Nutrient and Microbiome-Mediated Plant–Soil Feedback in Domesticated and Wild Andropogoneae: Implications for Agroecosystems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study System
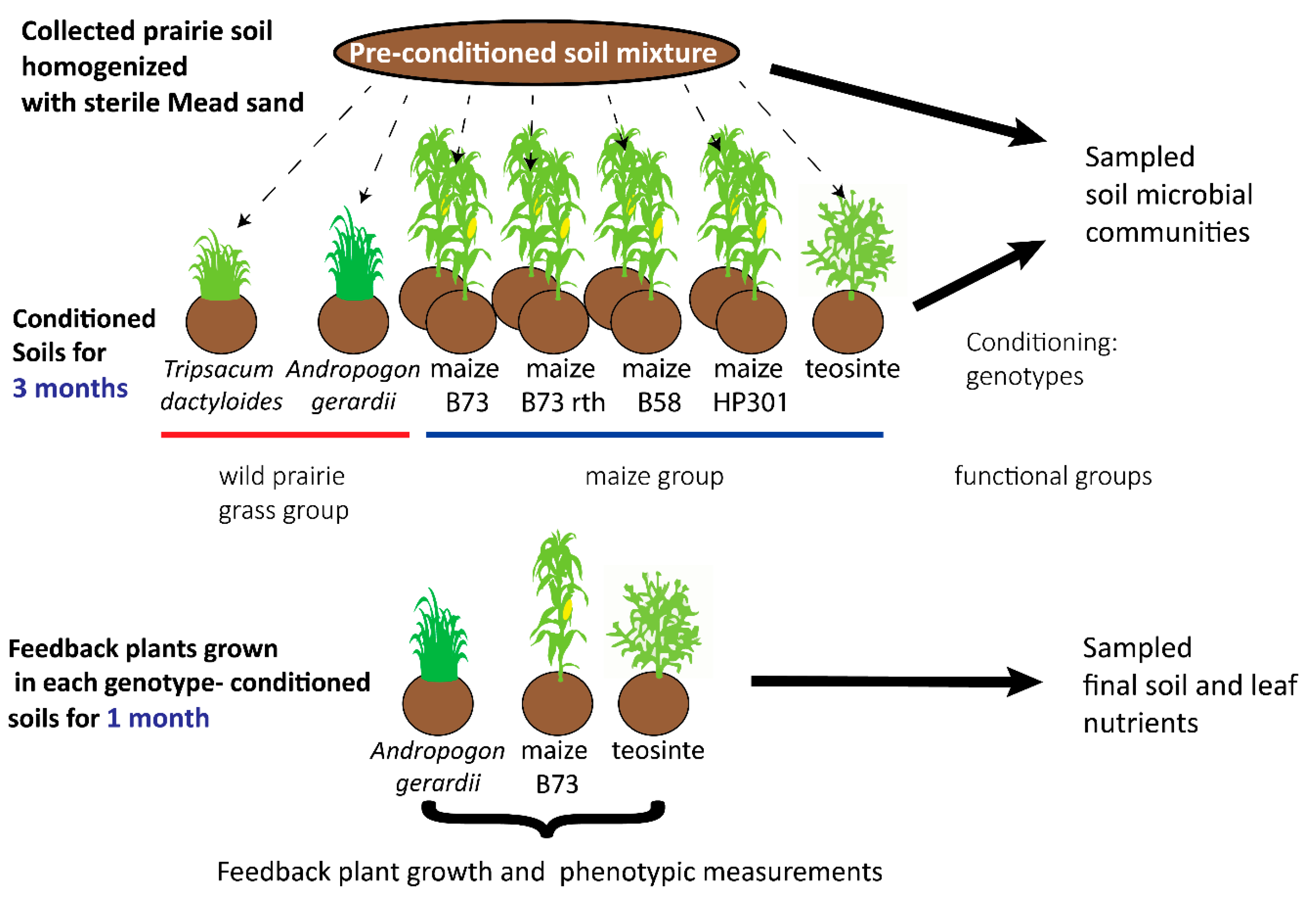
2.2. Overview of the Plant–Soil Feedback Experimental Design
2.2.1. Conditioning Phase: Seedling Growth, Phenotyping, and Collection of Soil Samples
2.2.2. Conditioning Phase: Seedling Growth and Collection of Soil Samples
2.2.3. Conditioning Phase: DNA Extraction, qPCR, Amplicon Sequencing, and Bioinformatic Analysis
2.2.4. Feedback Phase: Seedling Growth and Phenotyping
2.2.5. Elemental Analysis of Final Soil and Leaf Nutrients
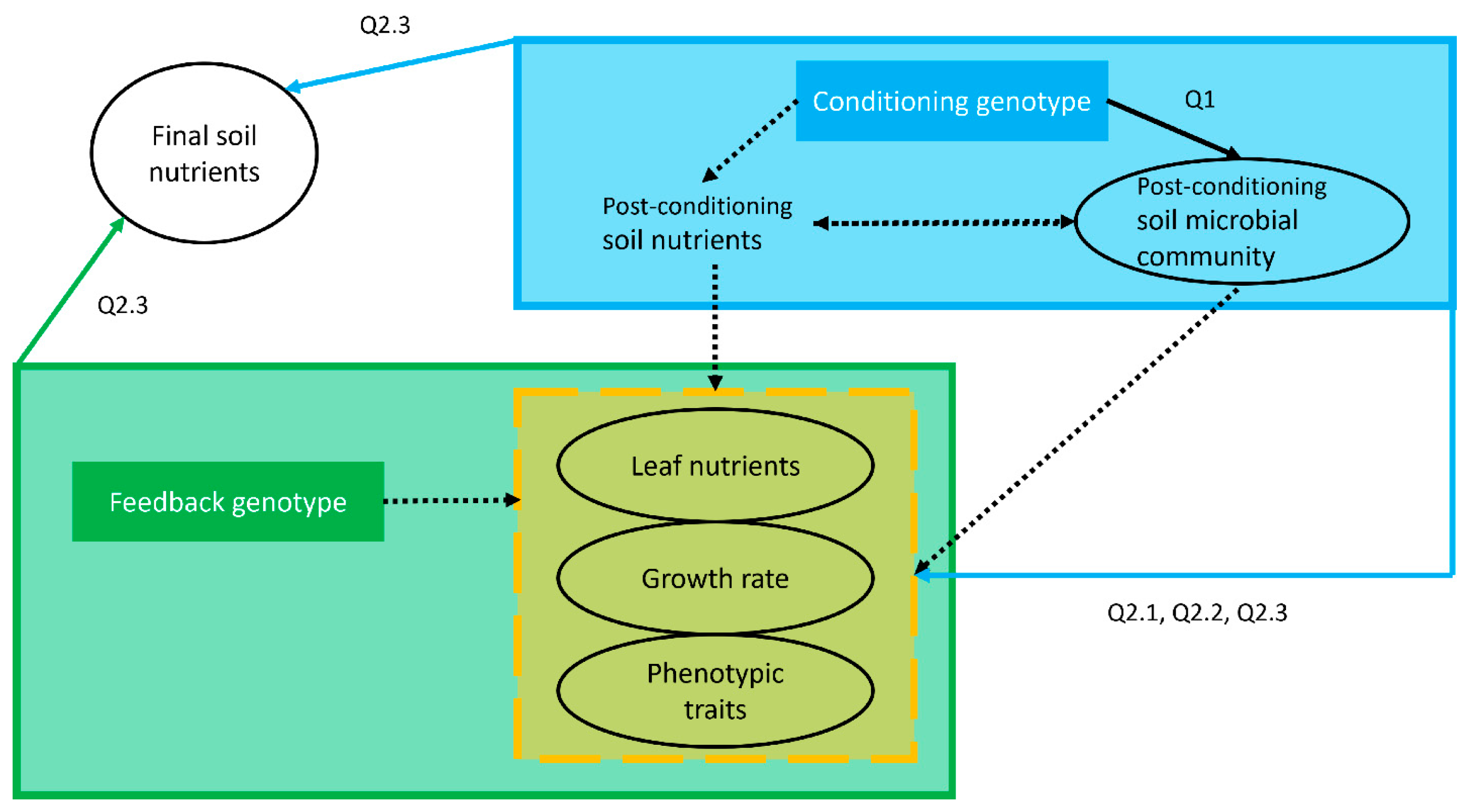
2.3. Statistical Analyses
2.3.1. Q1 Post-Conditioning Soil Microbial Community Structure
2.3.2. Q2.1 and 2.2 Phenotypic Traits and Plant Performance of the Feedback Plants
2.3.3. Q2.3 Variation in Soil Nutrient Concentrations across PSF Phases
3. Results
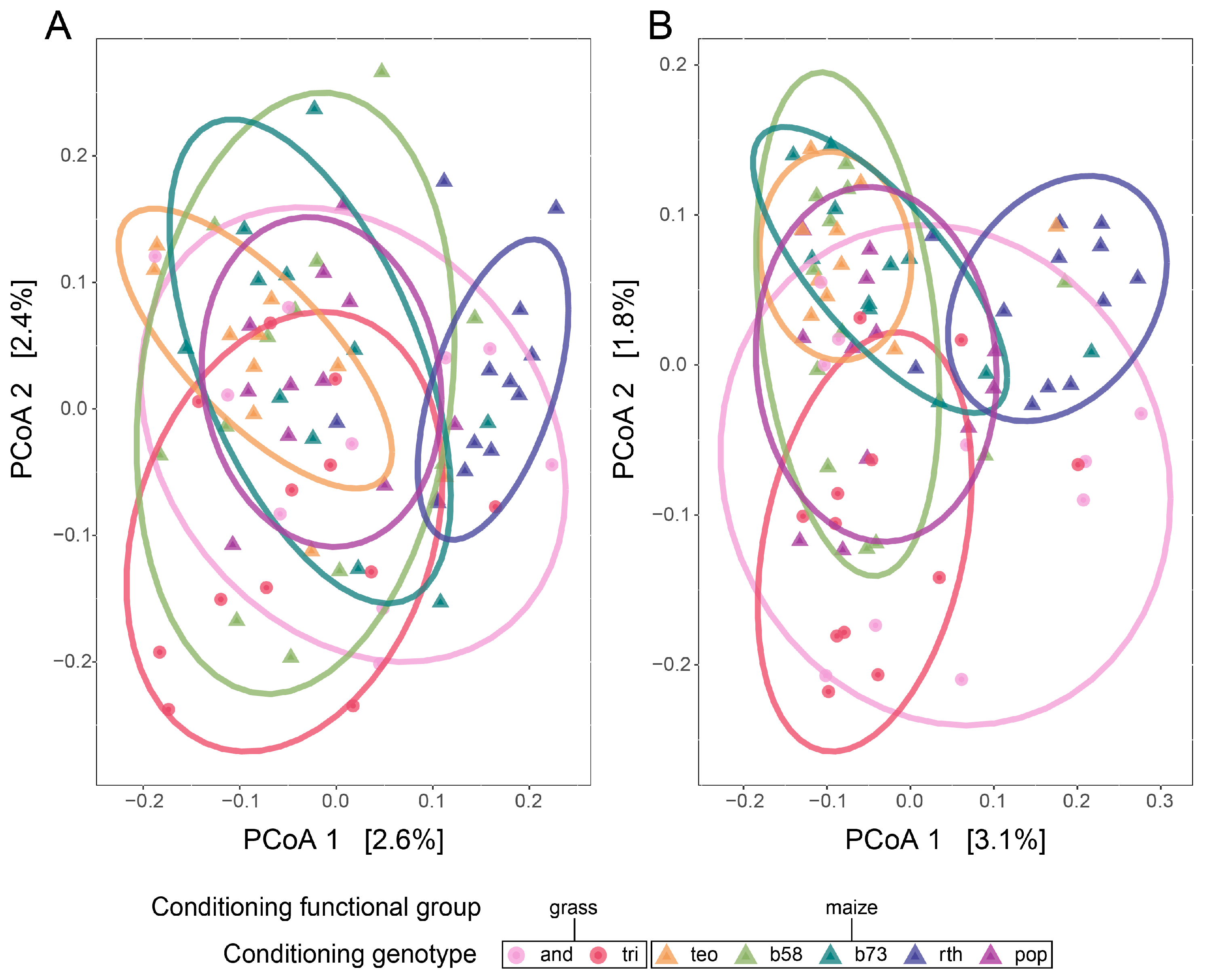
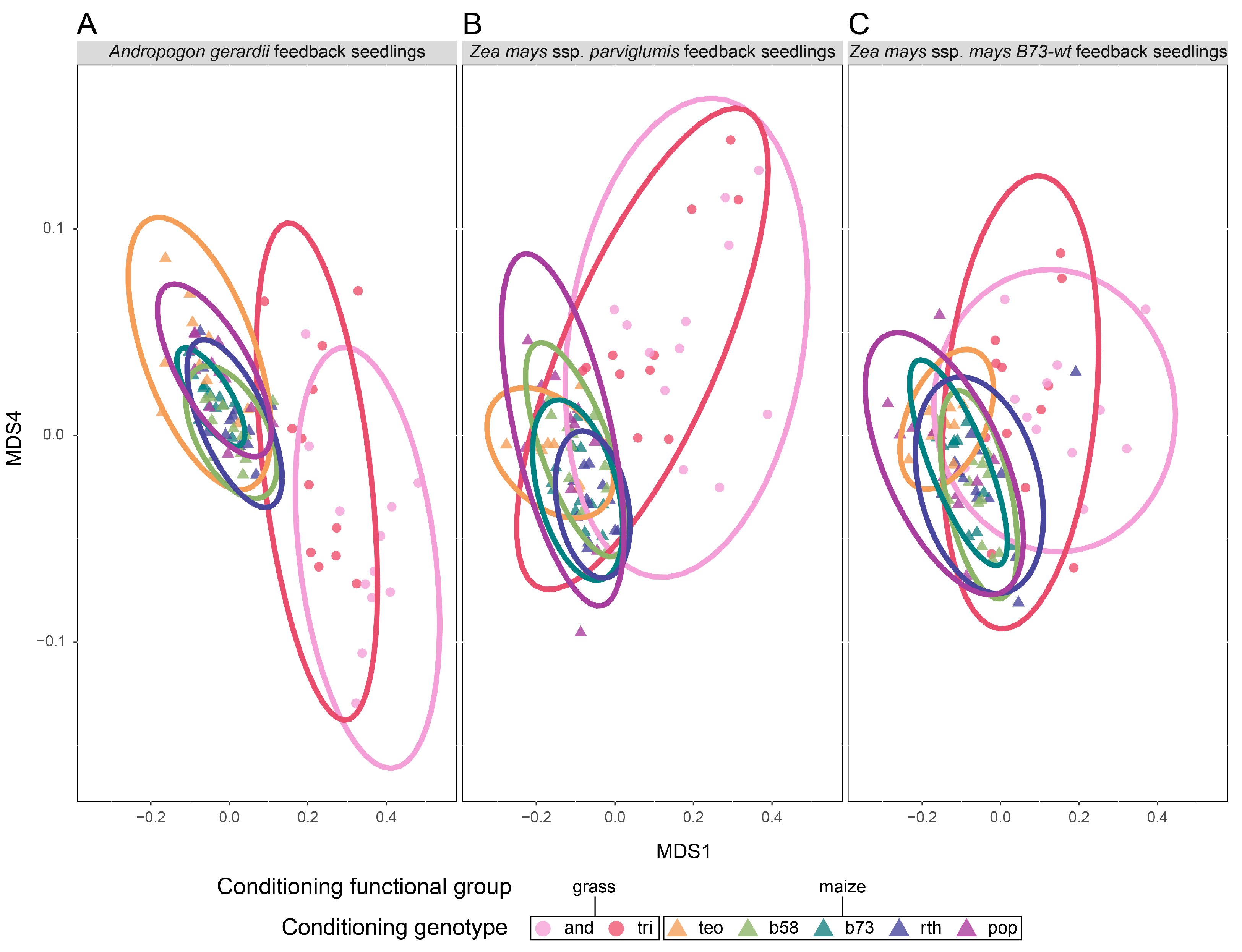
3.1. Q1 Shifts in the Soil Microbial Community Structure across Conditioning Genotypes
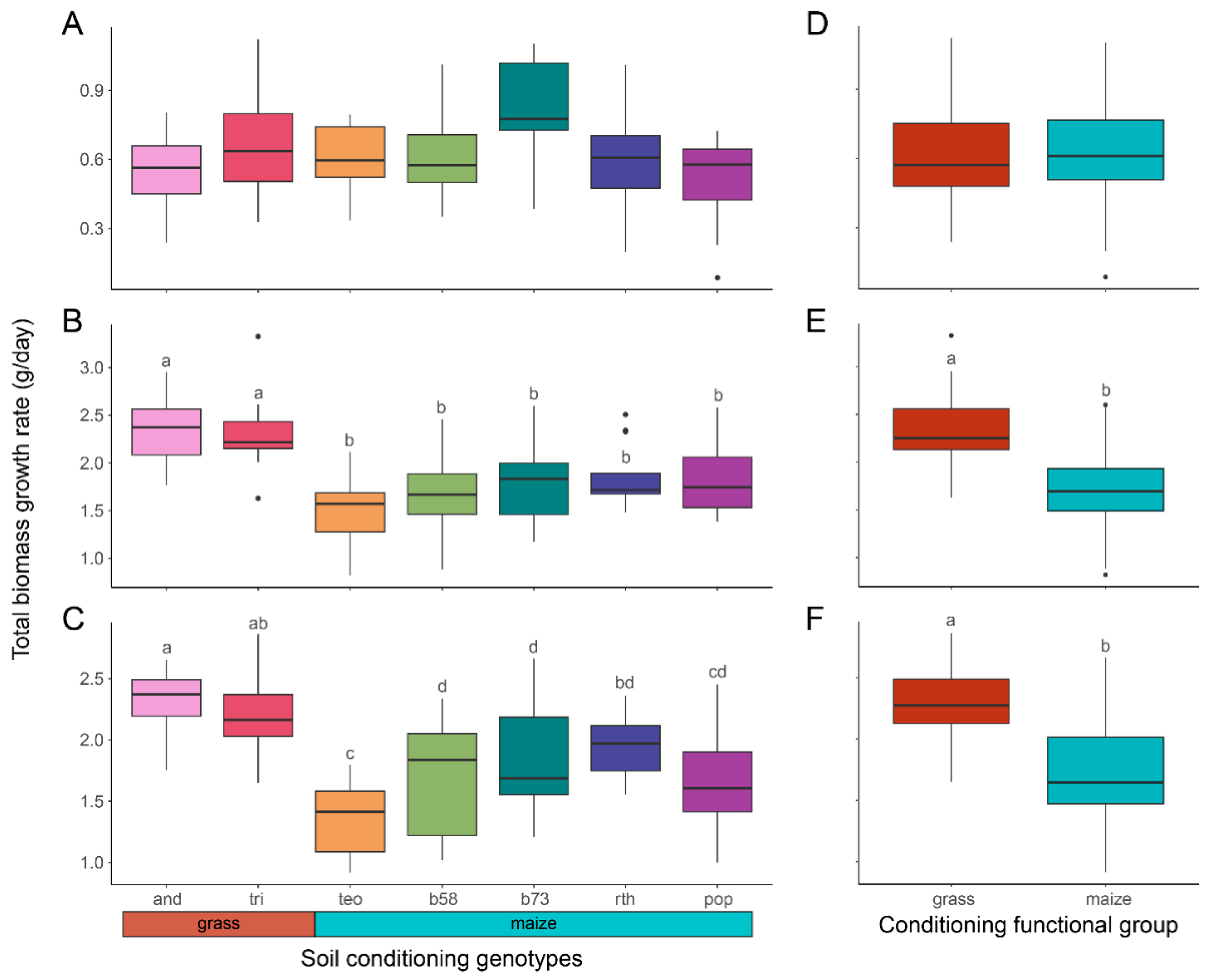
3.2. Q2.1 Variation of Feedback Plant Performance across Conditioning Genotypes
3.3. Q2.2 Variation in Phenotypic Traits and Leaf Nutrient Concentrations of Feedback Plants across the Conditioning Genotype
3.4. Q2.3 Effects of Conditioning and Feedback Genotypes on Nutrients
4. Discussion
4.1. Effects of Conditioning on Soil Microbial Communities
4.2. Plant–Soil Feedback on Plant Growth, Phenotypic Traits, and Soil Nutrients
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kearney, J. Food Consumption Trends and Drivers. Philos. Trans. R. Soc. B 2010, 365, 2793–2807. [Google Scholar] [CrossRef] [PubMed]
- Bommarco, R.; Kleijn, D.; Potts, S.G. Ecological Intensification: Harnessing Ecosystem Services for Food Security. Trends Ecol. Evol. 2013, 28, 230–238. [Google Scholar] [CrossRef]
- Amundson, R.; Berhe, A.A.; Hopmans, J.W.; Olson, C.; Sztein, A.E.; Sparks, D.L. Soil and Human Security in the 21st Century. Science 2015, 348, 1261071. [Google Scholar] [CrossRef] [PubMed]
- Bruns, H.A. Southern Corn Leaf Blight: A Story Worth Retelling. Agron. J. 2017, 109, 1218–1224. [Google Scholar] [CrossRef]
- Kopittke, P.M.; Menzies, N.W.; Wang, P.; McKenna, B.A.; Lombi, E. Soil and the Intensification of Agriculture for Global Food Security. Environ. Int. 2019, 132, 105078. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Marsh, E.L.; Kruger, G.; Lorenz, A.; Schachtman, D.P. Belowground Microbial Communities Respond to Water Deficit and Are Shaped by Decades of Maize Hybrid Breeding. Environ. Microbiol. 2020, 22, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Bybee-Finley, K.A.; Ryan, M.R. Advancing Intercropping Research and Practices in Industrialized Agricultural Landscapes. Agriculture 2018, 8, 80. [Google Scholar] [CrossRef]
- Li, C.; Hoffland, E.; Kuyper, T.W.; Yu, Y.; Zhang, C.; Li, H.; Zhang, F.; van der Werf, W. Syndromes of Production in Intercropping Impact Yield Gains. Nat. Plants 2020, 6, 653–660. [Google Scholar] [CrossRef]
- Mariotte, P.; Mehrabi, Z.; Bezemer, T.M.; De Deyn, G.B.; Kulmatiski, A.; Drigo, B.; Veen, G.F.; van der Heijden, M.G.A.; Kardol, P. Plant–Soil Feedback: Bridging Natural and Agricultural Sciences. Trends Ecol. Evol. 2018, 33, 129–142. [Google Scholar] [CrossRef]
- Wortman, S.E.; Drijber, R.A.; Francis, C.A.; Lindquist, J.L. Arable Weeds, Cover Crops, and Tillage Drive Soil Microbial Community Composition in Organic Cropping Systems. Appl. Soil Ecol. 2013, 72, 232–241. [Google Scholar] [CrossRef]
- Bever, J.D.; Platt, T.G.; Morton, E.R. Microbial Population and Community Dynamics on Plant Roots and Their Feedbacks on Plant Communities. Annu. Rev. Microbiol. 2012, 66, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Miller, Z.J.; Menalled, F.D. Impact of Species Identity and Phylogenetic Relatedness on Biologically-Mediated Plant-Soil Feedbacks in a Low and a High Intensity Agroecosystem. Plant Soil 2015, 389, 171–183. [Google Scholar] [CrossRef]
