA) modulates the development of coeliac disease in patients with the high risk heterodimer DQA1*0501/DQB1*0201. Gut 2002, 50, 336–340. [Google Scholar] [CrossRef]
- Bilbao, J.R.; Martín-Pagola, A.; Vitoria, J.C.; Zubillaga, P.; Ortiz, L.; Castaño, L. HLA-DRB1 and MHC class 1 chain-related A haplotypes in Basque families with celiac disease. Tissue Antigens 2002, 60, 71–76. [Google Scholar] [CrossRef]
- Bilbao, J.R.; Martín-Pagola, A.; Pérez de Nanclares, G.; Calvo, B.; Vitoria, J.C.; Vázquez, F.; Castaño, L. HLA-DRB1 and MICA in Autoimmunity. Ann. N. Y. Acad. Sci. 2003, 1005, 314–318. [Google Scholar] [CrossRef]
- Bravo, F.P.; Araya, M.; Mondragón, A.; Rios, G.; Alarcón, T.; Roessler, J.; Santos, J. Genetic differences in HLA-DQA1∗ and DQB1∗ allelic distributions between celiac and control children in Santiago, Chile. Hum. Immunol. 1999, 60, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Aaron, L.; Torsten, M.; Patricia, W. Autoimmunity in celiac disease: Extra-intestinal manifestations. Autoimmun. Rev. 2019, 18, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Matthias, T. Extra intestinal manifestations of CD: Common pathways in the gut- remote organs’ axes. Int. J. Celiac Dis. 2017, 5, 24–27. [Google Scholar]
- Lerner, A.; Matthias, T. GUT-the Trojan Horse in Remote Organs’ Autoimmunity. J. Clin. Cell. Immunol. 2016, 7, 10–4172. [Google Scholar] [CrossRef]
- Catassi, C.; Verdu, E.F.; Bai, J.C.; Lionetti, E. Coeliac disease. Lancet 2022, 399, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A. New therapeutic strategies for celiac disease. Autoimmun. Rev. 2010, 9, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Ghazanfar, H.; Kandhi, S.; Shin, D.; Muthumanickam, A.; Gurjar, H.; Qureshi, Z.A.; Shaban, M.; Farag, M.; Haider, A.; Budhathoki, P.; et al. Impact of COVID-19 on the Gastrointestinal Tract: A Clinical Review. Cureus 2022, 14, e23333. [Google Scholar] [CrossRef]
- Lerner, A. Are my patients with celiac disease at higher risk of COVID-19 virus. Int. J. Celiac Dis. 2020, 8, 35–38. [Google Scholar]
- Lerner, A. COVID-19 and the human gut: A new runner on the tract. Int. J. Celiac Dis. 2020, 8, 64–67. [Google Scholar]
- Lerner, A. The COVID-19 vaccination debate: CoV-2 in celiac disease: A Pathogen or just along for the ride? Int. J. Celiac Dis. 2021, 9, 6–9. [Google Scholar]
- Samasca, G.; Lerner, A. Celiac disease in the COVID-19 pandemic. J. Transl. Autoimmun. 2021, 4, 100120. [Google Scholar] [CrossRef] [PubMed]
- Clerbaux, L.-A.; Mayasich, S.A.; Muñoz, A.; Soares, H.; Petrillo, M.; Albertini, M.C.; Lanthier, N.; Grenga, L.; Amorim, M.-J. Gut as an Alternative Entry Route for SARS-CoV-2: Current Evidence and Uncertainties of Productive Enteric Infection in COVID-19. J. Clin. Med. 2022, 11, 5691. [Google Scholar] [CrossRef]
- Lerner, A.; McCarty, M.F. The Aging Bowel Dysfunction and Elderly Vulnerability towards COVID-19 Infection. Life 2021, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- McCarty, M.F.; Lerner, A. Perspective: Prospects for Nutraceutical Support of Intestinal Barrier Function. Adv. Nutr. Int. Rev. J. 2021, 12, 316–324. [Google Scholar] [CrossRef]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-W.; Wu, X.-X.; Jiang, X.-G.; Xu, K.-J.; Ying, L.-J.; Ma, C.-L.; Li, S.-B.; Wang, H.-Y.; Zhang, S.; Gao, H.-N.