- Segnitz, R.M.; Russo, S.E.; Davies, S.J.; Peay, K.G. Ectomycorrhizal Fungi Drive Positive Phylogenetic Plant–Soil Feedbacks in a Regionally Dominant Tropical Plant Family. Ecology 2020, 101, e03083. [Google Scholar] [CrossRef] [PubMed]
- van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant–Soil Feedbacks: The Past, the Present and Future Challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Ågren, G.I.; Wetterstedt, J.Å.M.; Billberger, M.F.K. Nutrient Limitation on Terrestrial Plant Growth—Modeling the Interaction between Nitrogen and Phosphorus. New Phytol. 2012, 194, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Augusto, L.; Achat, D.L.; Jonard, M.; Vidal, D.; Ringeval, B. Soil Parent Material—A Major Driver of Plant Nutrient Limitations in Terrestrial Ecosystems. Glob. Change Biol. 2017, 23, 3808–3824. [Google Scholar] [CrossRef] [PubMed]
- Colombi, T.; Chakrawal, A.; Herrmann, A.M. Carbon Supply–Consumption Balance in Plant Roots: Effects of Carbon Use Efficiency and Root Anatomical Plasticity. New Phytol. 2022, 233, 1542–1547. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cahill Jr, J.F. Plant Phenotypic Plasticity Belowground: A Phylogenetic Perspective on Root Foraging Trade-Offs. Am. Nat. 2005, 166, 216–230. [Google Scholar] [CrossRef]
- Lambers, H.; Poorter, H. Inherent Variation in Growth Rate Between Higher Plants: A Search for Physiological Causes and Ecological Consequences. In Advances in Ecological Research; Begon, M., Fitter, A.H., Eds.; Academic Press: Cambridge, MA, USA, 1992; Volume 23, pp. 187–261. [Google Scholar]
- Louw-Gaume, A.E.; Rao, I.M.; Gaume, A.J.; Frossard, E. A Comparative Study on Plant Growth and Root Plasticity Responses of Two Brachiaria Forage Grasses Grown in Nutrient Solution at Low and High Phosphorus Supply. Plant Soil 2010, 328, 155–164. [Google Scholar] [CrossRef]
- Perkins, L.B.; Johnson, D.W.; Nowak, R.S. Plant-Induced Changes in Soil Nutrient Dynamics by Native and Invasive Grass Species. Plant Soil 2011, 345, 365–374. [Google Scholar] [CrossRef]
- Isaac, M.E.; Nimmo, V.; Gaudin, A.C.M.; Leptin, A.; Schmidt, J.E.; Kallenbach, C.M.; Martin, A.; Entz, M.; Carkner, M.; Rajcan, I.; et al. Crop Domestication, Root Trait Syndromes, and Soil Nutrient Acquisition in Organic Agroecosystems: A Systematic Review. Front. Sustain. Food Syst. 2021, 5, 716480. [Google Scholar] [CrossRef]
- Rao, I.M.; Ayarza, M.A.; Garcia, R. Adaptive Attributes of Tropical Forage Species to Acid Soils I. Differences in Plant Growth, Nutrient Acquisition and Nutrient Utilization among C4 Grasses and C3 Legumes. J. Plant Nutr. 1995, 18, 2135–2155. [Google Scholar] [CrossRef]
- Tůma, I.; Holub, P.; Fiala, K. Soil Nutrient Heterogeneity and Competitive Ability of Three Grass Species (Festuca Ovina, Arrhenatherum Elatius and Calamagrostis Epigejos) in Experimental Conditions. Biologia 2009, 64, 694–704. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G.; Kourtev, P.; Huang, W. Changes in Soil Functions Following Invasions of Exotic Understory Plants in Deciduous Forests. Ecol. Appl. 2001, 11, 1287–1300. [Google Scholar] [CrossRef]
- García-Palacios, P.; Milla, R.; Delgado-Baquerizo, M.; Martín-Robles, N.; Álvaro-Sánchez, M.; Wall, D.H. Side-Effects of Plant Domestication: Ecosystem Impacts of Changes in Litter Quality. New Phytol. 2013, 198, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.J.; Quinton, J.N.; Weigelt, A.; De Deyn, G.B.; Bardgett, R.D. Plant Diversity and Root Traits Benefit Physical Properties Key to Soil Function in Grasslands. Ecol. Lett. 2016, 19, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Kulmatiski, A.; Beard, K.H.; Stark, J.M. Soil History as a Primary Control on Plant Invasion in Abandoned Agricultural Fields. J. Appl. Ecol. 2006, 43, 868–876. [Google Scholar] [CrossRef]
- Dotaniya, M.L.; Meena, V.D. Rhizosphere Effect on Nutrient Availability in Soil and Its Uptake by Plants: A Review. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2015, 85, 1–12. [Google Scholar] [CrossRef]
- Di Lonardo, D.P.; Manrubia, M.; De Boer, W.; Zweers, H.; Veen, G.F.; Van der Wal, A. Relationship between Home-Field Advantage of Litter Decomposition and Priming of Soil Organic Matter. Soil Biol. Biochem. 2018, 126, 49–56. [Google Scholar] [CrossRef]
- Tilman, D.; Wedin, D. Plant Traits and Resource Reduction for Five Grasses Growing on a Nitrogen Gradient. Ecology 1991, 72, 685–700. [Google Scholar] [CrossRef]
- Laungani, R.; Knops, J.M.H. Microbial Immobilization Drives Nitrogen Cycling Differences among Plant Species. Oikos 2012, 121, 1840–1848. [Google Scholar] [CrossRef]
- Marschner, H. Marschner’s Mineral Nutrition of Higher Plants; Academic Press: Waltham, MA, USA, 2011; ISBN 978-0-12-384906-9. [Google Scholar]
- Mathesius, U. Chapter 4.2—Soil-Root Interface. In Plants in Action; Munns, R., Schmidt, S., Beveridge, C., Eds.; Australian Society of Plant Scientists: Hobart, Australia; New Zealand Society of Plant Biologists: Otago, NZ, USA, 2015. [Google Scholar]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, Variation, and Assembly of the Root-Associated Microbiomes of Rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T. Assembly and Ecological Function of the Root Microbiome across Angiosperm Plant Species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [PubMed]
- Hines, J.; Pabst, S.; Mueller, K.E.; Blumenthal, D.M.; Cesarz, S.; Eisenhauer, N. Soil-Mediated Effects of Global Change on Plant Communities Depend on Plant Growth Form. Ecosphere 2017, 8, e01996. [Google Scholar] [CrossRef]
- Lund, M.; Agerbo Rasmussen, J.; Ramos-Madrigal, J.; Sawers, R.; Gilbert, M.T.P.; Barnes, C.J. Rhizosphere Bacterial Communities Differ among Traditional Maize Landraces. Environ. DNA 2022, 4, 1241–1249. [Google Scholar] [CrossRef]
- Tkacz, A.; Bestion, E.; Bo, Z.; Hortala, M.; Poole, P.S. Influence of Plant Fraction, Soil, and Plant Species on Microbiota: A Multikingdom Comparison. mBio 2020, 11, e02785-19. [Google Scholar] [CrossRef]
- Muenzbergova, Z.; Surinova, M. The Importance of Species Phylogenetic Relationships and Species Traits for the Intensity of Plant-Soil Feedback. Ecosphere 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Quattrone, A.; Lopez-Guerrero, M.; Yadav, P.; Meier, M.A.; Russo, S.E.; Weber, K.A. Interactions between Root Hairs and the Soil Microbial Community Affect the Growth of Maize Seedlings. Plant Cell Environ. 2023. [Google Scholar] [CrossRef]
- Dazzo, F.B.; Yanni, Y.G.; Jones, A.; Elsadany, A.Y.; Dazzo, F.B.; Yanni, Y.G.; Jones, A.; Elsadany, A.Y. CMEIAS Bioimage Informatics That Define the Landscape Ecology of Immature Microbial Biofilms Developed on Plant Rhizoplane Surfaces. AIMS Bioeng. 2015, 2, 469–486. [Google Scholar] [CrossRef]
- Rana, K.L.; Kour, D.; Yadav, A.N.; Yadav, N.; Saxena, A.K. Chapter 16—Agriculturally Important Microbial Biofilms: Biodiversity, Ecological Significances, and Biotechnological Applications. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms; Yadav, M.K., Singh, B.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 221–265. ISBN 978-0-444-64279-0. [Google Scholar]
- Szoboszlay, M.; Lambers, J.; Chappell, J.; Kupper, J.V.; Moe, L.A.; McNear, D.H. Comparison of Root System Architecture and Rhizosphere Microbial Communities of Balsas Teosinte and Domesticated Corn Cultivars. Soil Biol. Biochem. 2015, 80, 34–44. [Google Scholar] [CrossRef]
- Velmourougane, K.; Thapa, S.; Prasanna, R. Prospecting Microbial Biofilms as Climate Smart Strategies for Improving Plant and Soil Health: A Review. Pedosphere 2023, 33, 129–152. [Google Scholar] [CrossRef]
- Aslam, M.M.; Karanja, J.K.; Dodd, I.C.; Waseem, M.; Weifeng, X. Rhizosheath: An Adaptive Root Trait to Improve Plant Tolerance to Phosphorus and Water Deficits? Plant Cell Environ. 2022, 45, 2861–2874. [Google Scholar] [CrossRef] [PubMed]