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-CoV-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368, m606. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Ma, J.; Guan, J.; Wang, M.; Song, Y.; Tian, D.; Li, P. Manifestations of Digestive system in hospitalized patients with novel coronavirus pneumonia in Wuhan, China: A single-center, descriptive study. Chin. J. Dig. 2020, 40, 151–156. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Cui, X.; Xiao, J.; Meng, T.; Zhou, W.; et al. The digestive system is a potential route of 2019- nCov infection: A bioinformatics analysis based on single-cell transcriptomes. BioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.-B.; Lyu, J.-R.; Lei, X.-M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.-M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020, 96, 19–24. [Google Scholar] [CrossRef]
- Tian, Y.; Rong, L.; Nian, W.; He, Y. Review article: Gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. 2020, 51, 843–851. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, R.S.; Qu, G.Q. Gross Examination Report of a COVID-19 Death Autopsy. J. Forensic Med. 2020, 36, 21–23. [Google Scholar] [CrossRef]
- Yang, Z.; Li, G.; Dai, X.; Liu, G.; Li, G.; Jie, Y. Three cases of novel coronavirus pneumonia with viral nucleic acids still positive in stool after throat swab detection turned negative. Chin. J. Dig. 2020, 40, E002. [Google Scholar] [CrossRef]
- Zhang, J.C.; Wang, S.B.; Xue, Y.D. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020, 92, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Taki, K.; Gahlot, R.; Sharma, A.; Dhangar, K. A chronicle of SARS-CoV-2: Part-I—Epidemiology, diagnosis, prognosis, transmission and treatment. Sci. Total Environ. 2020, 734, 139278. [Google Scholar] [CrossRef]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med 2022, 3, 371–387.e9. [Google Scholar] [CrossRef]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef]
- Lavania, M.; Joshi, M.S.; Ranshing, S.S.; Potdar, V.A.; Shinde, M.; Chavan, N.; Jadhav, S.M.; Sarkale, P.; Mohandas, S.; Sawant, P.M.; et al. Prolonged Shedding of SARS-CoV-2 in Feces of COVID-19 Positive Patients: Trends in Genomic Variation in First and Second Wave. Front. Med. 2022, 9, 835168. [Google Scholar] [CrossRef]
- Gandhi, P.A.; Singh, T. Feco-Oral Transmission of SARS-CoV-2. Asia Pac. J. Public Health 2020, 32, 370. [Google Scholar] [CrossRef]
- Targoński, R.; Gąsecka, A.; Prowancki, A.; Targoński, R. An alternative to airborne droplet transmission route of SARS-CoV-2, the feco-oral route, as a factor shaping COVID-19 pandemic. Med. Hypotheses 2022, 166, 110903. [Google Scholar] [CrossRef]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; La Rosa, G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020, 743, 140444. [Google Scholar] [CrossRef]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef]
- Schiepatti, A.; Alimenti, E.; Maimaris, S.; Nicolardi, M.L.; La Barbera, F.M.; Baiardi, P.; Biagi, F. Prevalence, incidence and clinical features of SARS-CoV-2 infection in adult coeliac patients. Eur. J. Gastroenterol. Hepatol. 2021, 33, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Gokden, Y.; Hot, S.; Adas, M.; Ogutmen Koc, D.; Atak, S.; Hot, A.B. Celiac disease and COVID-19 pandemic: Should we worry? Acta Gastroenterol. Belg. 2020, 83, 517–525. [Google Scholar] [PubMed]