- Bengough, A.G.; Loades, K.; McKenzie, B.M. Root Hairs Aid Soil Penetration by Anchoring the Root Surface to Pore Walls. J. Exp. Bot. 2016, 67, 1071–1078. [Google Scholar] [CrossRef]
- Ma, Z.; Guo, D.; Xu, X.; Lu, M.; Bardgett, R.D.; Eissenstat, D.M.; McCormack, M.L.; Hedin, L.O. Evolutionary History Resolves Global Organization of Root Functional Traits. Nature 2018, 555, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Roumet, C.; Birouste, M.; Picon-Cochard, C.; Ghestem, M.; Osman, N.; Vrignon-Brenas, S.; Cao, K.; Stokes, A. Root Structure–Function Relationships in 74 Species: Evidence of a Root Economics Spectrum Related to Carbon Economy. New Phytol. 2016, 210, 815–826. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root Exudate Metabolites Drive Plant-Soil Feedbacks on Growth and Defense by Shaping the Rhizosphere Microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Okubo, A.; Matsusaka, M.; Sugiyama, S. Impacts of Root Symbiotic Associations on Interspecific Variation in Sugar Exudation Rates and Rhizosphere Microbial Communities: A Comparison among Four Plant Families. Plant Soil 2016, 399, 345–356. [Google Scholar] [CrossRef]
- Doan, T.H.; Doan, T.A.; Kangas, M.J.; Ernest, A.E.; Tran, D.; Wilson, C.L.; Holmes, A.E.; Doyle, E.L.; Durham Brooks, T.L. A Low-Cost Imaging Method for the Temporal and Spatial Colorimetric Detection of Free Amines on Maize Root Surfaces. Front. Plant Sci. 2017, 8, 1513. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Wen, T.-J.; Zimmermann, R.; Chimot-Marolle, P.; Da Costa e Silva, O.; Bruce, W.; Lamkey, K.R.; Wienand, U.; Schnable, P.S. The Maize (Zea mays L.) Roothairless3 Gene Encodes a Putative GPI-Anchored, Monocot-Specific, COBRA-like Protein That Significantly Affects Grain Yield. Plant J. 2008, 54, 888–898. [Google Scholar] [CrossRef]
- Horn, R.; Wingen, L.U.; Snape, J.W.; Dolan, L. Mapping of Quantitative Trait Loci for Root Hair Length in Wheat Identifies Loci That Co-Locate with Loci for Yield Components. J. Exp. Bot. 2016, 67, 4535–4543. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.; Feeney, D.S.; Brown, L.K.; Naveed, M.; Ruiz, S.; Koebernick, N.; Bengough, A.G.; Hallett, P.D.; Roose, T.; Puértolas, J.; et al. Significance of Root Hairs for Plant Performance under Contrasting Field Conditions and Water Deficit. Ann. Bot. 2021, 128, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Saengwilai, P.; Strock, C.; Rangarajan, H.; Chimungu, J.; Salungyu, J.; Lynch, J.P. Root Hair Phenotypes Influence Nitrogen Acquisition in Maize. Ann. Bot. 2021, 128, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Vivanco, J.M. Regulation and Function of Root Exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are Root Exudates More Important than Other Sources of Rhizodeposits in Structuring Rhizosphere Bacterial Communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Guerrero, M.G.; Wang, P.; Phares, F.; Schachtman, D.P.; Alvarez, S.; van Dijk, K. A Glass Bead Semi-Hydroponic System for Intact Maize Root Exudate Analysis and Phenotyping. Plant Methods 2022, 18, 25. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Klironomos, J.N. Feedback with Soil Biota Contributes to Plant Rarity and Invasiveness in Communities. Nature 2002, 417, 67–70. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef]
- Schmid, M.W.; van Moorsel, S.J.; Hahl, T.; De Luca, E.; De Deyn, G.B.; Wagg, C.; Niklaus, P.A.; Schmid, B. Effects of Plant Community History, Soil Legacy and Plant Diversity on Soil Microbial Communities. J. Ecol. 2021, 109, 3007–3023. [Google Scholar] [CrossRef]
- Casper, B.B.; Castelli, J.P. Evaluating Plant–Soil Feedback Together with Competition in a Serpentine Grassland. Ecol. Lett. 2007, 10, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Gehant, L.; Kotanen, P.M.; Johnson, M.T.J. Phylogenetic Relatedness, Phenotypic Similarity and Plant–Soil Feedbacks. J. Ecol. 2017, 105, 786–800. [Google Scholar] [CrossRef]
- Adak, A.; Prasanna, R.; Babu, S.; Bidyarani, N.; Verma, S.; Pal, M.; Shivay, Y.S.; Nain, L. Micronutrient Enrichment Mediated by Plant-Microbe Interactions and Rice Cultivation Practices. J. Plant Nutr. 2016, 39, 1216–1232. [Google Scholar] [CrossRef]
- Prasanna, R.; Kanchan, A.; Ramakrishnan, B.; Ranjan, K.; Venkatachalam, S.; Hossain, F.; Shivay, Y.S.; Krishnan, P.; Nain, L. Cyanobacteria-Based Bioinoculants Influence Growth and Yields by Modulating the Microbial Communities Favourably in the Rhizospheres of Maize Hybrids. Eur. J. Soil Biol. 2016, 75, 15–23. [Google Scholar] [CrossRef]
- Edwards, J.; Santos-Medellín, C.; Nguyen, B.; Kilmer, J.; Liechty, Z.; Veliz, E.; Ni, J.; Phillips, G.; Sundaresan, V. Soil Domestication by Rice Cultivation Results in Plant-Soil Feedback through Shifts in Soil Microbiota. Genome Biol. 2019, 20, 221. [Google Scholar] [CrossRef] [PubMed]
- Fanin, N.; Fromin, N.; Bertrand, I. Functional Breadth and Home-Field Advantage Generate Functional Differences among Soil Microbial Decomposers. Ecology 2016, 97, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Qin, J.; Tahmasbian, I.; Wang, F.; Zhou, S.; Xu, Z.; Cui, X. Litter Amendment Rather than Phosphorus Can Dramatically Change Inorganic Nitrogen Pools in a Degraded Grassland Soil by Affecting Nitrogen-Cycling Microbes. Soil Biol. Biochem. 2018, 120, 145–152. [Google Scholar] [CrossRef]
- Cantarel, A.A.M.; Pommier, T.; Desclos-Theveniau, M.; Diquélou, S.; Dumont, M.; Grassein, F.; Kastl, E.-M.; Grigulis, K.; Laîné, P.; Lavorel, S.; et al. Using Plant Traits to Explain Plant–Microbe Relationships Involved in Nitrogen Acquisition. Ecology 2015, 96, 788–799. [Google Scholar] [CrossRef]
- Conant, R.T.; Paustian, K.; Elliott, E.T. Grassland Management and Conversion into Grassland: Effects on Soil Carbon. Ecol. Appl. 2001, 11, 343–355. [Google Scholar] [CrossRef]
- Heinen, R.; Hannula, S.E.; De Long, J.R.; Huberty, M.; Jongen, R.; Kielak, A.; Steinauer, K.; Zhu, F.; Bezemer, T.M. Plant Community Composition Steers Grassland Vegetation via Soil Legacy Effects. Ecol. Lett. 2020, 23, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Milla, R.; García-Palacios, P.; Matesanz, S. Looking at Past Domestication to Secure Ecosystem Services of Future Croplands. J. Ecol. 2017, 105, 885–889. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of Plant Domestication on Rhizosphere Microbiome Assembly and Functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.C. Understanding Soil Processes: One of the Last Frontiers in Biological and Ecological Research. Australas. Plant Pathol. 2011, 40, 207–214. [Google Scholar] [CrossRef]
- Geisen, S.; Briones, M.J.I.; Gan, H.; Behan-Pelletier, V.M.; Friman, V.-P.; de Groot, G.A.; Hannula, S.E.; Lindo, Z.; Philippot, L.; Tiunov, A.V.; et al. A Methodological Framework to Embrace Soil Biodiversity. Soil Biol. Biochem. 2019, 136, 107536. [Google Scholar] [CrossRef]
- Nazli, F.; Najm-ul-Seher; Khan, M.Y.; Jamil, M.; Nadeem, S.M.; Ahmad, M. Soil Microbes and Plant Health. In Plant Disease Management Strategies for Sustainable Agriculture through Traditional and Modern Approaches; Ul Haq, I., Ijaz, S., Eds.; Sustainability in Plant and Crop Protection; Springer International Publishing: Cham, Switzerland, 2020; pp. 111–135. ISBN 978-3-030-35955-3. [Google Scholar]
- Bachle, S.; Griffith, D.M.; Nippert, J.B. Intraspecific Trait Variability in Andropogon Gerardii, a Dominant Grass Species in the US Great Plains. Front. Ecol. Evol. 2018, 6, 217. [Google Scholar] [CrossRef]
- Vogel, K.P. Improving Warm-Season Forage Grasses Using Selection, Breeding, and Biotechnology. In Native Warm-Season Grasses: Research Trends and Issues; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2000; pp. 83–106. ISBN 978-0-89118-609-0. [Google Scholar]
- Collins, G.N. The Phylogeny of Maize. Bull. Torrey Bot. Club 1930, 57, 199–210. [Google Scholar] [CrossRef]