- Gholam-Mostafaei, F.S.; Asri, N.; Parvani, N.; Khamene, E.A.; Barzegar, F.; Rostami-Nejad, M.; Rezaei-Tavirani, M.; Shahbazkhani, B.; Jahani-Sherafat, S.; Rostami, K.; et al. Prevalence and outcome of COVID-19 among Iranian celiac patients. Gastroenterol. Hepatol. Bed Bench 2022, 15, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Asri, N.; Mojarad, E.N.; Mirjalali, H.; Mohebbi, S.R.; Baghaei, K.; Rostami-Nejad, M.; Yadegar, A.; Rezaei-Tavirani, M.; Aghdaei, H.A.; Rostami, K.; et al. Toward finding the difference between untreated celiac disease and COVID-19 infected patients in terms of CD4, CD25 (IL-2 Rα), FOXP3 and IL-6 expressions as genes affecting immune homeostasis. BMC Gastroenterol. 2021, 21, 462. [Google Scholar] [CrossRef]
- Li, J.; Tian, A.; Yang, D.; Zhang, M.; Chen, L.; Wen, J.; Chen, P. Celiac Disease and the Susceptibility of COVID-19 and the Risk of Severe COVID-19: A Mendelian Randomization Study. Clin. Transl. Gastroenterol. 2022, 13, e00480. [Google Scholar] [CrossRef]
- Zhen, J.; Stefanolo, J.P.; Temprano, M.d.l.P.; Tedesco, S.; Seiler, C.; Caminero, A.F.; De-Madaria, E.; Huguet, M.M.; Vivas, S.; Niveloni, S.I.; et al. The Risk of Contracting COVID-19 Is Not Increased in Patients With Celiac Disease. Clin. Gastroenterol. Hepatol. 2020, 19, 391–393. [Google Scholar] [CrossRef]
- Hadi, Y.B.; Sohail, A.H.; Lakhani, D.A.; Naqvi, S.F.; Kupec, J.T.; Pervez, A. Outcomes of SARS-CoV-2 infection in patients with celiac disease: A multicenter research network study. Ann. Gastroenterol. 2022, 35, 164–168. [Google Scholar] [CrossRef]
- Rathore, S.S.; Velasquez-Botero, F.; Nieto-Salazar, M.A.; Flowers, T.C.; Hasan, J.; Parashar, A.K.; Tanveer, K.; Aneis, H.; Buremoh, A.I.; Yusuf, K.; et al. Prevalence and clinical outcomes of COVID-19 in patients with pre-existing celiac disease: A systematic review and meta-analysis. Rev. Med Virol. 2023, 33, e2433. [Google Scholar] [CrossRef]
- Crocco, M.; Calvi, A.; Canzoneri, F.; Malerba, F.; Zampatti, N.; Chiaro, A.; Arrigo, S.; Gandullia, P.; Proietti, S.; Bonassi, S. The Influence of SARS-CoV-2 Pandemic on the Diagnosis of Celiac Disease and Clinical Practice in Pediatric Gastroenterology. Nutrients 2023, 15, 559. [Google Scholar] [CrossRef]
- Elli, L.; Barisani, D.; Vaira, V.; Bardella, M.T.; Topa, M.; Vecchi, M.; Doneda, L.; Scricciolo, A.; Lombardo, V.; Roncoroni, L. How to manage celiac disease and gluten-free diet during the COVID-19 era: Proposals from a tertiary referral center in a high-incidence scenario. BMC Gastroenterol. 2020, 20, 387. [Google Scholar] [CrossRef] [PubMed]
- Monzani, A.; Lionetti, E.; Felici, E.; Fransos, L.; Azzolina, D.; Rabbone, I.; Catassi, C. Adherence to the Gluten-Free Diet during the Lockdown for COVID-19 Pandemic: A Web-Based Survey of Italian Subjects with Celiac Disease. Nutrients 2020, 12, 3467. [Google Scholar] [CrossRef]
- Catassi, G.N.; Vallorani, M.; Cerioni, F.; Lionetti, E.; Catassi, C. A negative fallout of COVID-19 lockdown in Italy: Life-threatening delay in the diagnosis of celiac disease. Dig. Liver Dis. 2020, 52, 1092–1093. [Google Scholar] [CrossRef] [PubMed]
- Cakir, M.; Guven, B.; Issi, F.; Ozkaya, E. New-onset celiac disease in children during COVID-19 pandemic. Acta Paediatr. 2022, 111, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, Y.; Prévost, J.; Ullah, I.; Lu, M.; Gong, S.Y.; Tauzin, A.; Gasser, R.; Vézina, D.; Anand, S.P.; et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2022, 38, 110210. [Google Scholar] [CrossRef] [PubMed]