- Wang, Q.; Dooner, H.K. Dynamic Evolution of Bz Orthologous Regions in the Andropogoneae and Other Grasses. Plant J. 2012, 72, 212–221. [Google Scholar] [CrossRef]
- Yao, Q.-L.; Chen, F.-B.; Liu, H.-F.; Fang, P. Evolution of Maize Landraces in Southwest China: Evidence from the Globulin1 Gene. Biochem. Syst. Ecol. 2015, 61, 54–61. [Google Scholar] [CrossRef]
- Yan, L.; Kenchanmane Raju, S.K.; Lai, X.; Zhang, Y.; Dai, X.; Rodriguez, O.; Mahboub, S.; Roston, R.L.; Schnable, J.C. Parallels between Natural Selection in the Cold-Adapted Crop-Wild Relative Tripsacum Dactyloides and Artificial Selection in Temperate Adapted Maize. Plant J. 2019, 99, 965–977. [Google Scholar] [CrossRef]
- González, J.D.J.S.; Corral, J.A.R.; García, G.M.; Ojeda, G.R.; Larios, L.D.L.C.; Holland, J.B.; Medrano, R.M.; Romero, G.E.G. Ecogeography of Teosinte. PLoS ONE 2018, 13, e0192676. [Google Scholar] [CrossRef]
- Guan, J.-C.; Li, C.; Flint-Garcia, S.; Suzuki, M.; Wu, S.; Saunders, J.W.; Dong, L.; Bouwmeester, H.J.; McCarty, D.R.; Koch, K.E. Maize Domestication Phenotypes Reveal Strigolactone Networks Coordinating Grain Size Evolution with Kernel-Bearing Cupule Architecture. Plant Cell 2023, 35, 1013–1037. [Google Scholar] [CrossRef] [PubMed]
- Hung, H.-Y.; Browne, C.; Guill, K.; Coles, N.; Eller, M.; Garcia, A.; Lepak, N.; Melia-Hancock, S.; Oropeza-Rosas, M.; Salvo, S.; et al. The Relationship between Parental Genetic or Phenotypic Divergence and Progeny Variation in the Maize Nested Association Mapping Population. Heredity 2012, 108, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, B.M. Diversity in Global Maize Germplasm: Characterization and Utilization. J. Biosci. 2012, 37, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M.; Ying, K.; Fu, Y.; Ji, T.; Yeh, C.-T.; Jia, Y.; Wu, W.; Richmond, T.; Kitzman, J.; Rosenbaum, H.; et al. Maize Inbreds Exhibit High Levels of Copy Number Variation (CNV) and Presence/Absence Variation (PAV) in Genome Content. PLoS Genet. 2009, 5, e1000734. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.-J.; Schnable, P.S. Analyses of Mutants of Three Genes That Influence Root Hair Development in Zea mays (Gramineae) Suggest That Root Hairs Are Dispensable. Am. J. Bot. 1994, 81, 833–842. [Google Scholar] [CrossRef]
- Barker, N.P.; Clark, L.G.; Davis, J.I.; Duvall, M.R.; Guala, G.F.; Hsiao, C.; Kellogg, E.A.; Linder, H.P.; Mason-Gamer, R.J.; Mathews, S.Y.; et al. Phylogeny and Subfamilial Classification of the Grasses (Poaceae). Ann. Mo. Bot. Gard. 2001, 88, 373–457. [Google Scholar]
- Mathews, S.; Spangler, R.E.; Mason-Gamer, R.J.; Kellogg, E.A. Phylogeny of Andropogoneae Inferred from Phytochrome B, GBSSI, and ndhF. Int. J. Plant Sci. 2002, 163, 441–450. [Google Scholar] [CrossRef]
- Brandt, A.J.; del Pino, G.A.; Burns, J.H. Experimental Protocol for Manipulating Plant-Induced Soil Heterogeneity. J. Vis. Exp. 2014, 85, e51580. [Google Scholar] [CrossRef]
- Rinella, M.J.; Reinhart, K.O. Toward More Robust Plant-Soil Feedback Research. Ecology 2018, 99, 550–556. [Google Scholar] [CrossRef]
- Wolf, D.C.; Skipper, H.D. Soil Sterilization. In Methods of Soil Analysis; Soil Sciences Society of America, Inc.: Madison, WI, USA, 1994; pp. 41–51. ISBN 978-0-89118-865-0. [Google Scholar]
- Mahboob, W.; Rehman, H.; Afzal, I.; Basra, S. Seed Priming Enhances Crop Stand of Spring Maize by Improving Temperature Resistance at Seedling Stage. Int. J. Agric. Biol. 2013. [Google Scholar] [CrossRef]
- Finneseth, C.H. Evaluation and Enhancement of Seed Lot Quality in Eastern Gamagrass [Tripsacum dactyloides (L.) L.]. Ph.D. Thesis, University of Kentucky, Lexington, KY, USA, 2010. [Google Scholar]
- Tian, X.; Knapp, A.D.; Moore, K.J.; Brummer, E.C.; Bailey, T.B. Cupule Removal and Caryopsis Scarification Improves Germination of Eastern Gamagrass Seed. Crop. Sci. 2002, 42, 185–189. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. 1950. Available online: https://www.cabdirect.org/cabdirect/abstract/19500302257 (accessed on 24 June 2022).