- Trovato, C.M.; Montuori, M.; Pietropaoli, N.; Oliva, S. COVID-19 and celiac disease: A pathogenetic hypothesis for a celiac outbreak. Int. J. Clin. Pract. 2021, 75, e14452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Faiq, M.A.; Pareek, V.; Raza, K.; Narayan, R.K.; Prasoon, P.; Kumar, P.; Kulandhasamy, M.; Kumari, C.; Kant, K.; et al. Relevance of SARS-CoV-2 related factors ACE2 and TMPRSS2 expressions in gastrointestinal tissue with pathogenesis of digestive symptoms, diabetes-associated mortality, and disease recurrence in COVID-19 patients. Med. Hypotheses 2020, 144, 110271. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef]
- Verhagen, J.; van der Meijden, E.D.; Lang, V.; Kremer, A.E.; Völkl, S.; Mackensen, A.; Aigner, M.; Kremer, A.N. Human CD4+ T cells specific for dominant epitopes of SARS-CoV-2 Spike and Nucleocapsid proteins with therapeutic potential. Clin. Exp. Immunol. 2021, 205, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Aboulaghras, S.; Piancatelli, D.; Taghzouti, K.; Balahbib, A.; Alshahrani, M.M.; Al Awadh, A.A.; Goh, K.W.; Ming, L.C.; Bouyahya, A.; Oumhani, K. Meta-Analysis and Systematic Review of HLA DQ2/DQ8 in Adults with Celiac Disease. Int. J. Mol. Sci. 2023, 24, 1188. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.B.; Kaabinejadian, S.; Yari, H.; Peters, B.; Barra, C.; Gragert, L.; Hildebrand, W.; Nielsen, M. Machine learning reveals limited contribution of trans-only encoded variants to the HLA-DQ immunopeptidome. Commun. Biol. 2023, 6, 442. [Google Scholar] [CrossRef]
- Obermair, F.-J.; Renoux, F.; Heer, S.; Lee, C.H.; Cereghetti, N.; Loi, M.; Maestri, G.; Haldner, Y.; Wuigk, R.; Iosefson, O.; et al. High-resolution profiling of MHC II peptide presentation capacity reveals SARS-CoV-2 CD4 T cell targets and mechanisms of immune escape. Sci. Adv. 2022, 8, eabl5394. [Google Scholar] [CrossRef]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Sciurti, M.; Fornaroli, F.; Gaiani, F.; Bonaguri, C.; Leandro, G.; Di Mario, F.; Angelis, G.L.D. Genetic susceptibilty and celiac disease: What role do HLA haplotypes play? Acta Biomed. 2018, 89, 17–21. [Google Scholar] [CrossRef]
- Sahin, Y.; Mermer, S. Frequency of celiac disease and distribution of HLA-DQ2/DQ8 haplotypes among siblings of children with celiac disease. World J. Clin. Pediatr. 2022, 11, 351–359. [Google Scholar] [CrossRef]
- Megiorni, F.; Pizzuti, A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: Practical implications of the HLA molecular typing. J. Biomed. Sci. 2012, 19, 88. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Cortez, A.; Melo, A.C.; Chaves, V.E.; Condino-Neto, A.; Camargos, P. Do HLA class II genes protect against pulmonary tuberculosis? A systematic review and meta-analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1567–1580. [Google Scholar] [CrossRef] [PubMed]
- Walker-Sperlin, V.; Digitale, J.C.; Viard, M.; Martin, M.P.; Bashirova, A.; Yuki, Y.; Ramsuran, V.; Kulkarni, S.; Naranbhai, V.; Li, H.; et al. Genetic variation that determines TAPBP expression levels associates with the course of malaria in an HLA allotype-dependent manner. Proc. Natl. Acad. Sci. USA 2022, 119, e2205498119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chikata, T.; Kuse, N.; Murakoshi, H.; Gatanaga, H.; Oka, S.; Takiguchi, M. Immunological Control of HIV-1 Disease Progression by Rare Protective HLA Allele. J. Virol. 2022, 96, e0124822. [Google Scholar] [CrossRef] [PubMed]
- Crux, N.B.; Elahi, S. Human Leukocyte Antigen (HLA) and Immune Regulation: How Do Classical and Non-Classical HLA Alleles Modulate Immune Response to Human Immunodeficiency Virus and Hepatitis C Virus Infections? Front. Immunol. 2017, 8, 832. [Google Scholar] [CrossRef] [PubMed]
- Ou, G.; Liu, X.; Xu, H.; Ji, X.; Liu, X.; Wang, J. Variation and expression of HLA-DPB1 gene in HBV infection. Immunogenetics 2021, 73, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; de Castro, M.V.; Naslavsky, M.S.; Scliar, M.O.; Silva, N.S.B.; Pereira, R.N.; Ciriaco, V.A.O.; Castro, C.F.B.; Mendes-Junior, C.T.; Silveira, E.d.S.; et al. MUC22, HLA-A, and HLA-DOB variants and COVID-19 in resilient super-agers from Brazil. Front. Immunol. 2022, 13, 975918. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, S.; Ismail, A.; Sabran, M.R.; Mohtarrudin, N.; Kaka, U.; Nadeem, M. In-vivo study of synthetic and natural food colors effect on biochemical and immunity parameters. Food Sci. Technol. 2021, 42, e41420. [Google Scholar] [CrossRef]
- Langton, D.J.; Bourke, S.C.; Lie, B.A.; Reiff, G.; Natu, S.; Darlay, R.; Burn, J.; Echevarria, C. The influence of HLA genotype on the severity of COVID-19 infection. HLA 2021, 98, 14–22. [Google Scholar] [CrossRef]
- Augusto, D.G.; Hollenbach, J.A. HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection. Curr. Opin. Immunol. 2022, 76, 102178. [Google Scholar] [CrossRef]
- Augusto, D.G.; Murdolo, L.D.; Chatzileontiadou, D.S.M.; Sabatino, J.J.; Yusufali, T.; Peyser, N.D.; Butcher, X.; Kizer, K.; Guthrie, K.; Murray, V.W.; et al. A common allele of HLA is associated with asymptomatic SARS-CoV-2 infection. Nature 2023, 620, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Barquera, R.; Collen, E.; Di, D.; Buhler, S.; Teixeira, J.; Llamas, B.; Nunes, J.M.; Sanchez-Mazas, A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA 2020, 96, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Purbey, P.K.; Roy, K.; Gupta, S.; Paul, M.K. Mechanistic insight into the protective and pathogenic immune-responses against SARS-CoV-2. Mol. Immunol. 2023, 156, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Buckley, P.R.; Lee, C.H.; Pinho, M.P.; Babu, R.O.; Woo, J.; Antanaviciute, A.; Simmons, A.; Ogg, G.; Koohy, H. HLA-dependent variation in SARS-CoV-2 CD8 + T cell cross-reactivity with human coronaviruses. Immunology 2022, 166, 78–103. [Google Scholar] [CrossRef] [PubMed]
- Karnaukhov, V.; Paes, W.; Woodhouse, I.B.; Partridge, T.; Nicastri, A.; Brackenridge, S.; Shcherbinin, D.; Chudakov, D.M.; Zvyagin, I.V.; Ternette, N.; et al. HLA variants have different preferences to present proteins with specific molecular functions which are complemented in frequent haplotypes. Front. Immunol. 2022, 13, 1067463. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.-S.; Lee, Y.-H.; Jo, H.-A.; Baek, I.-C.; Kim, S.-M.; Sohn, H.-J.; Kim, T.-G. Comprehensive Analysis of CD4+ T Cell Response Cross-Reactive to SARS-CoV-2 Antigens at the Single Allele Level of HLA Class II. Front. Immunol. 2022, 12, 774491. [Google Scholar] [CrossRef] [PubMed]