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Rapid Method for Coextraction of DNA and RNA from Natural Environments for Analysis of Ribosomal DNA- and rRNA-Based Microbial Community Composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [Google Scholar] [CrossRef] [PubMed]
- Westrop, J.P.; Yadav, P.; Nolan, P.; Campbell, K.M.; Singh, R.; Bone, S.E.; Chan, A.H.; Kohtz, A.J.; Pan, D.; Healy, O.; et al. Nitrate-Stimulated Release of Naturally Occurring Sedimentary Uranium. Environ. Sci. Technol. 2023, 57, 4354–4366. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- De Cáceres, M.; Legendre, P.; Moretti, M. Improving Indicator Species Analysis by Combining Groups of Sites. Oikos 2010, 119, 1674–1684. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Fox, J. Applied Regression Analysis and Generalized Linear Models; Sage Publications: Thousand Oaks, CA, USA, 2015. [Google Scholar]
- Lin, H.; Peddada, S.D. Analysis of Compositions of Microbiomes with Bias Correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R. Vegan: Community Ecology Package. R Package Version 2.5-7 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 22 June 2022).
- Shaffer, J.P. Multiple Hypothesis Testing. Annu. Rev. Psychol. 1995, 46, 561–584. [Google Scholar] [CrossRef]
- Shetty, S.A.; Lahti, L. Microbiomeutilities: Utilities for Microbiome Analytics 2020. Available online: https://microbiome.github.io/tutorials/ (accessed on 22 June 2022).
- Warton, D.I.; Wright, S.T.; Wang, Y. Distance-Based Multivariate Analyses Confound Location and Dispersion Effects. Methods Ecol. Evol. 2012, 3, 89–101. [Google Scholar] [CrossRef]
- Hutchinson, M.I.; Bell, T.A.S.; Gallegos-Graves, L.V.; Dunbar, J.; Albright, M. Merging Fungal and Bacterial Community Profiles via an Internal Control. Microb. Ecol. 2021, 82, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qu, Y.; Li, S.; Feng, K.; Wang, S.; Cai, W.; Liang, Y.; Li, H.; Xu, M.; Yin, H.; et al. Soil Bacterial Quantification Approaches Coupling with Relative Abundances Reflecting the Changes of Taxa. Sci. Rep. 2017, 7, 4837. [Google Scholar] [CrossRef]
- Andrew, D.R.; Fitak, R.R.; Munguia-Vega, A.; Racolta, A.; Martinson, V.G.; Dontsova, K. Abiotic Factors Shape Microbial Diversity in Sonoran Desert Soils. Appl. Environ. Microbiol. 2012, 78, 7527–7537. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Strauss, S.L. Insights into the Taxonomic and Functional Characterization of Agricultural Crop Core Rhizobiomes and Their Potential Microbial Drivers. Sci. Rep. 2021, 11, 10068. [Google Scholar] [CrossRef]
- Brisson, V.L.; Schmidt, J.E.; Northen, T.R.; Vogel, J.P.; Gaudin, A.C.M. Impacts of Maize Domestication and Breeding on Rhizosphere Microbial Community Recruitment from a Nutrient Depleted Agricultural Soil. Sci. Rep. 2019, 9, 15611. [Google Scholar] [CrossRef]
- Jang, W.S.; Neff, J.C.; Im, Y.; Doro, L.; Herrick, J.E. The Hidden Costs of Land Degradation in US Maize Agriculture. Earth’s Future 2021, 9, e2020EF001641. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Mazza Rodrigues, J.L.; Brisson, V.L.; Kent, A.; Gaudin, A.C.M. Impacts of Directed Evolution and Soil Management Legacy on the Maize Rhizobiome. Soil Biol. Biochem. 2020, 145, 107794. [Google Scholar] [CrossRef]
- York, L.M.; Galindo-Castañeda, T.; Schussler, J.R.; Lynch, J.P. Evolution of US Maize (Zea mays L.) Root Architectural and Anatomical Phenes over the Past 100 Years Corresponds to Increased Tolerance of Nitrogen Stress. J. Exp. Bot. 2015, 66, 2347–2358. [Google Scholar] [CrossRef]
- Weaver, J. North American Prairie. In Papers of John E. Weaver (1884–1956); University of Nebraska-Lincoln: Lincoln, NE, USA, 1954. [Google Scholar]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The Under-Recognized Dominance of Verrucomicrobia in Soil Bacterial Communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The Diversity and Biogeography of Soil Bacterial Communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Strap, J.L. Actinobacteria–Plant Interactions: A Boon to Agriculture. In Bacteria in Agrobiology: Plant Growth Responses; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 285–307. ISBN 978-3-642-20332-9. [Google Scholar]
- Heinze, J.; Sitte, M.; Schindhelm, A.; Wright, J.; Joshi, J. Plant-Soil Feedbacks: A Comparative Study on the Relative Importance of Soil Feedbacks in the Greenhouse versus the Field. Oecologia 2016, 181, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Smith-Ramesh, L.M.; Reynolds, H.L. The next Frontier of Plant–Soil Feedback Research: Unraveling Context Dependence across Biotic and Abiotic Gradients. J. Veg. Sci. 2017, 28, 484–494. [Google Scholar] [CrossRef]
- Wang, G.; Bei, S.; Li, J.; Bao, X.; Zhang, J.; Schultz, P.A.; Li, H.; Li, L.; Zhang, F.; Bever, J.D.; et al. Soil Microbial Legacy Drives Crop Diversity Advantage: Linking Ecological Plant–Soil Feedback with Agricultural Intercropping. J. Appl. Ecol. 2021, 58, 496–506. [Google Scholar] [CrossRef]
- Bonanomi, G.; Lorito, M.; Vinale, F.; Woo, S.L. Organic Amendments, Beneficial Microbes, and Soil Microbiota: Toward a Unified Framework for Disease Suppression. Annu. Rev. Phytopathol. 2018, 56, 1–20. [Google Scholar] [CrossRef]
- Loreau, M.; Hector, A. Partitioning Selection and Complementarity in Biodiversity Experiments. Nature 2001, 412, 72–76. [Google Scholar] [CrossRef]
- Kumar, A.; Shahbaz, M.; Koirala, M.; Blagodatskaya, E.; Seidel, S.J.; Kuzyakov, Y.; Pausch, J. Root Trait Plasticity and Plant Nutrient Acquisition in Phosphorus Limited Soil. J. Plant Nutr. Soil Sci. 2019, 182, 945–952. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef]
- Dijkstra, F.A.; Zhu, B.; Cheng, W. Root Effects on Soil Organic Carbon: A Double-Edged Sword. New Phytol. 2021, 230, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Capek, P.T.; Manzoni, S.; Kastovska, E.; Wild, B.; Diakova, K.; Barta, J.; Schnecker, J.; Blasi, C.; Martikainen, P.J.; Alves, R.J.E.; et al. A Plant-Microbe Interaction Framework Explaining Nutrient Effects on Primary Production. Nat. Ecol. Evol. 2018, 2, 1588–1596. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Ho, M.D. Rhizoeconomics: Carbon Costs of Phosphorus Acquisition. Plant Soil 2005, 269, 45–56. [Google Scholar] [CrossRef]
- Ma, X.; Li, X.; Ludewig, U. Arbuscular Mycorrhizal Colonization Outcompetes Root Hairs in Maize under Low Phosphorus Availability. Ann. Bot. 2021, 127, 155–166. [Google Scholar] [CrossRef]