- Naemi, F.M.A.; Al-Adwani, S.; Al-Khatabi, H.; Al-Nazawi, A. Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J. Med. Virol. 2021, 93, 4430–4437. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bautista, J.F.; Sampedro, A.; Gómez-Vicente, E.; Rodríguez-Granger, J.; Reguera, J.A.; Cobo, F.; Ruiz-Cabello, F.; López-Nevot, M. HLA Class II Polymorphism and Humoral Immunity Induced by the SARS-CoV-2 mRNA-1273 Vaccine. Vaccines 2022, 10, 402. [Google Scholar] [CrossRef]
- Vojdani, A.; Vojdani, E.; Melgar, A.L.; Redd, J. Reaction of SARS-CoV-2 antibodies with other pathogens, vaccines, and food antigens. Front. Immunol. 2022, 13, 5254. [Google Scholar] [CrossRef]
- Ellis, R.J.; Varela-Calvino, R.; Tree, T.I.M.; Peakman, M. HLA Class II molecules on haplotypes associated with type 1 diabetes exhibit similar patterns of binding affinities for coxsackievirus P2C peptides. Immunology 2005, 116, 337–346. [Google Scholar] [CrossRef]
- Adli, A.; Rahimi, M.; Khodaie, R.; Hashemzaei, N.; Hosseini, S.M. Role of genetic variants and host polymorphisms on COVID-19: From viral entrance mechanisms to immunological reactions. J. Med. Virol. 2022, 94, 1846–1865. [Google Scholar] [CrossRef] [PubMed]
- David, J.; Mody, B. Potential selective advantage mechanism for polymorphic genetics in Celiac Disease. Med. Hypotheses 2011, 77, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.C.; Schmidt, A.G.; Bölke, E.; Uhrberg, M.; Keitel, V.; Feldt, T.; Jensen, B.; Häussinger, D.; Adams, O.; Schneider, E.M.; et al. Association of HLA genotypes, AB0 blood type and chemokine receptor 5 mutant CD195 with the clinical course of COVID-19. Eur. J. Med. Res. 2021, 26, 107. [Google Scholar] [CrossRef] [PubMed]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front. Immunol. 2020, 11, 605688. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Partridge, T.; Wormald, C.; Kawahara, R.; Stalls, V.; Aggelakopoulou, M.; Parker, J.; Doherty, R.P.; Morejon, Y.A.; Lee, E.; et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 2021, 35, 109179. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bautista, J.F.; Rodriguez-Nicolas, A.; Rosales-Castillo, A.; López-Ruz, M.; Martín-Casares, A.M.; Fernández-Rubiales, A.; Anderson, P.; Garrido, F.; Ruiz-Cabello, F.; López-Nevot, M. Study of HLA-A, -B, -C, -DRB1 and -DQB1 polymorphisms in COVID-19 patients. J. Microbiol. Immunol. Infect. 2022, 55, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Copley, H.C.; Gragert, L.; Leach, A.R.; Kosmoliaptsis, V. Influence of HLA Class II Polymorphism on Predicted Cellular Immunity Against SARS-CoV-2 at the Population and Individual Level. Front. Immunol. 2021, 12, 669357. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, T.; Sivan, M.; Delaney, B.; Evans, R.; Milne, R. Long covid—An update for primary care. BMJ 2022, 378, e072117. [Google Scholar] [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Piazza, M.; Di Cicco, M.; Pecoraro, L.; Ghezzi, M.; Peroni, D.; Comberiati, P. Long COVID-19 in Children: From the Pathogenesis to the Biologically Plausible Roots of the Syndrome. Biomolecules 2022, 12, 556. [Google Scholar] [CrossRef]
- Lerner, A.; O’Bryan, T.; Matthias, T. Navigating the Gluten-Free Boom: The Dark Side of Gluten Free Diet. Front. Pediatr. 2019, 7, 414. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Matthias, T. Gluten-free Diet—Tough Alley in Torrid Time. Int. J. Celiac Dis. 2017, 5, 50–55. [Google Scholar] [CrossRef]
- Vojdani, A.; Vojdani, E.; Saidara, E.; Maes, M. Persistent SARS-CoV-2 Infection, EBV, HHV-6 and Other Factors May Contribute to Inflammation and Autoimmunity in Long COVID. Viruses 2023, 15, 400. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Aslani, H.R.; Shayesteh, M.R.H.; Taghipour, A.; Nasser, A.; Safari, H.; Alizade-Sani, M.; Dehghan, A.; Azimi, T. Are Viruses and Parasites Linked to Celiac Disease? A Question that Still has no Definite Answer. Curr. Pharm. Biotechnol. 2019, 20, 1181–1193. [Google Scholar] [CrossRef]
- Calabretto, M.; Di Carlo, D.; Falasca, F.; Mazzuti, L.; Meacci, A.; Donato, G.; Greco, N.; Mezzatesta, L.; Morrone, A.; Turriziani, O.; et al. Analysis of viral nucleic acids in duodenal biopsies from adult patients with celiac disease. Eur. J. Gastroenterol. Hepatol. 2022, 34, 1107–1110. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]

| Protective HLA Alleles | Country | References |
|---|---|---|
| HLA-DPB1*04:01 | Germany, Finland, Sweden, and USA | [42] |
| HLA C14, DR11, DR15, DQ3 | Turkey | [43] |
| Cw4 and DQ1 | Spain | [44] |
| HLADQB1*06 | Brazil | [45] |
| DRB1*13, DQA1*0102, DQB1*06 | Tunisia | [46] |
| DQB1*0502/DQA1*0102 | Sardinia | [47] |
| MICA-A5.1 | Spain | [48] |
| MICA-A9 | Basque country, Spain | [49,50] |
| DQA1*0101, DQA1*0201, DQB1*0301 | Chile | [51] |
| Gastrointestinal Aspects | Enteric Features | References |
|---|---|---|
| Symptoms | Diarrhea, nausea, vomiting | [65,66,67] |
| ACE2 expression | All along the GIT | [68,69] |
| Inflammation and damage | Lymphocytic infiltration, edema, necrosis, degeneration, cellular shedding | [70,71] |
| Viral particles, nucleocapsid proteins | In the stomach, duodenum, and colonic cells | [70,71,72,73] |
| Stool shading | SARS-CoV-2 RNA in stool | [74,75,76,77] |
| Gut replication | SARS-CoV-2 multiplication in the gut | [76] |
| Feco–Oral transmission | Infectious virus is recovered from stool and urine samples. | [76,77,78,79] |
| Sewage, wastewater transmission | SARS-CoV-2 particles, RNA, and infectivity | [80,81] |
| SARS-CoV-2 Parent Protein | CCC Protein ID | Epitope Sequence CCC vs. SARS-CoV-2 ** | Identity | % Identity | Start | End | HLA-DQA10301-DQB10302 * Rank | HLA-DQA10501-DQB10201 * Rank |
|---|---|---|---|---|---|---|---|---|
| NSP3 replicase polyprotein 1ab P0DTD1 | P0C6X5 | SKDYISSNGPLKVG SDDYIATNGPLKVG | 11/15 | 73.4% | 1086 | 1100 | 4.28 | |
| NSP12 replicase polyprotein 1ab P0DTD1 | P0C6X2 P0C6X6 P0C6X1 P0C6X5 | VVGVLTLDNQDLNGN IVGVLTLDNQDLNGN | 14/15 | 93.3% | 4593 | 4607 | 4.33 | |
| P0C6X2 | DFIQTAPGFGVAVAD DFIQTTPGSGVPVVD | 11/15 | 73.3% | 4613 | 4627 | 3.38 | ||
| FIQTAPGFGVAVADS FIQTTPGSGVPVVDS | 11/15 | 73.3% | 4614 | 4628 | 1.15 | 4.18 | ||
| IQTAPGFGVAVADSY IQTTPGSGVPVVDSY | 11/15 | 73.3% | 4615 | 4629 | 0.76 | 2.75 | ||
| QTAPGFGVAVADSYY QTTPGSGVPVVDSYY | 11/15 | 73.3% | 4616 | 4630 | 0.63 | 2.24 | ||
| P0C6X2 P0C6X6 | RQIFVDGVPFVVSIG RKIFVDGVPFVVSTG | 13/15 | 86.7% | 4723 | 4737 | 4.34 | ||
| KDLLLYAADPAMHVA KELLVYAADPAMHAA | 12/15 | 80.0% | 4761 | 4775 | 4.3 | |||
| P0C6X6 | TSGVKFQTVKPGNFN TNNVAFQTVKPGNFN | 12/15 | 80.0% | 4794 | 4808 | 3.03 | ||
| SGVKFQTVKPGNFNQ NNVAFQTVKPGNFNK | 11/15 | 73.3% | 4795 | 4809 | 1.6 | |||