- Oliverio, A.M.; Bissett, A.; McGuire, K.; Saltonstall, K.; Turner, B.L.; Fierer, N. The Role of Phosphorus Limitation in Shaping Soil Bacterial Communities and Their Metabolic Capabilities. mBio 2020, 11, e01718-20. [Google Scholar] [CrossRef]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Hunter, M.C.; Smith, R.G.; Schipanski, M.E.; Atwood, L.W.; Mortensen, D.A. Agriculture in 2050: Recalibrating Targets for Sustainable Intensification. BioScience 2017, 67, 386–391. [Google Scholar] [CrossRef]


| Abbreviation | Description | Units |
|---|---|---|
| Plant performance | ||
| Total bm gr | total biomass growth rate per month | g (harvested plant biomass—initial plant biomass)/month |
| Phenotypic traits | ||
| LMR | leaf mass ratio | g leaf dry weight/g plant biomass |
| LAR | leaf area ratio | cm2/g leaf dry weight |
| SLA | specific leaf area | cm2 leaf area/g leaf dry weight |
| LDMC | leaf dry matter content | g leaf dry weight/g leaf fresh weight |
| LTD | leaf tissue density | g leaf dry weight/cm3 leaf volume |
| Leaf thick | mean leaf thickness | mm |
| LA | mean area of a leaf | cm2 |
| RMR | root mass ratio | g root biomass/g plant biomass |
| RLR | root length ratio | cm root length/g plant biomass |
| SRL | specific root length | cm root/g root dry weight |
| RTD | root tissue density | g total root dry weight/cm3 root volume |
| Leaf nutrient concentrations | ||
| Leaf C | leaf carbon | Percent carbon |
| Leaf N | leaf nitrogen | Percent nitrogen |
| Leaf P | leaf phosphorus | Percent phosphorus |
| Leaf K | leaf potassium | Percent potassium |
| Leaf Ca | leaf calcium | Percent calcium |
| Leaf Mg | leaf magnesium | Percent magnesium |
| Soil nutrient concentrations | ||
| Soil C | soil carbon | Percent carbon |
| Soil N | soil nitrogen | Percent nitrogen |
| Soil P | soil phosphorus | mg phosphorus/kg soil |
| Soil K | soil potassium | mg potassium/kg soil |
| Soil Ca | soil calcium | mg calcium/kg soil |
| Soil Mg | soil magnesium | mg magnesium/kg soil |
| perMANOVA | Test of Dispersion | ||||
|---|---|---|---|---|---|
| Factor | F | Probability (p) | R2 | F | Probability (p) |
| Abundance-weighted | |||||
| Functional group | 1.32 | 0.001 | 0.02 | 1.37 | 0.246 |
| Functional group: conditioning genotype | 1.10 | 0.001 | 0.07 | - | - |
| Presence–absence | |||||
| Functional group | 1.23 | 0.001 | 0.02 | 0.68 | 0.411 |
| Functional group: conditioning genotype | 1.12 | 0.001 | 0.07 | - | - |
| Response Variable | Andropogon gerardii | Z. mays ssp. mays B73-wt | Z. mays ssp. parviglumis |
|---|---|---|---|
| Plant performance | |||
| Total bm gr | F6,75 = 3.04 padj = 0.235 | F6,63 = 9.88 padj < 0.001 | F6,65 = 16.9 padj < 0.001 |
| Phenotypic traits | |||
| LMR | F6,62 = 0.48 padj = 1.000 | F6,64 = 0.07 padj = 1.000 | F6,65 = 0.55 padj = 1.000 |
| SLA | F6,74 = 0.54 padj = 1.000 | F6,62 = 4.21 padj = 0.029 | F6,65 = 2.59 padj = 0.594 |
| LDMC | F6,64 = 1.54 padj = 1.000 | F6,63 = 0.82 padj = 1.000 | F6,74 = 0.93 padj = 1.000 |
| LTD | F6,74 = 0.53 padj = 1.000 | F6,64 = 0.58 padj = 1.000 | F6,72 = 0.52 padj = 1.000 |
| LAR | F6,74 = 0.61 padj = 1.000 | F6,63 = 1.21 padj = 1.000 | F6,65 = 2.21 padj = 1.000 |
| LA | F6,63 = 1.70 padj = 1.000 | F6,61 = 21.85 padj < 0.001 | F6,65 = 3.22 padj = 0.182 |
| RMR | F6,62 = 1.03 padj = 1.000 | F6,63 = 8.80 padj < 0.001 | F6,66 = 0.88 padj = 1.000 |
| RLR | F6,63 = 3.96 padj = 0.046 | F6,74 = 1.21 padj = 1.000 | F6,65 = 1.69 padj = 1.000 |
| SRL | F6,58 = 3.11 padj = 0.239 | F6,62 = 1.74 padj = 1.000 | F6,53 = 1.71 padj = 1.000 |
| RTD | F6,74 = 3.00 padj = 0.257 | F6,63 = 0.64 padj = 1.000 | F6,64 = 0.97 padj = 1.000 |
| Leaf nutrient concentrations | |||
| Leaf C | F6,70 = 0.57 padj = 1.000 | F6,74 = 1.72 padj = 1.000 | F6,74 = 3.10 padj = 0.211 |
| Leaf N | F6,74 = 2.05 padj = 1.000 | F6,63 = 4.22 padj = 0.029 | F6,64 = 2.67 padj = 0.516 |
| Leaf P | F6,60 = 1.21 padj = 0.315 | F6,63 = 0.70 padj = 1.000 | F6,64 = 1.58 padj = 1.000 |
| Leaf K | F6,70 = 1.96 padj = 0.083 | F6,63 = 54.68 padj < 0.001 | F6,64 = 22.17 padj < 0.001 |
| Leaf Ca | F6,57 = 2.80 padj = 0.110 | F6,63 = 14.46 padj < 0.001 | F6,74 = 8.90 padj < 0.001 |
| Leaf Mg | F6,61 = 0.91 padj = 0.493 | F6,63 = 28.94 padj < 0.001 | F6,74 = 13.05 padj < 0.001 |
| Soil nutrient concentrations | |||
| Soil C | F6,60 = 3.52 padj = 0.036 | F6,61 = 4.62 padj = 0.005 | F6,60 = 2.61 padj = 0.194 |
| Soil N | F6,60 = 2.43 padj = 0.165 | F6,61 = 3.49 padj = 0.023 | F6,61 = 1.59 padj = 0.630 |
| Soil P | F6,60 = 11.43 padj < 0.001 | F6,51 = 18.88 padj < 0.001 | F6,63 = 25.24 padj < 0.001 |
| Soil K | F6,70 = 120.38 padj < 0.001 | F6,74 = 24.50 padj < 0.001 | F6,72 = 46.59 padj < 0.001 |
| Soil Ca | F6,70 = 1.69 padj = 0.521 | F6,50 = 1.09 padj = 0.417 | F6,61 = 2.42 padj = 0.194 |
| Soil Mg | F6,70 = 3.12 padj = 0.053 | F6,74 = 3.14 padj = 0.015 | F6,61 = 2.34 padj = 0.194 |
| perMANOVA | Test of Dispersion | ||||
|---|---|---|---|---|---|
| Factor | F | Probability (p) | R2 | F | Probability (p) |
| Functional group | 75.56 | 0.001 | 0.20 | 30.36 | <0.001 |
| Feedback genotype | 9.93 | 0.001 | 0.05 | 0.51 | 0.599 |
| Functional group: conditioning genotype | 6.55 | 0.001 | 0.09 | - | - |
| Feedback × conditioning genotype | 2.26 | 0.001 | 0.07 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quattrone, A.; Yang, Y.; Yadav, P.; Weber, K.A.; Russo, S.E. Nutrient and Microbiome-Mediated Plant–Soil Feedback in Domesticated and Wild Andropogoneae: Implications for Agroecosystems. Microorganisms 2023, 11, 2978. https://doi.org/10.3390/microorganisms11122978
Quattrone A, Yang Y, Yadav P, Weber KA, Russo SE. Nutrient and Microbiome-Mediated Plant–Soil Feedback in Domesticated and Wild Andropogoneae: Implications for Agroecosystems. Microorganisms. 2023; 11(12):2978. https://doi.org/10.3390/microorganisms11122978
Chicago/Turabian StyleQuattrone, Amanda, Yuguo Yang, Pooja Yadav, Karrie A. Weber, and Sabrina E. Russo. 2023. "Nutrient and Microbiome-Mediated Plant–Soil Feedback in Domesticated and Wild Andropogoneae: Implications for Agroecosystems" Microorganisms 11, no. 12: 2978. https://doi.org/10.3390/microorganisms11122978
APA StyleQuattrone, A., Yang, Y., Yadav, P., Weber, K. A., & Russo, S. E. (2023). Nutrient and Microbiome-Mediated Plant–Soil Feedback in Domesticated and Wild Andropogoneae: Implications for Agroecosystems. Microorganisms, 11(12), 2978. https://doi.org/10.3390/microorganisms11122978