| P0C6X2 P0C6X6 | VKFQTVKPGNFNQD VAFQTVKPGNFNKD | 12/15 | 80.0% | 4796 | 4810 | 0.7 | ||
| VKFQTVKPGNFNQDF VAFQTVKPGNFNKDF | 13/15 | 86.7% | 4797 | 4811 | 2.98 | |||
| FFFTQDGNAAITDYN FFFAQDGNAAISDYD | 12/15 | 80.0% | 4832 | 4846 | 2.9 | |||
| FFTQDGNAAITDYNY FFAQDGNAAISDYDY | 12/15 | 80.0% | 4833 | 4847 | 1.98 | |||
| FTQDGNAAITDYNYY FAQDGNAAISDYDYY | 12/15 | 80.0% | 4834 | 4848 | 2.12 | |||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 | SSGDATTAFANSVFN SSGDATTAYANSVFN | 14/15 | 93.3% | 5073 | 5087 | 4.91 | 2.73 | |
| P0C6X2 | DYVYLPYPDPS DYVYLPYPDPS | 11/15 | 73.3% | 5213 | 5227 | 4.02 | ||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 | LLIERFVSLAIDAYP LMIERFVSLAIDAYP | 14/15 | 93.3% | 5246 | 5260 | 3.36 | 1.42 | |
| LIERFVSLAIDAYPL MIERFVSLAIDAYPL | 14/15 | 93.3% | 5247 | 5261 | 4.39 | 1.75 | ||
| IERFVSLAIDAYPL IERFVSLAIDAYPL | 14/15 | 93.3% | 5248 | 5262 | 1.53 | |||
| ERYVSLAIDAYPLSK ERFVSLAIDAYPLTK | 13/15 | 86.7% | 5249 | 5263 | 3.4 | |||
| NSP13 replicase polyprotein 1ab P0DTD1 | P0C6X5 P0C6X1 | PEVNADIVVVDEVSM PETTADIVVFDEISM | 11/15 | 73.3% | 5688 | 5702 | 1.08 | |
| P0C6X2 P0C6X6 | RAKHYVYIGDPAQLP RAKHYVYIGDPAQLP | 15/15 | 100.0% | 5716 | 5730 | 3.24 | ||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 | AKHYVYIGDPAQLPA AKHYVYIGDPAQLPA | 15/15 | 100.0% | 5717 | 5731 | 2.45 | ||
| KHYVYIGDPAQLPAP KHYVYIGDPAQLPAP | 15/15 | 100.0% | 5718 | 5732 | 3.52 | |||
| CPKEIVDTVSALVYE CPAEIVDTVSALVYD | 13/15 | 86.7% | 5768 | 5782 | 3.27 | |||
| PKEIVDTVSALVYEN PAEIVDTVSALVYDN | 13/15 | 86.7% | 5769 | 5783 | 2.67 | 1.3 | ||
| EIVDTVSALVYENK EIVDTVSALVYDNK | 13/15 | 86.7% | 5770 | 5784 | 1.09 | 0.65 | ||
| EIVDTVSALVYENKL EIVDTVSALVYDNKL | 14/15 | 93.3% | 5771 | 5785 | 1.61 | 2.77 | ||
| P0C6X6 P0C6X2 | IVETVSALVYDNKLK IVDTVSALVYDNKLK | 14/15 | 93.3% | 5772 | 5786 | 4.2 | ||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 | EYDYVIYSQTAETAH EYDYVIFTQTTETAH | 12/15 | 80.0% | 5864 | 5878 | 3.76 | ||
| P0C6X1 P0C6X2 P0C6X6 | YDYVIYSQTAETAHS YDYVIFTQTTETAHS | 12/15 | 80.0% | 5865 | 5879 | 3.07 | ||
| P0C6X1 P0C6X5 P0C6X2 P0C6X6 | TAETAHSVNVNRFNV TTETAHSCNVNRFNV | 13/15 | 86.7% | 5873 | 5887 | 3.43 | ||
| ETAHSVNVNRFNVA ETAHSCNVNRFNVA | 13/15 | 86.7% | 5874 | 5888 | 2.98 | |||
| Spike glycoprotein P0DTC2 | Q5MQD0 | FGAISSSLQEILSR FGAISSVLNDILSR | 11/15 | 73.4% | 969 | 983 | 4.86 | |
| P36334 Q5MQD0 | DALEAQVQIDRLING DKVEAEVQIDRLITG | 11/15 | 73.3% | 985 | 999 | 4.45 | 4.48 | |
| DALEAQVQIDRLING DPPEAEVQIDRLITG | 11/15 | 73.3% | 985 | 999 | 2.93 | 3.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lerner, A.; Benzvi, C.; Vojdani, A. HLA-DQ2/8 and COVID-19 in Celiac Disease: Boon or Bane. Microorganisms 2023, 11, 2977. https://doi.org/10.3390/microorganisms11122977
Lerner A, Benzvi C, Vojdani A. HLA-DQ2/8 and COVID-19 in Celiac Disease: Boon or Bane. Microorganisms. 2023; 11(12):2977. https://doi.org/10.3390/microorganisms11122977
Chicago/Turabian StyleLerner, Aaron, Carina Benzvi, and Aristo Vojdani. 2023. "HLA-DQ2/8 and COVID-19 in Celiac Disease: Boon or Bane" Microorganisms 11, no. 12: 2977. https://doi.org/10.3390/microorganisms11122977
APA StyleLerner, A., Benzvi, C., & Vojdani, A. (2023). HLA-DQ2/8 and COVID-19 in Celiac Disease: Boon or Bane. Microorganisms, 11(12), 2977. https://doi.org/10.3390/microorganisms11122977








